Abstract
Located only a short distance off the southernmost shore of the Greater Indian subcontinent, the island of Sri Lanka has long been inhabited by various ethnic populations. Mainly comprising the Vedda, Sinhalese (Up- and Low-country) and Tamil (Sri Lankan and Indian); their history of settlements on the island and the biological relationships among them have remained obscure. It has been hypothesized that the Vedda was probably the earliest inhabitants of the area, followed by Sinhalese and Tamil from the Indian mainland. This study, in which 271 individuals, representing the Sri Lankan ethnic populations mentioned, were typed for their mitochondrial DNA (mtDNA) hypervariable segment 1 (HVS-1) and part of hypervariable segment 2 (HVS-2), provides implications for their settlement history on the island. From the phylogenetic, principal coordinate and analysis of molecular variance results, the Vedda occupied a position separated from all other ethnic people of the island, who formed relatively close affiliations among themselves, suggesting a separate origin of the former. The haplotypes and analysis of molecular variance revealed that Vedda people’s mitochondrial sequences are more related to the Sinhalese and Sri Lankan Tamils’ than the Indian Tamils’ sequences. MtDNA haplogroup analysis revealed that several West Eurasian haplogroups as well as Indian-specific mtDNA clades were found amongst the Sri Lankan populations. Through a comparison with the mtDNA HVS-1 and part of HVS-2 of Indian database, both Tamils and Sinhalese clusters were affiliated with Indian subcontinent populations than Vedda people who are believed to be the native population of the island of Sri Lanka.
Similar content being viewed by others
Introduction
With its close proximity to the Greater Indian subcontinent, separated from the southern tip of the mainland only by the Palk Strait and the Gulf of Mannar (with the width varying between 24 and 140 km), the island of Sri Lanka, made accessible by sea from all parts of coastal India, has long been inhabited by various ethnic people. The mainland origins for the majority of these people have been hypothesized, but without their specific migration and settlement history on the island, they are yet to be fully elucidated. Of approximately the total size of 20 million, the population of Sri Lanka is heterogeneous on the bases of ethnicity, languages and religious faiths.1, 2
The Buddhist Sinhalese who speak Sinhala, affiliated with the North Indian Prakit,3 a branch of the Indo-European language family, contribute to the majority on the island, accounting for 73.8% of the total population. Their division into Up- and Low-country ethnic counterparts was a recent phenomenon after the European colonization, with people in the coastal provinces, formerly the subjects under Western domination, recognized as the Low-country Sinhalese, and those living in the inland mountainous area, ruled by the Sinhalese kings, later known as the Up-country Sinhalese. With their history on the island stretching back into the remote past, the Sri Lankan Tamils who speak Tamil, a language of the Dravidian family, and profess Hinduism, comprise 13.9% of the total population. Their ethnic counterpart of the same religious faith, the Indian Tamils, of probably more recent origin from the mainland, contributes 4.6% of the island population. Muslims who mainly speak Tamil account for 7.2% of the total population. Other minorities on the island, with each contributing less than 0.5% of the total population, comprise the Muslim Malays whose language belonging to the Austronesian linguistic family, the Christian who speak English, and the Vedda, believed to be the most indigenous people on the island,4, 5 whose present dialects identified as a hybrid between older Vedda language and Sinhala.6
Paleoclimates of Sri Lanka were made relatively stable under the influences of Southwestern monsoons that have strengthened since the terminal Pleistocene and early Holocene, the tropical cyclones and the two intermonsoons.7 Its climatic stability would have favored human migrations for settlements in the area since the distant past. Archeological records of human settlements on the island were conventionally attributed to four consecutive periods: the Paleolithic (125 000–37 000 YBP), the Mesolithic (37 000–2900 YBP), the protohistorical (2900–2500 YBP) and the historical (after 2500 YBP).8, 9 Interestingly, the oldest skeletal remains of anatomically modern man (Homo sapiens) reported from the South Asian region, and dated tentatively to 37 000 YBP, were discovered from the cave site, Fahien-lena,8 on the island, with their association with the present-day Vedda people proposed on a comparative anatomical ground.10 With the molecular analyses provided on genetic structure of the island populations, achievement for more insights into the history of human settlements in the area is truly promising.
The molecular genetic studies on Sri Lankan ethnic people have been relatively scant so far, with only a few autosomal and Y chromosome results accumulated in the forensic databases.11, 12, 13 This present study provides the first opportunity under which the higher resolution mitochondrial DNA (mtDNA) genetic structure is elucidated, based on sequencing of the hypervariable segment 1 (HVS-1) and part of hypervariable segment 2 (HVS-2), of the majority of the Sri Lankan ethnic populations, the Vedda, Sinhalese (Up- and Low-country) and Tamil (Sri Lankan and Indian), providing an insight into the understanding of the history of human settlements on the island.
