Abstract
Human hereditary deafness at the DFNB29 locus on chromosome 21q22.1 is caused by recessive mutations of CLDN14, encoding claudin 14. This tight junction protein is tetramembrane spanning that localizes to the apical tight junctions of organ of Corti hair cells and in many other tissues. Typically, the DFNB29 phenotype is characterized by prelingual, bilateral, sensorineural hearing loss. The goal of this study was to define the identity and frequency of CLDN14 mutations and associated inner ear phenotypes in a cohort of 800 Pakistani families segregating deafness. Hearing loss in 15 multi-generational families was found to co-segregate with CLDN14-linked STR markers. The sequence of the six exons and regions flanking the introns of CLDN14 in these 15 families revealed five likely pathogenic alleles. Two are novel missense substitutions (p.Ser87Ile and p.Ala94Val), whereas p.Arg81His, p.Val85Asp and p.Met133ArgfsX23 have been reported previously. Haplotype analyses indicate that p.Val85Asp and p.Met133ArgfsX23 are founder mutations. The p.Val85Asp accounts for ∼67% of the mutant alleles of CLDN14 in our cohort. Combined with the previously reported data, CLDN14 mutations were identified in 18 of 800 Pakistani families (2.25; 95% CI, 1.4–3.5). Hearing loss in the affected individuals homozygous for CLDN14 mutations varied from moderate to profound. This phenotypic variability may be due to environmental factors (for example drug and noise exposure) and/or genetic modifiers.
Similar content being viewed by others
Introduction
Mutations of CLDN14 cause autosomal recessive nonsyndromic deafness at the DFNB29 locus. To date, six different pathogenic variants of human CLDN14 have been identified in families segregating severe to profound hearing loss, but no obvious vestibular phenotype.1, 2, 3, 4 Similarly, a Cldn14 knockout mouse is also deaf.5 Although claudin 14 is expressed in the mouse vestibular sensory epithelium, Cldn14 knockout mice appear to have no obvious vestibular disorder such as circling behavior or head bobbing.5
The mammalian claudin family of 27 genes encodes tight junction proteins that function to maintain integrity of the apical and basolateral membrane domains and prevent diffusion of solutes and solvent molecules through intercellular spaces within epithelial sheets.6, 7, 8, 9 The claudin proteins are predicted to have four transmembrane domains and short cytosolic amino and carboxy termini.10, 11 Although the first and the fourth transmembrane regions as well as the extracellular loops are highly conserved among the different claudin species, the second and the third transmembrane regions are variable.12 The first extracellular loop of these proteins has an important role in homophilic interactions (Figure 1a).13, 14 To date, most of the known mutations of claudin 14 are within or close to the second or third transmembrane domains (Figure 1a) and some of them have been shown to affect membrane localization. For example, p.Val85Asp impairs the ability of claudin 14 to form tight junction strands.4
Eight different pathogenic alleles of CLDN14 are associated with hearing loss in human. (a) Schematic of human claudin 14. Topology of claudin 14 was predicted by TMpred software. Yellow and blue amino acids indicate negatively and positively charged residues, respectively. The positions of eight residues mutated in all reported DFNB29 families are red. (b) Localization of claudin 14 in the apical bicellular tight junctions between the outer hair cells and Deiters’ cells (green). (c) Localization of claudin 14 (green) in the tight junctions between inner hair cells and pillar cells and between two adjacent pillar cells. Filamentous actin is highlighted by rhodamine–phalloidin (red). Scale bar=5 μm.
The structure of most intercellular tight junctions in the inner ear is similar to that reported in other epithelia.15, 16, 17, 18 However, the structure of the bicellular junctions between hair cells and supporting cells, especially between an outer hair cell and adjacent Deiter’s cell are more elaborate and highly specialized to maintain the ionic barrier between endolymph and perilymph.16, 17, 18 These tight junctions contain a high amount of claudin 14 and are prominently stained with anticlaudin 14 antibody (Figures 1b and c). The apical junctional complexes between the cells of the organ of Corti lack desmosomes and gap junctions and have a combination of tight junction and adherens junction features, and extend down the depth of the reticular lamina, a region spanning 3–5 μm.18
The goal of this study was to determine the spectrum of mutant alleles, and the frequencies of these alleles in 800 Pakistani families segregating nonsyndromic deafness, and to measure variability in the clinical phenotype of CLDN14 pathogenic variants. We found that pathogenic alleles of CLDN14 are associated with hearing loss that ranges from moderate to profound, and that mutant alleles of this gene appear to be a common cause of heritable hearing loss among Pakistanis.
