Abstract
Kabuki syndrome (KS) is a congenital syndrome with an estimated prevalence of 1 in 32 000. Individuals with the syndrome have multiple malformations, but remain identifiable by the presence of the distinctive craniofacial anomalies associated with the condition. Discovered in 1981 by two independent groups of Japanese scientists, spearheaded by Yoshikazu Kuroki and Norio Niikawa, much ambiguity relating to the syndrome persisted for over 30 years after it was initially discovered, with no definitive conclusions about its etiology having ever been established. Recently, mutations within the MLL2 gene have been identified as potentially implicative. Mutations within the MLL2 gene in KS patients have been promising not only because of their relatively high presence in affected individuals, but also because of pre-existing information in the literature having validated mutant MLL2 genes in KS as a highly significant finding. Although found to be present in the majority of cases, the absence of MLL2 mutations in all patients with the syndrome is suggestive that the condition may still display a degree of genetic heterogeneity, and further still, present with more complex inter genomic interactions than initially proposed.
Similar content being viewed by others
Introduction
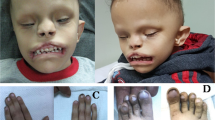
Kabuki syndrome (KS) is a congenital disorder with an estimated incidence of 1 in 32 000.1 Contrary to initial suggestions, KS has since been documented across multiple ethnicities, including those of Northern European, Brazilian, Filipino, Vietnamese, Arab, East Indian, Chinese, Mexican and African descent2 with approximately 400 cases having been reported globally.3 The syndrome presents with multiple malformations, and among the most commonly observed signs include distinctive craniofacial features, involving eversion of the lower lateral eyelid, arched eyebrows with the lateral one-third dispersed or sparse, depressed nasal tip and prominent ears.2 Cardiac defects are reported in approximately 32% of cases,4 although hypotonia5 and growth-related abnormalities1 are also commonly observed, with intellectual disability being present in >90% of cases.6
A limited number of organizations have arisen devoted to supporting individuals with the syndrome, but are known to be present in, Japan, Australia, France and the United Kingdom. Research with definitive conclusions is limited in the literature, and although the condition has been characterized since 1981, only over 30 years later had some ambiguity surrounding the etiology been resolved, which came in 2010 with the identification of potential causative defects within the ‘MLL2 gene’ (myeloid/lymphoid or mixed-lineage leukemia 2), further validated in July 2011.7 The identification of such mutations in KS patients were not only promising because of its relatively high presence 66–74%,7 but pre-existing information on the MLL2 gene,8, 9 were already able to partly validate mutant MLL2 genes as a strong candidate. Although further research will undoubtedly be required, owing to the significant phenotypic overlap of KS with other congenital syndromes10, 11 these latest developments are able to potentially offer at least one test for accurate diagnoses; taking diagnosis from the potentially speculative,7 to the molecularly validated. Currently, KS provides an example of how recent mass innovation in genome sequencing technologies and research into human genetics have been, and still continue to aid uncovering much uncertainty pertaining some of the rarest conditions known to be present globally.
History of discovery
KS, also known as Kabuki make-up syndrome, or Niikawa–Kuroki syndrome, was initially discovered in 1981 by two independent groups of Japanese scientists, spearheaded by Yoshikazu Kuroki and Norio Niikawa. In the first of two papers published on the condition, Kuroki gave an account of how in 1968 he was presented with a case of ‘an odd looking boy, with an array of malformations that did not fit into any known syndromes’. Kuroki, 10 years later, was presented with another patient with similar features, and over the following 2 years, an additional three patients were found. In his investigations of the five cases, Kuroki successfully identified a group of distinct ‘peculiar phenotypes’, which included moderate intellectual disabilities, postnatal growth retardation, susceptibility to infections, peculiar craniofacial features, and hearing anomalies.12 All five cases were found to share at least these phenotypes, which Kuroki acknowledged as sufficient to conclude that the five patients had the same malformation syndrome. A ‘new’ congenital syndrome had thus been characterized.