Materials and methods
Samples collection
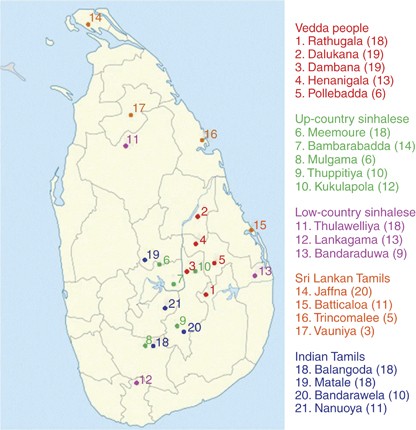
A total of 271 unrelated individuals belonging to five ethnic groups—Vedda people, Up-country Sinhalese and Low-country Sinhalese, Sri Lankan Tamils and Indian Tamils, were recruited in the study (Supplementary Table S1). The sample collection sites are shown in Figure 1. With informed consent, 3–5 hair follicles from each individual were collected. The study was carried out with the approval of the Ethics Committee of the Faculty of Medicine, University of Kelaniya, Sri Lanka. DNA was extracted using standard protocol.14
MtDNA sequence data
MtDNA HVS-1 (nt16024–nt16383) and part of HVS-2 (nt57–nt309) were amplified using primers L15904: 5′-CTAATACACCAGTCTTGTAAACCGGAG-3′ and H 16417: 5′-TTTCACGGAGGATGGTGGTC-3′, and L 16453: 5′-CCGGGCCCATAACACTTGGG-3′ and H 545: 5′-CGGGGTATGGGGTTAGCAGC-3′, respectively. The PCR products were purified and sequenced using DNA Analyzer (model 3730XL, Applied Biosystems, Foster, CA, USA) by Macrogen (Seoul, Republic of Korea). The sequencing data have been checked by a 4-eye principle and the low quality data were resequenced, otherwise excluded from the analysis. The samples to which definite haplogroup status could not be assigned were additionally checked for positions C5178A, T14783C and 9 bp deletion (8281–8289) for haplogroup D, M and B, respectively using PCR–RFLP method. MtDNA was amplified at positions 4476–5482 (L4476: 5′-CCC CTG GCC CAA CCC GTC ATC TAC-3′and H5482:5′-GGT AGG AGT AGC GTG GTA AGG GCG-3′) and positions 14444–15360 (L14444: 5′-TCC TCA ATA GCC ATC GCT G-3′ and H15360: 5′-GAT CCC GTT TCG TGC AAG-3′), the PCR product was digested by restriction enzyme AluI for the presence of C5178A (haplogroup D), and by AseI for the presence of T14783C (haplogroup M), respectively. For 9 bp deletion, mtDNA were amplified from position 8211–8311 (L8211: 5′-TCG TCC TAG AAT TAA TTC CCC-3′ and H8311: 5′-AAG TTC GCT TTA CAG-3′), and the size of PCR product was electrophoresed and visualized under ultraviolet light.
Data analysis
The sequences of HVS-1 and part of HVS-2 were aligned and compared with rCRS using ClustalW software in the BioEdit version 7.0.9 and ChromasLite program, respectively. The haplogroup assignment was done using Haplogrep (http://www.haplogrep.uibk.ac.at/)15 based on HVS-1 and part of HVS-2 sequences and manually checked according to the criteria of Phylotree Build 15.16 To further justify the haplogroup classification, the mitochondrial haplogroup was also assigned using MitoTool (http://www.mitotool.org).17 The software package DnaSP version 5 (Universitat de Barcelona, Barcelona, Spain) was used to calculate the number of polymorphic sites, haplotype diversity, nucleotide diversity and average number of nucleotide differences.18 The number of unique haplotypes for each population was evaluated based on the calculation of the total number of haplotypes and the number of shared haplotypes by ARLEQUIN version 3.5.1.219 (Swiss Institute of Bioinformatics, Bern, Switzerland) using the same strategy as presented in Yao et al.20
The software program MEGA 4.0.221 was used to draw the unrooted neighbor-joining trees of the Sri Lankan populations using net genetic distances (dA), which are defined as dA=dXY – (dX+dY)/2, where dXY is the mean pairwise difference between individuals from population X and Y, and dX (dY) is the mean pairwise difference between individuals within population X (or Y).22 As a tree presentation of the distance matrix might be misread as a succession of population splits, principal component analysis (PCA) was employed on the distance matrix. Two principal component analyses were performed on 21 Sri Lankan groups using their respective net genetic distances from HVS-1 and part of HVS-2 sequences and from haplogroup distribution frequencies by means of GenAlEx6.23 In order to compare with the Indian mainland, PCA was also performed on the Sri Lankan groups and 34 groups (both tribes and castes) from India (Supplementary Table S2) using their respective net genetic distances from HVS-1 sequences. Genetic structure was investigated using analyses of molecular variance by ARLEQUIN version 3.5.1.2 with a significance of variance components tested with 1000 permutations.19 The Mantel test was performed to assess the significance of the correlation between genetic and geographic distances of the Sri Lankan populations with 1000 random permutations using GenAlEx6.23 Two Phylogenetic networks of Haplogroups R and U from five Sri Lanka ethnic populations, and from Sri Lankan and Indian populations24, 25 were constructed using Network 4.6.1.1 (Fluxus Technology, Suffolk, UK).26
Results
MtDNA polymorphisms and shared haplotypes of Sri Lanka
The mtDNA HVS-1 (np. 16 024 to np. 16 383 of the rCRS) and part of HVS-2 (np. 57 to np. 309 of the rCRS) sequences were obtained from 271 individuals belonging to five Sri Lankan ethnic populations: 75 Vedda people, 60 Up-country Sinhalese and 40 Low-country Sinhalese, 39 Sri Lankan Tamils and 57 Indian Tamils. The polymorphisms observed in the study are provided in Supplementary Table S3. Deletions were observed at nucleotide positions 16 166, 16 258, and 249 whereas insertions were encountered at 16 188, 16 380 and 284.