Methods
Family participation and clinical evaluation
This study was approved by IRBs at the National Centre of Excellence in Molecular Biology (NCEMB), Lahore, Pakistan (FWA00001758), at the National Institutes of Health, USA (Combined Neuroscience IRB; OH-93-N-016), and at the Cincinnati Children’s Hospital Research Foundation, USA (2009-0684; 2010-0291). Written informed consent was obtained from adult subjects and parents of minor subjects. Hearing was evaluated in audiology clinics by pure tone audiometry at octave frequencies with intensities up to 110 dBHL. Vestibular function was evaluated by tandem gait and Romberg testing.19
Genotype and mutational analysis
Genomic DNA was extracted from 10 ml of peripheral venous blood as described.20, 21 Three fluorescently labeled microsatellite markers (D21S2078, D21S1252 and D21S2080) linked to CLDN14 were PCR-genotyped as described.20 Primers for PCR amplification and CLDN14 sequencing were designed using Primer3 (http://frodo.wi.mit.edu/).
Co-segregation of the mutations with hearing loss in each family was demonstrated for all subjects participating in this study. Control DNA samples from ethnically matched Pakistanis were sequenced to ascertain novel variants of CLDN14. Three prediction programs, SIFT,22 Polyphen-2 (Adzhubei et al.23) and MutationTaster24 were used to evaluate the potential effect of each novel missense mutation.
Immunolocalization of claudin 14 in the mouse organ of Corti
Immunolocalization of claudin 14 using tissue from C57BL/6 mouse organ of Corti was performed as described previously using a custom rabbit polyclonal PB108 anti-claudin 14 antibody with validated specificity. There was no immunolocalization signal observed when tissue from a Cldn14 knockout mouse was used.5
Results
Pathogenic variants of CLDN14
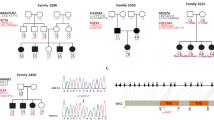
We reported that mutations of CLDN14 cause DFNB29 deafness.1 Subsequently, deafness segregating in 15 additional families (Figures 2 and 3) was found to be linked to STR markers for CLDN14 (Table 1). Sequence analysis of CLDN14 revealed four likely pathogenic variants. Among the five variants of CLDN14, two were novel missense substitutions p.Ser87Ile (c.259_260TC>AT) and p.Ala94Val (c.281C>T). In family PKDF361, we detected three nearly adjacent nucleotides changes (c.256A>G and c.259_260TC>AT; Figure 3b), which are predicted to result in two substitutions (p.Ile86Val and p.Ser87Ile; Table 1). SIFT, Polyphen-2 and MutationTaster predicted that p.Ile86Val is a benign polymorphism. Furthermore, claudin 14 orthologs in Armadillo, cow and hedgehog have a valine residue at position 86. Therefore, we considered p.Ile86Val as a non-pathogenic substitution although no carriers of c.256A>G were found in our 184 control subjects or in 1000 Genome and NHLBI-ESP databases. These data indicate that c.256A>G is a rare and benign variant whereas p.Ser87Ile is predicted to be deleterious (Table 1).
Pakistani DFNB29 families. Filled symbols represent individuals with prelingual, sensorineural hearing loss and double horizontal lines indicate a consanguineous marriage. Half filled symbols in family PKDF108 are individuals (ages 72 and 75 years) that have age-related hearing loss. Numbers represent unaffected siblings.