As part of his investigations, Kuroki found there to be no identifiable chromosomal abnormalities, as results from karyotype analysis of the five patients and their parents were normal when tested via conventional Giemsa staining. Kuroki’s results lead him to conclude that although the definitive etiology of the syndrome was largely unknown, the absence of familial occurrence and consanguinity seemed to exclude recessive inheritance, and the absence of a definite mutagen or teratogen common to all pregnancies made environmental causes even less of a likelihood. Among his five cases the sex ratio observed was M4:F1, leading Kuroki to suggest autosomal dominant or X-linked inheritance as the likeliest mode of transmission.
The second of two main papers published in determining the existence of the syndrome was headed by the Japanese scientist Norio Niikawa. Niikawa’s first patient presented as a female infant in 1967, which lead him to search for others with similar features. Niikawa also collated a total of five patients, on whom he based his investigations. Niikawa proceeded to identify possible common genetic errors in his patient sample; however, his analysis also uncovered no genetic abnormalities. Owing to the multitude of phenotypic expressions presented, Niikawa sort to conduct a differential diagnosis on all patients included, so as to ensure KS was in fact an uncharacterized condition. Niikawa noted that some of the features his patients presented with such as eversion of the lower eyelid, short stature and scoliosis were also present in individuals suffering from Fetal face (Rainbow) syndrome, however, the absence of macrocephaly and mental retardation in those suffering from Fetal face syndrome lead him to conclude that these were in fact two distinct conditions.11 In 2008, Utine et al.13 carried out a study on 10 patients presenting with KS and other patients affected by various other chromosomal imbalances. In their direct comparisons of KS patients and two related individuals with Trisomy 10, Utine et al.13 were able to highlight ‘significant overlaps’ in the clinical picture of KS and trisomy 10p, concluding chromosomal analysis to be an important step (by the process of elimination) in distinguishing KS as a distinct clinical entity.
Early research
Following discovery of the syndrome, further investigations were carried out into the initial suggestions by Kuroki et al.12 with regards to the possibility of autosomal dominant inheritance. Matsumoto et al.14 carried out a review on 251 affected individuals, 14 of whom were familial; with a case reflecting maternal transmission from mother to both son and daughter, as wells as an observation of father to daughter transmission. On identifying the possibility of transmission from both parents, Matsumoto et al.14 concluded it least likely that genomic imprinting were a cause, and that the findings of their study were ‘not inconsistent with the hypothesis that KS was an autosomal dominant disorder’. Further research by the likes of Poottinger et al.15 added more evidence to this hypothesis via their investigations of a particular family where a daughter was believed to have inherited the condition from her mother. In analysis of their cases’ extended family, no evidence for the condition was noted in the mothers three sibling, nor in her parents. Inconsistencies in familial transmission had further complicated matters in determining etiologies, but it nevertheless suggested the possibility that a new dominant mutation had arisen in the mother of the affected daughter.
The association between KS and chromosome X abnormalities was another possibility that had been widely investigated. Turner syndrome, a chromosomal abnormality in females presents with X chromosome monosomy (45X). The syndrome manifests itself via several phenotypes, including short stature, webbed neck and numerous congenital heart conditions. In 1994, Wellesley et al.10 identified a case in which a 2-year-old girl presented with clinical features of both Turner syndrome and KS. The diagnosis of Turner syndrome was confirmed by cytogenetic analysis, but KS was suspected as a result of the presence of characteristic Kabuki dysmorphic facial features, as well as the presence of microcephaly. In their findings, Wellesley et al.10 identified a range of overlapping phenotypes between the two syndromes, including primary ovarian dysfunction. The occurrence of both conditions in Wellesley’s patient, as well as overlapping of phenotypes, suggested a possible common etiological cause, opening yet another area for potential investigations.