There were a total of 147 haplotypes observed in the five Sri Lankan populations of this study. Thirty of them were shared between at least two populations. The Vedda population has the lowest proportion of shared haplotypes among their subgroups (63%) indicating their greater genetic diversity among subgroups. Sri Lankan Tamils and Indian Tamils possessed similar shared proportion (85%) whereas Up-country Sinhalese has a little higher number of population specific haplotypes (73%) than Low-country Sinhalese (70%) (Table 1). Interestingly, highest number of haplotype sharing was found between Vedda with Up-country Sinhalese and with Low-country Sinhalese. On the other hand, there was no haplotype sharing between the Vedda people with any of the Tamils (Table 1).
Diversity indices
Genetic diversity within the subgroups of Sri Lankan ethnic populations was assessed by haplotype diversity (H) and nucleotide diversity (π). The results are summarized in Supplementary Table S4. H ranged from 0.503–1.000 and π from 0.006–0.019. In general, Sinhalese (Up-country and Low-country) and Tamil (Sri Lankan and Indian) subgroups exhibited relatively higher haplotype diversity (0.861–1.000) than did those of the Vedda (0.503–0.965). The trend of nucleotide diversity follows the haplotype diversity. Higher nucleotide diversities (0.009–0.019) were observed among Sinhalese and Tamils. Notably, lower nucleotide diversity (0.006–0.009) was observed in two Vedda subgroups (VA-Rat and VA-Dal) than in the rest of the Vedda subgroups (0.012–0.014).
Pattern of genetic variation as revealed by genetic distance and phylogenetic analyses
Genetic distances among 21 subgroups of five ethnic populations of Sri Lanka were calculated from HVS-1 and part of HVS-2 sequences employing the Tajima-Nei method.27 The result is shown in Supplementary Table S5. The Mantel test for correspondence between genetic and geographic minimal distances was also performed, from which the significant correlation between the two distance matrices (r=0.15; P=0.02) was obtained; the result suggested the pattern of genetic differentiation observed among studied populations to be at least partly explicable in the light of the isolation-by-distance model.
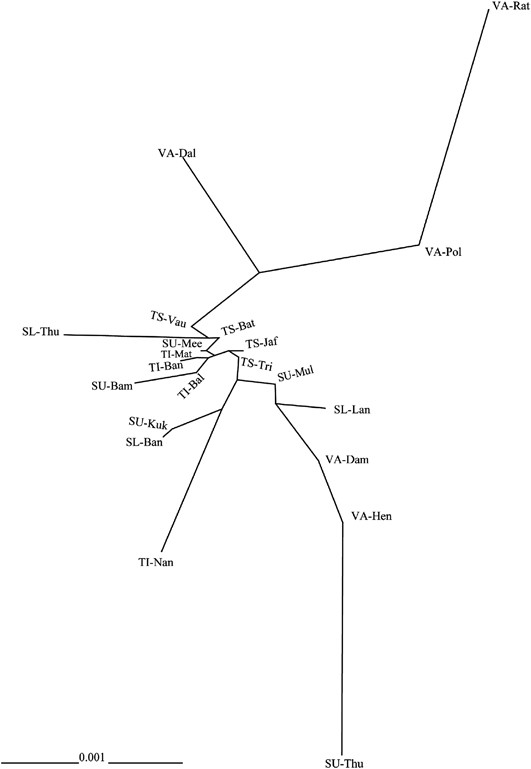
An unrooted neighbor-joining tree was constructed for phylogenetic relationships among 21 subgroups of five ethnic populations of Sri Lanka as illustrated in Figure 2. Another phylogenetic construction was also performed for the five ethnic populations when all subgroups within a population were collapsed; the result is shown in Supplementary Figure S1.
Unrooted neighbor-joining (NJ) tree of the 21 Sri Lankan populations based on the net genetic distances. (Abbreviations are given in Supplementary Table S1).
It is quite clear from Supplementary Figure S1 that the Vedda population was genetically separated from other Sri Lankan ethnic populations, with genetic distance being less between them. Indian Tamils established the closest genetic relationship with their Sri Lankan ethnic counterparts. Up-country Sinhalese formed close genetic affiliations with Sri Lankan Tamils and Low-country Sinhalese.