Pedigree of three DFNB29 families, sequencing chromatograms, pure tone audiograms and ClustalW alignment of 36 claudin 14 orthologs. (a) Pedigrees of families PKDF361, PKDF315 and PKDF488. Filled symbols represent affected individuals. (b) Wild-type and homozygous mutant nucleotide sequence chromatograms of exon 3 of CLDN14 illustrating homozygosity for the c.259_260TC>AT (p.Ser87Ile) and c.281C>T (p.Ala94Val) mutations (arrows). Shown in green are the amino acids that are mutated (red) in the DFNB29 families, while blue color represent the non-deleterious change found in family PKDF361. (c) ClustalW multiple sequence alignment of the 36 amino acids of claudin 14 shows that p.Ser87 and p.Ala94 residues are conserved across species (shaded background). For comparison, three previously reported mutated residues, p.Arg81, p.Val85 and p.Gly101 are also shown. Amino acids are numbered with reference to GenBank Accession number NP_036262. B&R: birds & reptiles; A: amphibians. (d) Pure tone air and bone conduction thresholds for family PKDF361 individuals V:1 (19 yo female), V:2 (24 yo male). Individual V:1 has severe to profound hearing loss in her left ear, whereas right ear showed profound deafness. Right ear air conduction: O; Left ear air conduction: X; Right ear bone conduction: >; Left ear bone conduction: <;↓indicates the threshold level beyond the measurable range. (e) Pure tone air and bone conduction thresholds for family PKDF488 individuals V:6 (42 yo male) revealed moderate sensorineural hearing loss in his left ear, whereas the right ear showed a severe degree of hearing loss. In contrast, individual V:7 (52 yo female) had severe to profound, bilateral, sensorineural hearing impairment.
In addition, we detected p.Ala94Val in families PKDF315 and PKDF488 (Figures 3a and b). Both p.Ser87Ile and p.Ala94Val mutations affect amino acid residues that are conserved among 36 claudin 14 orthologs (Figure 3c). No carriers of c.259_260TC>AT and c.281C>T were found among 384 ethnically matched control chromosomes that we Sanger-sequenced, in the 1000 Genome database, or in 5400 individuals listed in the NHLBI-ESP variant database. Our data indicate that these variants are not common polymorphisms, and in each family homozygosity for the mutant allele of CLDN14 co-segregated with deafness, whereas carriers had normal hearing.
We observed the previously reported variants c.254T>A (p.Val85Asp) in 12 families, c.242G>A (p.Arg81His) in one family and c.398delT (p.Met133ArgfsX23) in one family. All of these mutations co-segregated with deafness. STR markers linked to CLDN14 were genotyped in unrelated affected individuals homozygous for the c.254T>A and c.398delT mutations and for both alleles the flanking haplotypes were consistent with a founder effect (Table 1).
DFNB29 hearing loss phenotype
Pure tone air and bone conduction audiometry revealed inter- and intra-familial variability in the severity of hearing loss (Figures 3d, e and 4) in these families. The affected individuals of family PKDF361 had prelingual severe to profound hearing loss across all the tested frequencies (Figure 3d). The 42-year-old affected individual (V:6) of family PKDF488 had bilateral moderate to severe, sensorineural hearing loss, whereas his sibling (V:7 age 52 years) had profound hearing loss across almost all the frequencies (Figure 3e). Phenotypic variability was associated with the known p.Val85Asp mutation (Figure 4), an allele we reported in two large multi-generation Pakistani families segregating prelingual, severe to profound hearing loss.1, 25 In this study, we identified 11 additional families segregating the p.Val85Asp allele of CLDN14. Hearing loss in members of these families ranged from moderate to profound with greater severity at higher frequencies (Figure 4), which is in agreement with degeneration of sensory hair cells from base to apex seen in the Cldn14 knockout mice.5
Pure tone air and bone conduction measurements from DFNB29 families segregating p.Val85Asp revealed intra- and inter-familial variability in thresholds. (a) Twenty-one-year-old affected individual of family PKDF505 has profound, bilateral, sensorineural hearing loss, whereas his 23-year-old sibling has moderate to severe, bilateral sensorineural hearing impairment. Shown also is their 48-year-old mother, a carrier of p.Val85Asp allele, with normal hearing thresholds across all frequencies. (b) Ten-year-old affected individual of family PKDF704 has moderate to profound, bilateral, sensorineural hearing loss, whereas her 11-year-old sibling has severe to profound, bilateral sensorineural hearing impairment. (c) Twenty-five-year-old affected individual of family PKDF797 has severe to profound hearing loss, whereas a 23-year-old affected individual of family PKDF1092 has moderate to profound, sensorineural hearing loss.
Discussion
CLDN14 mutations are a common cause of recessive hearing loss in the Pakistani population as mutations of this gene account for 2.25% (18 of 800 families; 95% CI, 1.4–3.5) of deafness in the NCEMB Pakistani study cohort (Table 2).1, 2, 3, 25 The probands of the NCEMB deafness cohort are usually students in schools for the hearing impaired and are usually profoundly deaf from birth. We may have underestimated the contribution of mutations of CLDN14 to hearing loss by overlooking individuals with mild or delayed-onset hearing loss due to mutations of CLDN14.