More recently, Chen et al.16 investigated the case of a 6-year-old girl who presented with characteristic KS facial dysmorphologies, including long palpebral fissures, everted lateral third of the lower eyelids, arched eyebrows, a depressed nasal tip, as well as aortic coarctation and patent ductus arteriosus. Cytogenetic analysis confirmed Turner syndrome, and revealed a 45X/47XXX mosaicism, with 45X present in seven times greater proportion. Their findings led them to conclude that their case presented evidence for the association between KS and mosaicism for a high level of 45X and a low level of 47,XXX, and thus added mosaic 45,X/47,XXX to the list of cytogenetic abnormalities associated with KS. However, in light of their conclusions, Bianca et al.17 carried out a review of data from 8 KS patients, 32 TS and cases with 16 other sex chromosomal abnormalities, including 47,XXX; 47,XXY and 47,XYY. Their findings suggested there to be ‘no association’ between the two conditions, but did agree that further evaluations into the insights of overlapping phenotypes and co expression of both syndromes, could enable identification of the molecular basis and therefore cause of KS. Although results from the studies carried out by Chen et al.16 were not sufficient enough to make such definitive conclusions, such findings ultimately resulted in further research into the X chromosome potential for being at least partly involved in the KS phenotype.
In approximately 50% of Turner syndrome cases 45,X/46,XX mosaicism is known to be present.18 X Chromosome inactivation, initiated by the XIST gene, is a dosage compensating mechanism, as the difference between XX females and XY males is the ability to be functionally equalized by silencing one of the two X chromosomes in female.19 A ring X chromosome is an example of an X chromosomal structural abnormality, in which the two ends of the chromosome fuse.20 In approximately 6% of Turner syndrome cases, a ring X chromosome has been detected, and in a small group of such individuals, clinical manifestations resembling KS have also been documented.21
Rodriguez et al.21 carried out investigations on another female case displaying 45,X/46,X, r(X) mosaicism who had characteristics of both Turner syndrome and clinical features resembling KS. The patient was found to ultimately lack the presence of the XIST gene on the X ring and wild type X chromosomes. Rodriguez et al.21 suggested that the absence of the XIST gene and subsequent activity of genes localized to the r(X) resulted in the functional disomy of the contained genes on the r(X), thus being responsible for the patient’s phenotype. They also noted that the patient suffered from Congenital contractural arachnodactyly, which may have been attributed to the functional disomy of the IGBP1 gene (immunoglobulin-binding protein 1) present on the X chromosome; a gene associated with various other phenotypes, including mental retardation and micrognathia. Rodriguez et al.21 concluded that the phenotype associated with r(X) would therefore be able to be partially predicted by the grade of mosaicism, presence of the XIST gene and status of the level of inactivation concerning the r(X). The findings by Rodriguez et al.21, were further supported with research by Su et al.22, as they were also able to identify features of KS in a 13-year-old female with Turner syndrome mosaicism 45,X/46,X,r(X), and incomplete XIST gene expression on her ring X chromosome. Limitations to this theory, however, included the inability to extrapolate these findings to the cause of KS in male patients; with a potential argument reflected in the similar prevalence observed between males and females,23 thus suggesting a common etiological cause to be most likely. In light of recent findings, however, pre-existing data pertaining to the significance of the X chromosome in this etiology may indeed still be of great relevance. The recent identification of the MLL2 gene believed to be implicative in KS,3 and the more recently proposed X chromosomal interactions with this gene in a small subset of patients 24 have shed light on this ambiguity, and revealed the search into more complexed genomic interactions as a plausible line for further investigation.