Figure 2 illustrates more insights into the genetic relationships among the studied populations, with the description of genetic variation among subgroups within each ethnic population. From this unrooted neighbor-joining tree, it was confirmed that there was a greater genetic distance between the Vedda people and the rest of the populations. Two Vedda subgroups (VA-Dam and VA-Hen) were intermingled with the Sinhalese, both Up-country and Low-country, but not with any of Tamils. The Tamils, both Sri Lankan and Indian, clustered together. The genetic matrix in which the Tamil and Sinhalese subgroups, that cannot be clearly separated from each other, were observed towards one major branch of the tree, with the majority of the Vedda people towards the other. Interestingly, some Sinhalese groups (SU-Mul, SU-Mee and SL-Lan) were relatively closer to Tamils than to the rest of Sinhalese subgroups.
Principal component analysis
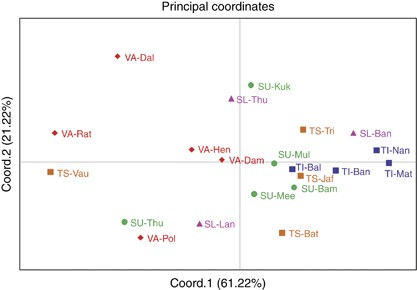
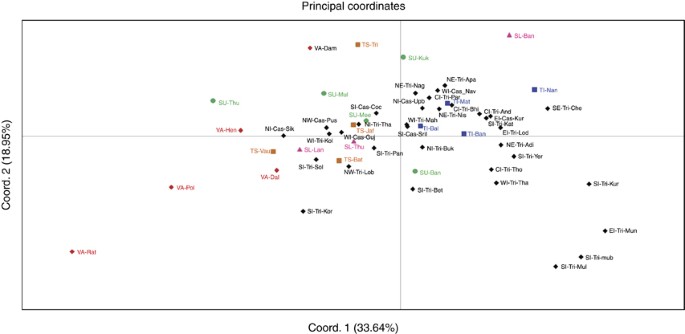
The net genetic distances from HVS-1 and part of HVS-2 sequences (Supplementary Table S5), and from haplogroup distribution frequencies (Supplementary Table S6), among 21 subgroups of five ethnic populations of Sri Lanka were treated as input vectors for PCA. Figure 3 displays the PCA map constructed from haplogroup distribution frequencies, for the first two principal components, which together account for 82.44% of the total variance. The majority of Vedda subgroups (except VA-Dam) were well separated from other ethnic populations of Sri Lanka on the first PC axis. Their separation from other ethnic populations is further extended on the second PC axis. The majority of Sinhalese and Tamil subgroups form close genetic proximities among themselves on both PC axes. Major exception to this clustering is found in SU-Thu. It was evident that Up-country Sinhalese are genetically closer to Sri Lankan Tamils. On the other hand, Sri Lankan Tamil subgroups were closer to each other when compared with Indian Tamils. Generally speaking, Vedda subgroups were more dispersed on the PCA map than within any other ethnic population, reflecting greater diversity among them. PCA map constructed from the net genetic distances from HVS-1 and part of HVS-2 sequences, which is in agreement with the PCA map constructed from haplogroup distribution frequencies, is also shown in Supplementary Figure S2.
Principal component analysis (PCA) map of the 21 Sri Lankan subpopulations based on net genetic distances derived from haplogroup distribution frequencies. (Abbreviations are given in Supplementary Table S1). A full color version of this figure is available at the Journal of Human Genetics journal online.
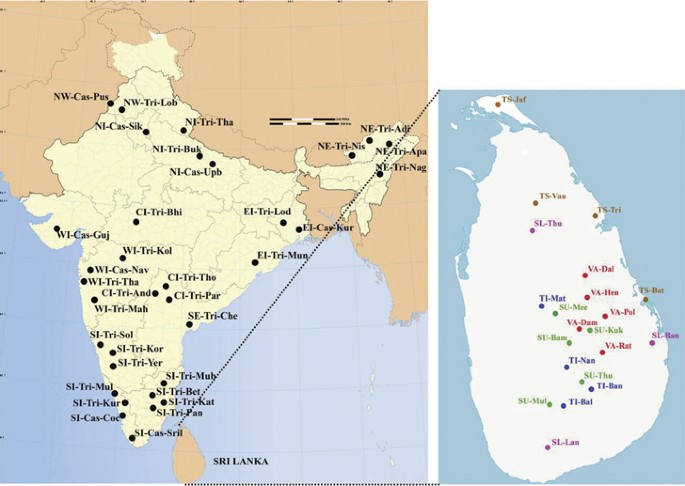
The PCA is extended further to include various other ethnic populations from the Indian subcontinent (Supplementary Table S2) Figure 4. The result shown in Figure 5 accounted for 52.59% of the total variation. All the Sinhalese and Tamil subgroups intermingle well with the majority of the Indian subcontinental populations, forming a large genetic matrix. However, Indian Tamils were separated from the rest of the Sri Lankan subgroups, except SU-Bam and SL-Ban, on the first PC axis. This is further strengthening of the hypothesis that Indian Tamils are genetically distinct from the rest of the Sri Lankan ethnic groups. Some Vedda groups (VA-Dal, VA-Hen and VA-Dam) are located at the periphery of this genetic matrix, whereas others (VA-Pol and VA-Rat) established only a remote relationship with the matrix.