Severity of hearing thresholds in our DFNB29 families does not seem to be directly correlated with age of the subject (Figure 4). For example, although similar in age, a 23-year-old affected woman of family PKDF505 has significantly better hearing, especially at low frequencies, than her 21-year-old sister (Figure 4). Similarly, in our previous study, audiograms from multiple affected individuals of family PKDF009 did not show any correlation between hearing thresholds and age of the affected individuals.25 It is possible that an environmental factor (for example, drugs, noise and so on) may be the cause of this inter- and intra-familial phenotypic variability in hearing thresholds. We hypothesized that a genetic modifier is the cause of this inter- and intra-familial phenotypic variability in hearing thresholds, especially in individual harboring the same CLDN14 mutation. Similar phenotypic variability has been documented for many other deafness-causing mutations in humans.26, 27, 28 The sensory epithelium of mouse inner ear expresses claudin family members 1, 2, 3, 9, 10, 12, 14 and 18,29 and variation in expression of these other claudins in the auditory system may be modulating the severity of the hearing loss phenotype.
Mutations in other tight junction proteins are also known to cause deafness in humans and mice.28, 30, 31 Claudin 11-deficient mice are deaf, demonstrating that this tight junction protein is also necessary for maintenance of the intra-strial compartment and generation of the endocochlear potential.30 Both claudin 14 and claudin 9 mutant mice also display deafness with no vestibular defects.5, 31 Claudin 14 is expressed specifically by the cells forming the reticular lamina (Figures 1b and c) and the vestibular sensory epithelia, whereas claudin 9 is present in nearly all of the epithelia lining the scala media and the vestibular organs.5, 31, 32 Mouse mutants of Cldn14 and Cldn9 both display cochlear hair cell loss by the second week of life, which progresses rapidly to include the entire cochlea within the next few weeks.5, 31 Loss of either of these claudins results in increased paracellular permeability of K+ in the reticular lamina and an elevation in the K+ concentration around the basolateral regions of hair cells, which is toxic.5, 31, 33, 34 Thus, both claudins are required to form a permeability barrier against cations.5, 31 The two novel mutations identified in this study, p.Ser87Ile and p.Ala94Val, are within the second transmembrane domain and in the vicinity of p.Val85, which has been shown to affect the membrane localization of claudin 14.4 Therefore, these two new mutations might also impair the trafficking of claudin 14 to the plasma membrane.
Two of the mutations (p.Arg81His; p.Ser87Ile) of CLDN14 identified in this study were found only once. However, unlike these two rare mutations, three other mutations (p.Val85Asp; p.Met133ArgfsX23; p.Ala94Val) account for ∼89% of the CLDN14 alleles we found in our cohort of Pakistani families segregating deafness (Table 1). In conclusion, there is considerable genetic and allelic heterogeneity that accounts for recessively inherited deafness in Pakistan (Table 2). CLDN14 mutations are a frequent cause of genetic deafness in this population and are associated with marked inter- and intra-familial variability in hearing thresholds.
Electronic database information
NHLBI-ESP variant database, http://evs.gs.washington.edu/EVS/
Primer3, http://frodo.wi.mit.edu/
1000 Genome, http://browser.1000genomes.org/
TMpred, http://www.ch.embnet.org/software/TMPRED_form.html
SIFT, http://sift.jcvi.org/
Polyphen-2, http://genetics.bwh.harvard.edu/pph2/
MutationTaster, http://www.mutationtaster.org/index.html
Accession codes
References
Wilcox, E. R., Burton, Q. L., Naz, S., Riazuddin, S., Smith, T. N., Ploplis, B. et al. Mutations in the gene encoding tight junction claudin-14 cause autosomal recessive deafness DFNB29. Cell 104, 165–172 (2001).
Lee, K., Ansar, M., Andrade, P. B., Khan, B., Santos-Cortez, R. L., Ahmad, W. et al. Novel CLDN14 mutations in Pakistani families with autosomal recessive non-syndromic hearing loss. Am. J. Med. Genet. A 158A, 315–321 (2012).
Bashir, R., Fatima, A. & Naz, S. Mutations in CLDN14 are associated with different hearing thresholds. J. Hum. Genet. 55, 767–770 (2010).
Wattenhofer, M., Reymond, A., Falciola, V., Charollais, A., Caille, D., Borel, C. et al. Different mechanisms preclude mutant CLDN14 proteins from forming tight junctions in vitro. Hum. Mutat. 25, 543–549 (2005).