The MLL2 gene association
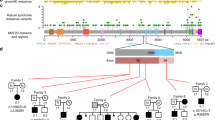
In the summer of 2010, Ng et al.3 published a paper on the newly identified gene related to KS. Mutations in the MLL2 gene were identified by exome sequencing, with the use of a next-generation sequencer, the Illumina Genome Analyzer II (GAII, Illumina, San Diego, CA, USA), and further validated by conventional Sanger sequencing. With this approach, the exons of interest were able to be extracted from the genomic DNA using microarrays in an initial hybridization step, after which the data were sequenced.25 Ng et al.3 proceeded with their investigations by sequencing the exome of ten unrelated individuals with KS, seven of whom were of European ancestry, two of Hispanic ancestry and one of mixed European and Haitian ancestry. In determining a target exome, the group focused their efforts on nonsynonymous variants, splice acceptor and donor site mutation, as well as coding indels. The mutations were eventually excluded from progressive analysis, as the identified mutations were considered to be minimally disruptive and unlikely to be reflective of the KS phenotype. The team then narrowed down to the chosen exome by consulting databases and excluding all other known variants. An average of 6.3 Gb of DNA per sample was sequenced, achieving 40 × coverage of the targeted exome.
The result from these investigations identified a single candidate gene (MUC16), which was shared across all ten exomes. However, the MUC16 gene was suggested to be a likely false positive owing to its extremely large size (14 507 amino acids). From their findings, Ng et al.3 postulated the theory that their groups’ failures in identifying a compelling candidate gene could have been attributed to the possible genetic heterogeneity of KS. Other possibilities listed were in the potential inaccuracies of sequencing machinery and the possibility of mutations outside the scope of the targeted exome.
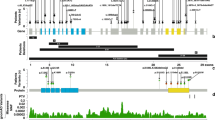
To accommodate for the possibility of genetic heterogeneity, the team conducted a ‘less stringent’ analysis, and began searching for candidate genes shared across subsets of affected individuals. Exome sequencing identified nonsense substitutions or frameshift indels within the MLL2 gene in seven of the ten individuals with KS. All seven loss-of-function variants in MLL2 were validated by Sanger sequencing, but ultimately, loss-of-function mutations in MLL2 were identified in nine out of ten cases. The group then proceeded to validate their findings by screening for MLL2 mutations in 53 additional cases. Overall, 32 distinct MLL2 mutations were identified in 35 of 53 families (66%) with KS. These mutations included 20 nonsense mutations, 7 indels and 5 amino acid substitutions. De novo mutations in the MLL2 gene were found to have arisen in 12 of the cases. There were also two familial cases of KS displaying parent to child transmission, where the MLL2 mutations resulting in p.4527K>X and p.5464T>M were identified. Ng et al.3 concluded stating the possibility of non-coding or missed mutations within MLL2 as a potential explanation for the 18 of 53 cases in whom no protein-altering variants were identifiable.
Hannibal et al.7 in 2011 screened 110 families with KS for mutations within the MLL2 gene by CGH (comparative genomic hybridization) Microarray analysis. The findings of the study revealed MLL2 mutations in 74% (81/110) of cases, although also identifying MLL2 transmission in two of three familial cases. The majority of mutations identified were de novo, and predicted to cause pathology on the basis of haploinsufficiency, a conclusion also supported by Micale et al.26 who identified 45 patients with MLL2 nucleotide variants in their cohort of 62 patients.