Populations of India that were used in this study. (Abbreviations are given in Supplementary Table S1 and Supplementary Table S2). A full color version of this figure is available at the Journal of Human Genetics journal online.
Principal component analysis (PCA) map of the 21 Sri Lankan subpopulations with Indian populations based on net genetic distances. (Abbreviations are given in Supplementary Table S1 and Supplementary Table S2). A full color version of this figure is available at the Journal of Human Genetics journal online.
MtDNA haplogroup in Sri Lanka
Although the mitochondrial coding region contains several phylogenetically relevant sites that are useful in assigning haplotypes to a haplogroup, the control region is also promising in putative haplogroup affiliation.28, 29 According to the haplogroup assignment based on HVS-1 and part of HVS-2 sequences of Sri Lankan population using Mitotool (http://www.mitotool.org)17 and Haplogrep 1515 and then manually rechecked it again with phylotree build 15 (http://www.phylotree.org)16 (Supplementary Table S3), the overall haplogroup analysis indicated that almost 50% of the individuals from all the studied populations belonged to haplogroup M lineages (including haplogroup M, D and G) followed by about 25% of R lineages (including haplogroup R, P and T) and 20% of U lineages. Other less frequent lineages were almost 4% of R0 (including haplogroup HV and H) and almost 2% of N lineages (including N, and W) (Table 2).
Haplogroup M was the most common haplogroup in Indian Tamils (70.18%), which was contributed mainly by sub-haplogroups M5a (14.03%) and M2a (12.28%). These sub-haplogroups were rarely found in other populations. Up-country Sinhalese, Low-country Sinhalese and Sri Lankan Tamils exhibited similar frequencies of haplogroup M (41.67–43.59%), though they possessed different sub-haplogroups frequencies. This might indicate that the later mentioned, especially Up-country Sinhalese and Low-country Sinhalese are more closely related to each other than to Indian Tamils who have a known migration history from India. Meanwhile, Vedda people had the lowest frequency of haplogroup M (17.33%). It is quite astonishing to see such a lower frequency of M haplogroup in the Vedda population when compared with southern Indian tribal groups (70–80%) as well as southern Indian caste populations (65%).30 This is probably due to the effect of genetic drift in the smaller population of Vedda. This is supported by other observation of reduced intrapopulation diversity among the subgroups of Vedda people.
On the other hand, Vedda people and Low-country Sinhalese showed relatively high frequencies of haplogroup R (45.33 and 25%, respectively) which was contributed mainly by sub-haplogroup R30b (38.67 and 20%). The haplogroup was less frequent in Up-country Sinhalese, Sri Lankan Tamils and Indian Tamils. Haplogroup U was mostly found in Vedda (29.33%) and Up-country Sinhalese (23.33%), with highest contribution from sub-haplogroups U1a’c (12 and 5%, respectively) and U7a (13.33 and 11.67%, respectively).
The haplogroup frequency of Vedda people from each site is shown in Supplementary Table S7. Low frequency of M haplogroup and high frequencies of R and U haplogroups were found to be the unique characteristics of Vedda. However, the frequencies of these haplogroups varied among Vedda from different sites. Two Vedda groups (VA-Dam and VA-Hen) posses the frequency of M haplogroup close to that of Up-country Sinhalese, Low-country Sinhalese and Sri Lankan Tamils, indicating the genetic admixture between these two Vedda groups and the other three populations. The Vedda subgroups shared haplogroup R30b/R8a1a3 at relatively high frequencies, the characteristic not found among subgroups of other ethnic populations on the island, suggested a common shared origin of the Vedda population. Median Joining network of HVS-1 and part of HVS-2 sequence of haplogroups R (62 individuals, 21 haplotypes) and U (52 individuals, 25 haplotypes) from five Sri Lankan populations were constructed (Supplementary Figure S3). In general, the network was in agreement with the mtDNA haplogroup analysis. Although posses less frequency of both haplogroups, the haplotypes, belonging to these haplogroups, of the other four Sri Lankan populations were more diverse than Vedda haplotypes, which were also highly derived within the tree. The Median Joining network incorporating data of HVS-1 and part of HVS-2 sequences of haplogroups R and U24, 25 from Indian populations was also performed (Supplementary Figure S4). The Median Joining network map does not reveal a basal status of the Vedda's sequences for the genetic differentiation of haplogroups R and U. It is more likely that these two haplogroups, found to be particularly prevalent in the Vedda, were derived from ancestors on the Indian subcontinent.
Three haplogroups, M2, U2i (U2a, U2b and U2c) and R5, recognized as a package of Indian-specific mtDNA clades harboring an equally deep coalescent age of about 50 000–70 000 years,30 were present in the ethnic populations of Sri Lanka. All the ethnic populations studied possess R5 with its highest frequency (10%) observed in Up-country Sinhalese. Haplogroup U2 was found in all the studied populations with its marked high frequency (10.25%) observed in Sri Lankan Tamils. Interestingly, all the types of haplogroups in Vedda people, except sub-haplogroups M36d and M73’79, are presented in other ethnic groups as well.