Ben-Yosef, T., Belyantseva, I. A., Saunders, T. L., Hughes, E. D., Kawamoto, K., Van Itallie, C. M. et al. Claudin 14 knockout mice, a model for autosomal recessive deafness DFNB29, are deaf due to cochlear hair cell degeneration. Hum. Mol. Genet. 12, 2049–2061 (2003).
Anderson, J. M. & Van Itallie, C. M. Physiology and function of the tight junction. Cold Spring Harb. Perspect. Biol. 1, a002584 (2009).
Schulzke, J. D. & Fromm, M. Tight junctions: molecular structure meets function. Ann. N. Y. Acad. Sci. 1165, 1–6 (2009).
Tsukita, S., Yamazaki, Y., Katsuno, T. & Tamura, A. Tight junction-based epithelial microenvironment and cell proliferation. Oncogene 27, 6930–6938 (2008).
Steed, E., Rodrigues, N. T., Balda, M. S. & Matter, K. Identification of MarvelD3 as a tight junction-associated transmembrane protein of the occludin family. BMC Cell Biol. 10, 95 (2009).
Tsukita, S. & Furuse, M. Overcoming barriers in the study of tight junction functions: from occludin to claudin. Genes Cells 3, 569–573 (1998).
Furuse, M., Fujita, K., Hiiragi, T., Fujimoto, K. & Tsukita, S. Claudin-1 and -2: novel integral membrane proteins localizing at tight junctions with no sequence similarity to occludin. J. Cell Biol. 141, 1539–1550 (1998).
Morita, K., Furuse, M., Fujimoto, K. & Tsukita, S. Claudin multigene family encoding four-transmembrane domain protein components of tight junction strands. Proc. Natl Acad. Sci. USA 96, 511–516 (1999).
Lim, T. S., Vedula, S. R., Hunziker, W. & Lim, C. T. Kinetics of adhesion mediated by extracellular loops of claudin-2 as revealed by single-molecule force spectroscopy. J. Mol. Biol. 381, 681–691 (2008).
Lim, T. S., Vedula, S. R., Kausalya, P. J., Hunziker, W. & Lim, C. T. Single-molecular-level study of claudin-1-mediated adhesion. Langmuir 24, 490–495 (2008).
Jahnke, K. [Intercellular junctions in the guinea pig stria vascularis as shown by freeze-etching (author’s transl)]. Anat. Embryol. 147, 189–201 (1975).
Jahnke, K. The fine structure of freeze-fractured intercellular junctions in the guinea pig inner ear. Acta Otolaryngol. Suppl. 336, 1–40 (1975).
Gulley, R. L. & Reese, T. S. Intercellular junctions in the reticular lamina of the organ of Corti. J. Neurocytol. 5, 479–507 (1976).
Nunes, F. D., Lopez, L. N., Lin, H. W., Davies, C., Azevedo, R. B., Gow, A. et al. Distinct subdomain organization and molecular composition of a tight junction with adherens junction features. J. Cell Sci. 119, 4819–4827 (2006).
Khasnis, A. & Gokula, R. M. Romberg’s test. J. Postgrad Med. 49, 169–172 (2003).
Ahmed, Z. M., Riazuddin, S., Bernstein, S. L., Ahmed, Z., Khan, S., Griffith, A. J. et al. Mutations of the protocadherin gene PCDH15 cause Usher syndrome type 1F. Am. J. Hum. Genet. 69, 25–34 (2001).
Grimberg, J., Nawoschik, S., Belluscio, L., McKee, R., Turck, A. & Eisenberg, A. A simple and efficient non-organic procedure for the isolation of genomic DNA from blood. Nucleic Acids Res. 17, 8390 (1989).
Kumar, P., Henikoff, S. & Ng, P. C. Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat. Protoc. 4, 1073–1081 (2009).
Adzhubei, I. A., Schmidt, S., Peshkin, L., Ramensky, V. E., Gerasimova, A., Bork, P. et al. A method and server for predicting damaging missense mutations. Nat. Methods 7, 248–249 (2010).
Schwarz, J. M., Rodelsperger, C., Schuelke, M. & Seelow, D. MutationTaster evaluates disease-causing potential of sequence alterations. Nat. Methods 7, 575–576 (2010).