Paulussen et al.27 in 2011 directly sequenced all 54 exons of the MLL2 gene in 45 KS patients. They identified 34 (75.6%) different mutations with all but one having never been previously identified, but all significant enough to cause a high frequency of absent or non-functional MLL2 protein. On further analysis of phenotype, the group were also able to identify statistically significant variances in both craniofacial abnormalities (P=0.0143), and growth retardation (P=0.0040) when comparing KS patients with an MLL2 mutation (n=34) to those without a identified MLL2 mutation (n=11), a finding also observed by Li et al.28 They sequenced the MLL2 exons of 34 patients with KS also identifying mainly de novo mutations within the MLL2 gene. In their study, MLL2 mutation carriers presented significantly more often with short stature (P=0.026) than those without.28 Such findings may now broadly give explanation to the variances identified in earlier studies that aimed to characterize the syndrome, and have thus reiterated initial suggestions for genetic heterogeneity. In 2005 Armstrong et al.23 investigated 48 ‘well defined’ cases of KS. Associated with KS were a group of distinct craniofacial features observed in the majority, which were the basis for identifying the syndrome as a distinct clinical entity in its first instance.11, 12 The features included microcephaly in 65% of cases, a forehead described as ‘tall’ in 92% of patients, and long palpebral fissures in 78% of patients. The points of distinct variance in overall phenotype included a fissuring of the tongue in only 2% of their cases, variability in palate deformities having been described as cleft (46%), high arched (31%), narrow (17%), or incompetent (6%) and hearing loss in 27% of individuals. Such findings may now be partly explainable on the basis of potentially ‘MLL2’-mediated KS and other such KS causative factors and/or genes yet to be identified.
Lederer et al.24 most recently published findings on data from two female and one male patient with KS. De novo partial or complete deletions of an X chromosome gene, KDM6A (lysine (K)-specific demethylase 6A, also known as UTX) were identified in all three patients. The pre-existing data on KDM6A identified its protein as one that interacts with MLL2, and the absence of such mutations within a cohort of 411 normal controls of a previous study, suggested KDM6A involvement as a significant finding. The KDM6A gene encodes for a histone demethylase, and is known to physiologically escape X inactivation.29, 30 In their study, a skewed inactivation pattern for the gene was observed in the majority of cells of all three patients. The KDM6A and MLL2 gene functioned together in the epigenetic control of transcriptionally active chromatin and were also previously identified, in combination as well as independently, to have significant roles in the regulation of muscle-specific genes during embryogenesis and development.31, 32, 33 A broad phenotypic spectrum was also observed between these patients, all three of whom did, however, posses many of the characteristic facial features.
MLL2 and the KS phenotype
The wild-type MLL2 gene is known to code for a large protein associated with the ‘SET’ family of proteins. The MLL2 SET domain is known to have an important role in the epigenetic control of chromatin states via the modulation of histone 3 and lysine 4 methyltransferase activity,3 and the majority of variants determined by Ng, were predicted to truncate the polypeptide chain of the protein before translation of the SET domain, suggesting loss of function, and therefore haploinsufficiency as the likely mechanism for the KS phenotype.3
Discovered in 1999 by FitzGerald et al.,9 the MLL2 gene (previously known as ALR, augmenter of liver regeneration), was determined to code for a protein with great similarity to the previously identified MLL gene located at the chromosome band 19q13.1. MLL had been found to be involved in chromosome rearrangements associated with leukemia in mammals and, although no MLL2 rearrangements with leukemia have been recorded, Tumino et al.34, did carry out an investigation into two KS patients, a child of 6 years with hepatoblastoma and an infant of 6 months with neuroblastoma; and concluded that, although the relationship between KS and such malignancies remained unclear, constitutional molecular defects must have had a significant role in oncogenesis, as shown by the increased incidence of cancers in infants with many molecular defined syndromes.