There are several West Eurasian haplogroups, belonging to the HV, W, T, U1, U5 and U7 lineages, found in Sri Lankan ethnic populations (Table 2). The western Eurasian contribution to the Sri Lankan maternal gene pool was about 19.94%, which is consistent with the previous report.30 Interestingly, West Eurasian contributions of 28.19, 25.33, 25 and 20% were detected in the Sri Lankan Tamils, Vedda people, Up-country Sinhalese and Low-country Sinhalese respectively, whereas only a 1.75% contribution was evident in the Indian Tamils. This again reflected the close genetic relationship among the two Sinhalese groups and Sri Lankan Tamils when compared with Indian Tamils. Haplogroup U1a and U7a were the only West Eurasian lineage observed in the Vedda people. Haplogroup T was present in two populations; Low-country Sinhalese (5%) and Sri Lankan Tamils (2.56%), whereas Haplogroup W was present only in Up-country Sinhalese (3.33%).
Lower frequencies than the West Eurasean haplogroups were observed for the East Asian haplogroups (M12 and G), which accounted for 5.91% of the total variation.
The genetic structure of the Sri Lankan populations
Grouping of the Sri Lankan populations according to different criterion was performed and statistically tested, using an analysis of molecular variance, to reveal the best model representing natural population differentiation. Beside the ethnic criteria adopted all through this study, populations were classified into groups according to linguistic, geographic and putative racial criterion. Results are shown in Table 3. When populations were classified into two groups, Vedda people probably representing earliest inhabitants of the island and others for newcomers, this grouping gave the minimum variance among subgroups within a population (87.85, P<0.001) and maximum variance among populations (8.15, P<0.001), representing the best model for population differentiation. This model of population differentiation is compatible with a deeper root of genetic divergence between the Vedda and non-Vedda populations than between subgroups within each population.
Discussion
This study demonstrates the mtDNA genetic relationships among five main recognized ethnic groups on the island of Sri Lanka, as well as their affiliations with several ethnic people of the Greater Indian subcontinent. All the island populations, except some subgroups of the Vedda, form close genetic affiliations among themselves and with majority of the groups from the mainland suggesting the origin of the majority of the island population on the Indian mainland. No definite association of the Sinhalese with any specific ethnic or linguistic groups of India was, however, detected in this study; thus, their exact immediate origin on the mainland remains yet to be confirmed.
There is no clear genetic separation based on the PCA map between Sinhalese and Tamils, and between Up- and Low-country Sinhalese of Sri Lanka. The latter phenomenon suggests a recent division of the Sinhalese into Up- and Low-country, the fact confirmed on a historical ground.31 For the groups represented in this study, majority of the Up-country Sinhalese formed closer association among themselves than did their Low-country ethnic counterparts. This is to a certain degree explicable in a light of the isolation-by-distance; the Up-country Sinhalese groups are more geographically proximal with each other than do their Low-country counterparts. However, the closer association of the Up-country Sinhalese with the Sri Lankan Tamils than with the Indian Tamils is not in agreement with the geographic distances among them. Despite recent habitation of the Indian Tamils in proximity of the Up-country Sinhalese, the Indian Tamils might have admixed, during the long distant past, more with Sri Lankan Tamils, who have lived on the island longer than their Indian ethnic counterparts.32, 33
The genetic distinctiveness of the Vedda people on the island of Sri Lanka, as reported in this study, confirm previous results based on the analyses of nuclear markers.11, 12, 13 The markedly higher frequencies of the haplogroup R30b/R8a1a3 in all Vedda subgroups than in other Sri Lankan populations is compatible with a hypothesis that all the Vedda subgroups would have shared a common origin. The greatest inter-population genetic diversity observed among the Vedda subgroups coupled with their relatively low haplotype and nucleotide diversity would reflect greater effect of the genetic drift in the Vedda than in other ethnic groups of Sri Lanka. The pattern of genetic differentiation observed in the Vedda is a characteristic also observed in various other aboriginal populations of the world with their relatively small subgroups experiencing a long history of separation.34, 35, 36, 37, 38, 39 Such population history was also proposed for the Vedda based on the anatomical analysis.10 Much greater genetic similarities with Sinhalese, and to a lesser degree with Sri Lankan Tamils, observed in some Vedda subgroups (VA-Dam, VA-Hen and VA-Pol) in comparison with other subgroups of the same ethnic category (VA-Rat and VA-Dal) suggests that the pattern of genetic admixture between older inhabitants (Vedda) and more recent newcomers (Sinhalese and Tamils) on the island was truly heterogeneous. Advance admixture of VA-Dam, VA-Hen and VA-Pol with other ethnic populations on the island is confirmed by the presence of several shared sub-haplogroups (M33a1, D, R5a and U7a) among them that are not found in VA-Rat and Va-Dal. The reduced intrapopulation genetic diversity observed among subgroups of the Vedda is most likely a result of severe genetic drift associated with the practice of endogamy among small-sized villages during the long distant past, a phenomenon with firm historical evidence.4, 5
References
Bandaranayake, S. The peopling of Sri Lanka: the national question and some problems of history and ethnicity. Ethn. Soc. Change Sri Lanka 1–19 (1985).