Ahmed, Z. M., Riazuddin, S., Friedman, T. B., Wilcox, E. R. & Griffith, A. J. Clinical manifestations of DFNB29 deafness. Adv. Otorhinolaryngol. 61, 156–160 (2002).
Schultz, J. M., Yang, Y., Caride, A. J., Filoteo, A. G., Penheiter, A. R., Lagziel, A. et al. Modification of human hearing loss by plasma-membrane calcium pump PMCA2. N. Engl. J. Med. 352, 1557–1564 (2005).
Riazuddin, S., Castelein, C. M., Ahmed, Z. M., Lalwani, A. K., Mastroianni, M. A., Naz, S. et al. Dominant modifier DFNM1 suppresses recessive deafness DFNB26. Nat. Genet. 26, 431–434 (2000).
Riazuddin, S., Ahmed, Z. M., Fanning, A. S., Lagziel, A., Kitajiri, S., Ramzan, K. et al. Tricellulin is a tight-junction protein necessary for hearing. Am. J. Hum. Genet. 79, 1040–1051 (2006).
Kitajiri, S., Miyamoto, T., Mineharu, A., Sonoda, N., Furuse, K., Hata, M. et al. Compartmentalization established by claudin-11-based tight junctions in stria vascularis is required for hearing through generation of endocochlear potential. J. Cell Sci. 117, 5087–5096 (2004).
Gow, A., Davies, C., Southwood, C. M., Frolenkov, G., Chrustowski, M., Ng, L. et al. Deafness in Claudin 11-null mice reveals the critical contribution of basal cell tight junctions to stria vascularis function. J. Neurosci. 24, 7051–7062 (2004).
Nakano, Y., Kim, S. H., Kim, H. M., Sanneman, J. D., Zhang, Y., Smith, R. J. et al. A claudin-9-based ion permeability barrier is essential for hearing. PLoS Genet. 5, e1000610 (2009).
Kitajiri, S. I., Furuse, M., Morita, K., Saishin-Kiuchi, Y., Kido, H., Ito, J. et al. Expression patterns of claudins, tight junction adhesion molecules, in the inner ear. Hear Res. 187, 25–34 (2004).
Hibino, H. & Kurachi, Y. Molecular and physiological bases of the K+ circulation in the mammalian inner ear. Physiology 21, 336–345 (2006).
Wangemann, P. Supporting sensory transduction: cochlear fluid homeostasis and the endocochlear potential. J. Physiol. 576, 11–21 (2006).
Anwar, S., Riazuddin, S., Ahmed, Z. M., Tasneem, S., Ateeq-ul-Jaleel, Khan, S. Y. et al. SLC26A4 mutation spectrum associated with DFNB4 deafness and Pendred’s syndrome in Pakistanis. J. Hum. Genet. 54, 266–270 (2009).
Park, H. J., Shaukat, S., Liu, X. Z., Hahn, S. H., Naz, S., Ghosh, M. et al. Origins and frequencies of SLC26A4 (PDS) mutations in east and south Asians: global implications for the epidemiology of deafness. J. Med. Genet. 40, 242–248 (2003).
Santos, R. L., Wajid, M., Khan, M. N., McArthur, N., Pham, T. L., Bhatti, A. et al. Novel sequence variants in the TMC1 gene in Pakistani families with autosomal recessive hearing impairment. Hum. Mutat. 26, 396 (2005).
Schultz, J. M., Khan, S. N., Ahmed, Z. M., Riazuddin, S., Waryah, A. M., Chhatre, D. et al. Noncoding mutations of HGF are associated with nonsyndromic hearing loss, DFNB39. Am. J. Hum. Genet. 85, 25–39 (2009).
Kurima, K., Peters, L. M., Yang, Y., Riazuddin, S., Ahmed, Z. M., Naz, S. et al. Dominant and recessive deafness caused by mutations of a novel gene, TMC1, required for cochlear hair-cell function. Nat. Genet. 30, 277–284 (2002).
Kitajiri, S. I., McNamara, R., Makishima, T., Husnain, T., Zafar, A. U., Kittles, R. A. et al. Identities, frequencies and origins of TMC1 mutations causing DFNB7/B11 deafness in Pakistan. Clin. Genet. 72, 546–550 (2007).
Nal, N., Ahmed, Z. M., Erkal, E., Alper, O. M., Lüleci, G., Dinç, O. et al. Mutational spectrum of MYO15A: the large N-terminal extension of myosin XVA is required for hearing. Hum. Mutat. 28, 1014–1019 (2007).