Issaeva et al.8 determined the MLL2 protein to have a significant role in promoter and transcription initiation sites of target genes via the role as a histone modifier. Issaeva found that in MLL2-knockout mice models, there were 20 genes most significantly downregulated, which comprised various functions, including cell migration, growth adhesion and transcriptional regulation. The translatability of Issaeva’s findings to the human model would undoubtedly require additional research however, this data is nevertheless significant when acknowledging the characteristic KS phenotype as the evident result of disruptions in embryogenesis. Of the 20 genes significantly downregulated in Issaeva’s MLL2 knockout mice models, included those related to musculoskeletal development, such as the ENO3 (enolase 3 beta-muscle), TNNT1, TNNT2 (troponin T type 2-cardiac), NPR3 (natriuretic peptide receptor C/guanylate cyclase C) and FHL1 genes. ENO3 is known to be responsible for glycolysis, muscle development and regeneration, TNNT2 for regulation of muscle contraction and development, whereas NPR3 involvement in skeletal development.8
Conclusion
Since its discovery in 1981, knowledge in the phenotypes and epidemiology of KS has improved immensely. Initially believed to have greatest incidence in the Japanese population,12 KS has since been identified across numerous ethnic groups2 with equal prevalence in both sexes.23 It has however, taken a long period of time to determine the molecular basis of the condition owing to the complexity of the condition itself, and recent discoveries are likely to be attributable to the availability of cost effective and efficient sequencing technologies. The numerous inconclusive attempts at determining the causative gene have surely not been in vain, as it is the findings from inconclusive studies that often formed the foundations for successive ones, and may still be relevant in the search for the ‘other’ implicated genes unaccounted for by MLL2 gene defects. The discovery of the MLL2 gene in recent times has, however, been the greatest of all developments for more accurate diagnosis of KS, as well as offers a greater understanding into the etiology of the condition. There are, however, no doubts that the current failure in identifying MLL2 mutations in all patients with KS has confirmed the possibility of genetic heterogeneity and continues to encourage more fine-scale searches within the genome.
References
Niikawa, N. et al Kabuki make-up (Niikawa-Kuroki) syndrome: a study of 62 patients. Am. J. Med. Genet. 31, 565–589 (1988).
Adam, M.P., Hudgins, L. Kabuki syndrome: a review. Clin. Genet. 67, 209–219 (2005).
Ng, S.B. et al Exome sequencing identifies MLL2 mutations as a cause of Kabuki syndrome. Nat. Genet. 42, 790–793 (2010).
Digilio, M.C. et al Congenital heart defects in Kabuki syndrome. Am. J. Med. Genet. 100, 269–274 (2001).
Schrander-Stumpel, C. et al The Kabuki (Niikawa-Kuroki) syndrome: further delineation of the phenotype in 29 non-Japanese patients. Eur. J. Pediatr. 153, 438–445 (1994).
Wessels, M.W. et al Kabuki syndrome: a review study of three hundred patients. Clin. Dysmorphol. 11, 95–102 (2002).
Hannibal, M.C. et al Spectrum of MLL2 (ALR) mutations in 110 cases of Kabuki syndrome. Am. J. Med. Genet. Part A 155, 1511–1516 (2011).
Issaeva, I. et al Knockdown of ALR (MLL2) reveals ALR target genes and leads to alterations in cell adhesion and growth. Mol. Cell. Biol. 27, 1889–1903 (2007).
FitzGerald, K.T., Diaz, M.O. MLL2: a new mammalian member of the trx/MLL family of genes. Genomics 59, 187–192 (1999).
Wellesley, D.G., Slaney, S. Kabuki make-up and Turner syndromes in the same patient. Clin. Dysmorphol. 3, 297–300 (1994).
Niikawa, N. et al Kabuki make-up syndrome: a syndrome of mentalretardation, unusual facies, large and protruding ears, and postnatal growth deficiency. J. Pediatr. 99, 565–569 (1981).
Kuroki, Y. et al A new malformation syndrome of long palpebralfissures, large ears, depressed nasal tip, and skeletal anomalies associated with postnatal dwarfism and mental retardation. J. Pediatr. 99, 570–573 (1981).
Utine, G.E. et al Kabuki syndrome and trisomy 10p. Genet. Couns. 19, 291–300 (2008).
Matsumoto, N., Niikawa, N. Kabuki make-up syndrome: a review. Am. J. Med. Genet. C. Semin. Med. Genet. 117C, 57–65 (2003).
Pottinger, C.C. et al Further evidence of dominant inheritance of Kabuki syndrome. Clin. Dysmorphol. 18, 215–217 (2009).
Chen, C.P. et al Kabuki syndrome in a girl with mosaic 45,X/47,XXX and aortic coarctation. Fertil. Steril. 89, 1826 e5–e7 (2008).