Statistical Abstract of the Democratic Socialist Republic of Sri Lanka, 2001 Department of Census and Statistics, Ministry of Finance, Colombo, Sri Lanka (2002).
De Silva, M. W. S. 3, (Gunter Narr Verlag, Tubingen (1979).
De Silva, C. R. in The Vanishing Aborigines ed. Samarasinghe D. A., 34–47 (ICES: Colombo, 1990).
Seligmann, C. G. & Seligmann, B. Z. The Veddas, (Cambridge University Press: Cambridge, 1911).
De Silva, M. W. S. Current Trends in Linguistics v, (Mouton: The Hague, 1969).
Premathilake, R. Human responses to late quaternary climatic changes in Central Sri Lanka. J. Natn. Sci. Found. Sri Lanka 33, 157–159 (2005).
Deraniyagala, S. U. Prehistory of Sri Lanka: An Ecological Perspective, (Department of Archaeology survey, Government of Sri Lanka Colombo, 1992).
Hawkey, D. In Spolia Zeylanica, Vol. 39 ed. Wikramasinghe N., 13–27 (Department of National Museum: Colombo, 2002).
Kennedy, K. A. R. Recent Discoveries and Studies of the Human Skeletal Remains of Ancient Sri Lankans: A palaeoanthropological update, (MD Publications Pvt. Ltd.: New Delhi, 1993).
Illeperuma, R. J., Markalanda, D., Mountain, J. L., Ratnasooriya, W. D., Fernandopulle, N. D. & Bamshad, M. J. Haplotype data for 12 Y-chromosome STR loci of Sri Lankans. Forensic Sci. Int. Genet. 4, e119–e120 (2010).
Illeperuma, R. J., Markalanda, D. A., Ratnasooriya, W. D. & Fernandopulle, N. D. Genetic variation at 11 autosomal STR loci in the aboriginal people, the Veddahs of Sri Lanka. Forensic Sci. Int. Genet. 4, 142 (2010).
Illeperuma, R. J., Mohotti, S. N., De Silva, T. M., Fernandopulle, N. D. & Ratnasooriya, W. D. Genetic profile of 11 autosomal STR loci among the four major ethnic groups in Sri Lanka. Forensic Sci. Int. Genet. 3, e105–e106 (2009).
Moore, D. D (ed.). Preparation and analysis of DNA, (John Wiley and Son: USA, 1995).
Kloss-Brandstatter, A., Pacher, D., Schonherr, S., Weissensteiner, H., Binna, R., Specht, G. et al. HaploGrep: a fast and reliable algorithm for automatic classification of mitochondrial DNA haplogroups. Hum. Mutat. 32, 25–32 (2011).
van Oven, M. & Kayser, M. Updated comprehensive phylogenetic tree of global human mitochondrial DNA variation. Hum. Mutat. 30, E386–E394 (2009).
Fan, L. & Yao, Y. G. An update to MitoTool: using a new scoring system for faster mtDNA haplogroup determination. Mitochondrion 13, 360–363 (2013).
Librado, P. & Rozas, J. DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics 25, 1451–1452 (2009).
Excoffier, L. & Lischer, H. E. Arlequin suite ver 3.5: a new series of programs to perform population genetics analyses under Linux and Windows. Mol. Ecol. Resour. 10, 564–567 (2010).
Yao, Y. G., Nie, L., Harpending, H., Fu, Y. X., Yuan, Z. G. & Zhang, Y. P. Genetic relationship of Chinese ethnic populations revealed by mtDNA sequence diversity. Am. J. Phys. Anthropol. 118, 63–76 (2002).
Tamura, K., Dudley, J., Nei, M. & Kumar, S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24, 1596–1599 (2007).
Nei, M. (ed.) Molecular Evolutionary Genetics, (Columbia University Press: New York, 1987).
Peakall, R. & Smouse, P. E. GENALEX6: genetic analysis in Excel. Population genetic software for teaching and research. Mol. Ecol. Notes. 6, 288–295 (2006).
Palanichamy, M. G., Sun, C., Agrawal, S., Bandelt, H. J., Kong, Q. P., Khan, F. et al. Phylogeny of mitochondrial DNA macrohaplogroup N in India, based on complete sequencing: implications for the peopling of South Asia. Am. J. Hum. Genet. 75, 966–978 (2004).
Sharma, G., Tamang, R., Chaudhary, R., Singh, V. K., Shah, A. M., Anugula, S. et al. Genetic affinities of the central Indian tribal populations. PLoS One 7, e32546 (2012).
Bandelt, H. J., Forster, P. & Rohl, A. Median-joining networks for inferring intraspecific phylogenies. Mol. Biol. Evol. 16, 37–48 (1999).
Tajima, F. & Nei, M. Estimation of evolutionary distance between nucleotide sequences. Mol. Biol. Evol. 1, 269–285 (1984).