Liburd, N., Ghosh, M., Riazuddin, S., Naz, S., Khan, S., Ahmed, Z. et al. Novel mutations of MYO15A associated with profound deafness in consanguineous families and moderately severe hearing loss in a patient with Smith-Magenis syndrome. Hum. Genet. 109, 535–541 (2001).
Choi, B. Y., Ahmed, Z. M., Riazuddin, S., Bhinder, M. A., Shahzad, M., Husnain, T. et al. Identities and frequencies of mutations of the otoferlin gene (OTOF) causing DFNB9 deafness in Pakistan. Clin. Genet. 75, 237–243 (2009).
Chishti, M. S., Bhatti, A., Tamim, S., Lee, K., McDonald, M. L., Leal, S. M. et al. Splice-site mutations in the TRIC gene underlie autosomal recessive nonsyndromic hearing impairment in Pakistani families. J. Hum. Genet. 53, 101–105 (2008).
Kitajiri, S., Sakamoto, T., Belyantseva, I. A., Goodyear, R. J., Stepanyan, R., Fujiwara, I. et al. Actin-bundling protein TRIOBP forms resilient rootlets of hair cell stereocilia essential for hearing. Cell 141, 786–798 (2010).
Riazuddin, S., Khan, S. N., Ahmed, Z. M., Ghosh, M., Caution, K., Nazli, S. et al. Mutations in TRIOBP, which encodes a putative cytoskeletal-organizing protein, are associated with nonsyndromic recessive deafness. Am. J. Hum. Genet. 78, 137–143 (2006).
Borck, G., Ur Rehman, A., Lee, K., Pogoda, H. M., Kakar, N., von Ameln, S. et al. Loss-of-function mutations of ILDR1 cause autosomal-recessive hearing impairment DFNB42. Am. J. Hum. Genet. 88, 127–137 (2011).
Ahmed, Z. M., Morell, R. J., Riazuddin, S., Gropman, A., Shaukat, S., Ahmad, M. M. et al. Mutations of MYO6 are associated with recessive deafness, DFNB37. Am. J. Hum. Genet. 72, 1315–1322 (2003).
Rehman, A. U., Gul, K., Morell, R. J., Lee, K., Ahmed, Z. M., Riazuddin, S. et al. Mutations of GIPC3 cause nonsyndromic hearing loss DFNB72 but not DFNB81 that also maps to chromosome. Hum. Genet. 130, 759–765 19p. (2011).
Rehman, A. U., Morell, R. J., Belyantseva, I. A., Khan, S. Y., Boger, E. T., Shahzad, M. et al. Targeted capture and next-generation sequencing identifies C9orf75, encoding taperin, as the mutated gene in nonsyndromic deafness DFNB79. Am. J. Hum. Genet. 86, 378–388 (2010).
Khan, S. Y., Ahmed, Z. M., Shabbir, M. I., Kitajiri, S., Kalsoom, S., Tasneem, S. et al. Mutations of the RDX gene cause nonsyndromic hearing loss at the DFNB24 locus. Hum. Mutat. 28, 417–423 (2007).
Acknowledgements
We thank the families for the participation and cooperation, and R Bhatti and T Kausar for technical assistance and Drs D Drayna, G Nayak and K Kurima for critiques of the manuscript. This work was also supported by an Action on Hearing Loss grant and National Institute on Deafness and Other Communication Disorders (NIDCD/NIH) research grants R01 DC011803 and R01 DC011748 to SR and intramural funds from NIDCD DC000039-15 to TBF.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Rights and permissions
About this article
Cite this article
Bashir, ZeH., Latief, N., Belyantseva, I. et al. Phenotypic variability of CLDN14 mutations causing DFNB29 hearing loss in the Pakistani population. J Hum Genet 58, 102–108 (2013). https://doi.org/10.1038/jhg.2012.143
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/jhg.2012.143
Keywords
This article is cited by
-
Molecular genetic landscape of hereditary hearing loss in Pakistan
Human Genetics (2022)
-
Multiple claudin–claudin cis interfaces are required for tight junction strand formation and inherent flexibility
Communications Biology (2018)
-
A common variant in CLDN14 causes precipitous, prelingual sensorineural hearing loss in multiple families due to founder effect
Human Genetics (2017)