Bianca, S. et al Kabuki syndrome and sex chromosomal anomalies: is it really an association? Fertil. Steril. 91, e6 (2009).
Wiktor, A.E., Van Dyke, D.L. Detection of low level sex chromosome mosaicism in Ullrich-Turner syndrome patients. Am. J. Med. Genet. A. 138A, 259–261 (2005).
Ng, K. et al Xist and the order of silencing. EMBO. Rep. 8, 34–39 (2007).
Shchelochkov, O. et al Mosaicism for r(X) and der(X)del(X)(p11.23)dup(X)(p11.21p11.22) provides insight into the possible mechanism of rearrangement. Mol. Cytogenet. 1, 16 (2008).
Rodriguez, L. et al A small and active ring X chromosome in a female with features of Kabuki syndrome. Am. J. Med. Genet. A. 146A, 2816–2821 (2008).
Su, P.H. et al Kabuki make-up (Niikawa-Kuroki) syndrome with mosaicism ring chromosome X and incomplete XIST gene expression. Acta. Paediatr. Taiwan 48, 28–31 (2007).
Armstrong, L. et al Further delineation of Kabuki syndrome in 48 well-defined new individuals. Am. J. Med. Genet. Part A 132A, 265–272 (2005).
Lederer, D. et al Deletion of KDM6A, a histone demethylase interacting with MLL2, in three patients with Kabuki syndrome. Am. J. Hum. Genet. 90, 119–124 (2012).
Cirulli, E.T. et al Screening the human exome: a comparison of whole genome and whole transcriptome sequencing. Genome Biol. 11, R57 (2010).
Micale, L. et al Mutation spectrum of MLL2 in a cohort of Kabuki syndrome patients. Orphanet J. Rare Dis. 6, 38 (2011).
Paulussen, A.D.C. et al MLL2 mutation spectrum in 45 patients with Kabuki syndrome. Hum. Mut. 32, E2018–E2025 (2011).
Li, Y. et al A mutation screen in patients with Kabuki syndrome. Hum. Genet. 130, 715–724 (2011).
Greenfield, A. et al The UTX gene escapes X inactivation in mice and humans. Hum. Mol. Genet. 7, 737–742 (1998).
Lan, F. et al A histone H3 lysine 27 demethylase regulates animal posterior development. Nature 449, 689–694 (2007).
Aziz, A., Liu, Q.C., Dilworth, F.J. Regulating a master regulator: establishing tissue-specific gene expression in skeletal muscle. Epigenetics 5, 691–695 (2010).
Herz, H.M. et al The H3K27me3 demethylase dUTX is a suppressor of Notch- and Rb-dependent tumors in Drosophila. Mol. Cell. Biol. 30, 2485–2497 (2010).
Seenundun, S. et al UTX mediates demethylation of H3K27me3 at muscle-specific genes during myogenesis. EMBO J. 29, 1401–1411 (2010).
Tumino, M. et al Kabuki syndrome and cancer in two patients. Am. J. Med. Genet. A. 152A, 1536–1539 (2010).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
I have no relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript. This includes employment, consultancies, honoraria, stock ownership or options, expert testimony, grants or patents received or pending, or royalties, and no writing assistance was used in the production of this manuscript.
Rights and permissions
About this article
Cite this article
Bokinni, Y. Kabuki syndrome revisited. J Hum Genet 57, 223–227 (2012). https://doi.org/10.1038/jhg.2012.28
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/jhg.2012.28
Keywords
This article is cited by
-
“Gestalt diagnosis” for children with suspected genetic syndromes
Italian Journal of Pediatrics (2015)
-
Autoimmune haematological disorders in two Italian children with Kabuki Syndrome
Italian Journal of Pediatrics (2014)
-
Identification of KMT2D and KDM6A mutations by exome sequencing in Korean patients with Kabuki syndrome
Journal of Human Genetics (2014)