Cordaux, R., Saha, N., Bentley, G. R., Aunger, R., Sirajuddin, S. M. & Stoneking, M. Mitochondrial DNA analysis reveals diverse histories of tribal populations from India. Eur. J. Hum. Genet. 11, 253–264 (2003).
Dubut, V., Murail, P., Pech, N., Thionville, M. D. & Cartault, F. Inter- and extra-Indian admixture and genetic diversity in reunion island revealed by analysis of mitochondrial DNA. Ann. Hum. Genet. 73, 314–334 (2009).
Metspalu, M., Kivisild, T., Metspalu, E., Parik, J., Hudjashov, G., Kaldma, K. et al. Most of the extant mtDNA boundaries in south and southwest Asia were likely shaped during the initial settlement of Eurasia by anatomically modern humans. BMC Genet. 5, 26 (2004).
Deraniyagala, P. E. P. The physical anthropology of Ceylon. Ceylon MNational Museum's Ethnographic Series Publication No.2 (1961).
Geiger, W. The Mahavamsa, or the Great Chronicle of Ceylon, (Pali Text Society: Sri Lanka, 1964).
Law, B. C. The Dipavamsa, or the chronicle of the island of Ceylon. Ceylon Hist. J. 7, 1–266 (1957).
Vigilant, L., Stoneking, M., Harpending, H., Hawkes, K. & Wilson, A. C. African populations and the evolution of human mitochondrial DNA. Science 253, 1503–1507 (1991).
Watson, E., Bauer, K., Aman, R., Weiss, G., von Haeseler, A. & Paabo, S. mtDNA sequence diversity in Africa. Am. J. Hum. Genet. 59, 437–444 (1996).
Excoffier, L. & Schneider, S. Why hunter-gatherer populations do not show signs of pleistocene demographic expansions. Proc. Natl Acad Sci. USA 96, 10597–10602 (1999).
Chen, Y. S., Olckers, A., Schurr, T. G., Kogelnik, A. M., Huoponen, K. & Wallace, D. C. mtDNA variation in the South African Kung and Khwe-and their genetic relationships to other African populations. Am. J. Hum. Genet. 66, 1362–1383 (2000).
Endicott, P., Gilbert, M. T., Stringer, C., Lalueza-Fox, C., Willerslev, E., Hansen, A. J. et al. The genetic origins of the Andaman Islanders. Am. J. Hum. Genet. 72, 178–184 (2003).
Thangaraj, K., Singh, L., Reddy, A. G., Rao, V. R., Sehgal, S. C., Underhill, P. A. et al. Genetic affinities of the Andaman Islanders, a vanishing human population. Curr. Biol. 13, 86–93 (2003).
Acknowledgements
We are grateful to all the people who have donated their hair samples for making this study possible. We would like to thank Dr Upeksha Samaraweerachchi, Deepal Edirisinghe, Nirmala Ranaweera, Dayarathna Ranaweera, Sanjeewa Jayakody, G.G. Sirisena, U.S. Yapa, K. Sanath, Achala Chandradasa, T. Kumarasiri, M. Pushpawathi, Nimesha Palliyaguruge and H. Ranaweera for their excellent help in the field trips. Our special thanks extended to Professor Dr Senake Bandaranayake, Associate Professor Dr Suraphan Na Bangchang and Dr Bhoom Suktitipat and the three anonymous reviewers for their critical comments of the manuscript and their constructive discussions, to Dr Russell Thomson for the proof-reading of this manuscript and to Professor Drs Hans Jurgen Bandelt and Walther Parson for their valuable comments on the dataset validation. We also thank scholars who kindly provided their published mtDNA HVS-1 sequences. This work was partly supported by Siriraj Graduate Thesis Scholarship, Faculty of Medicine Siriraj Hospital Mahidol University, Thailand to LR. PL is supported by ‘Chalermphrakiat’ Grant, Faculty of Medicine Siriraj Hospital, Mahidol University, Thailand.
Author information
Authors and Affiliations
Corresponding author
Additional information
Supplementary Information accompanies the paper on Journal of Human Genetics website
Supplementary information
Rights and permissions
About this article
Cite this article
Ranaweera, L., Kaewsutthi, S., Win Tun, A. et al. Mitochondrial DNA history of Sri Lankan ethnic people: their relations within the island and with the Indian subcontinental populations. J Hum Genet 59, 28–36 (2014). https://doi.org/10.1038/jhg.2013.112
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/jhg.2013.112
Keywords
This article is cited by
-
The mitochondrial genomes of two Pre-historic Hunter Gatherers in Sri Lanka
Journal of Human Genetics (2023)
-
The maternal ancestry of the Kavaratti islanders and the last glacial maximum aftermath
Molecular Genetics and Genomics (2023)
-
Mitochondrial DNA (CA)n dinucleotide repeat variations in Sinhalese and Vedda populations in Sri Lanka
Genetica (2022)
-
X-chromosomal STR based genetic polymorphisms and demographic history of Sri Lankan ethnicities and their relationship with global populations
Scientific Reports (2021)
-
Indigenous and traditional foods of Sri Lanka
Journal of Ethnic Foods (2020)