Abstract
Tamsulosin hydrochloride is one of the most potent drugs for treatment of benign prostatic hyperplasia (BPH), however, the efficacy of tamsulosin hydrochloride varies among individuals. In this study, we measured the maximum serum concentration (Cmax) of tamsulosin hydrochloride in 182 of BPH patients and found remarkable individual variability. To investigate the genetic factors that regulate pharmacokinetics of tamsulosin hydrochloride, we conducted a genome-wide association study in these 182 BPH patients. As a result, rs16902947 on chromosome 5p13.2, rs7779057 on 7q22.3, rs35681285 on 7p21.2 and rs2122469 on 8p21.3 indicated possible associations with Cmax of tamsulosin hydrochloride (P=1.29 × 10−7, 2.15 × 10−7, 4.35 × 10−7 and 7.03 × 10−7, respectively), although these single-nucleotide polymorphisms (SNPs) did not reach the genome-wide significance threshold after Bonferroni correction. As these associated SNPs showed additive effects on serum tamsulosin hydrochloride concentration, we defined the ‘Cmax prediction index’ based on genotypes of these SNPs. This index clearly associated with Cmax values (P=4.5 × 10−6), indicating the possible roles of these four variants in tamsulosin hydrochloride pharmacokinetics. Our findings would partially explain the variability of the response to the tamsulosin hydrochloride treatment.
Similar content being viewed by others
Introduction
Lower urinary tract symptoms caused by benign prostatic hyperplasia (BPH) is one of the most common problems in elderly men.1 Though it is not a life-threatening disorder, BPH can affect quality of life by various symptoms such as urinary frequency, urinary urgency, nocturia and sensation of incomplete bladder emptying. In addition, the number of BPH patients has been significantly increasing in accordance with the aging of the population in developed countries.
Although the etiology of BPH is not fully understood, α1 adrenergic receptors (α1ARs) were shown to have pivotal roles in the development of BPH. α1ARs belong to the G-protein-coupled receptor super family and mediate the actions of the catecholamine, norepinephrine and epinephrine. Norepinephrine released from sympathetic nerves would regulate prostatic smooth muscle tone and, subsequently, cause mechanical bladder outlet obstruction.2, 3, 4, 5 Among three subtypes of α1ARs, α1A-AR was shown to be highly expressed in human prostate tissues.6, 7 Therefore, selective α1A-AR antagonists are used as the first-line medical treatment for patients with BPH-associated lower urinary tract symptoms.8, 9
As tamsulosin hydrochloride exhibits some degree of specificity for the α1A-AR over α1B-AR,10 tamsulosin hydrochloride is one of the most widely prescribed drugs for the treatment of BPH. In fact, tamsulosin hydrochloride could improve the clinical symptoms in two-third of BPH patients11 but not in the remaining one-third. Because the efficacy of tamsulosin hydrochloride varies among individuals, prediction of treatment response would improve the overall treatment outcome. In this study, we measured the serum concentration of tamsulosin hydrochloride in 182 of BPH patients and found remarkable inter-individual variability. We also conducted genome-wide association study (GWAS) to identify the genetic factors that regulate pharmacokinetics of tamsulosin hydrochloride.
Materials and methods
Subjects
In this study, newly diagnosed patients with BPH at four Japanese hospitals (Iwate Medical University, Nagoya City University, Kyoto prefectural University of Medicine and Kochi Medical School) were enrolled. All BPH patients were diagnosed by urologist on the basis of clinical features and laboratory test.12 The following patients were excluded from the analysis: (1) drug-induced or neurogenic bladder dysfunction; (2) prostate cancer; and (3) urethrostenosis. All subjects were orally administrated with 0.2 mg of tamsulosin hydrochloride once a day. Written informed consents had been obtained from all the subjects to participate in this study. This study was approved by the ethics committee of each Institute.
Evaluation of serum tamsulosin hydrochloride concentration.
All study subjects were evaluated for serum tamsulosin hydrochloride concentration during the period of stable prescription at 8 weeks or later after the first administration. Each patient took 0.2 mg of tamsulosin hydrochloride (without water tablet) soon after breakfast every day. Blood samples were drawn from patients around 4–8 h after drug administration. Serum tamsulosin hydrochloride concentration was determined by high performance liquid chromatography at Mitsubishi Kagaku Co. (Tokyo, Japan). We estimated Cmax of the serum tamsulosin based on the result in the previous pharmacokinetic study,13 in which serum concentration was peaked at 8 h after oral administration of 0.2 mg tamsulosin. Estimated serum half-life was 9 h. According to these findings, we estimated the Cmax based on the serum drug concentration at 4–8 h after drug oral administration.
Genotyping
Genomic DNA of all the patients was extracted from whole blood by standard method. We applied 750 ng DNA of each sample to Illumina Human610-Quad Beadchip (San Diego, CA, USA) according to the manufacturer’s protocols at Center for Genomic Medicine, RIKEN. Samples with call rates <0.98 were excluded from the analysis. Then, we excluded single-nucleotide polymorphisms (SNPs) with call rates <0.99 or ambiguous intensity plots from the following analyses. Subsequently, we excluded the SNP with minor allele frequency of <0.01, or with an exact P-value of the Hardy–Weinberg equilibrium test <1.0 × 10−7. Finally, 481 678 SNPs passed our quality control process.
Statistical analysis
Associations of the SNPs with Cmax of tamsulosin hydrochloride were assessed by linear regression, assuming the additive effects of the alleles on the Cmax, using PLINK version 1.06.14 Significance level after Bonferroni correction for multiple testing was P=1.03 × 10−7 (0.05/481 678) in this analysis. We also calculated false discovery rate-based q-values to measure the statistical significance at the genome-wide level by using R program.15 Differences in Cmax of tamsulosin hydrochloride in the different genotype groups were evaluated by Kruskal–Wallis tests using PASW statistics software (version 18.0.0; SPSS, Chicago, IL, USA).
Results
This study investigated the genetic factors that influence the serum level of tamsulosin hydrochloride in patients orally taking 0.2 mg of tamsulosin hydrochloride once per day. All subjects were newly diagnosed with BPH on the basis of clinical information and laboratory test. All patients were Japanese, and median age was 71.0 ranging 55–87 years old. Cmax was successfully determined in 182 patients by HLPC as described previously.13 The result indicated that Cmax ranged from 1.2 to 65.1 ng ml−1 and varied enormously among individuals (Figure 1). Actually, patients who were ranked in the bottom 25% showed the average Cmax of 3.8 ng ml−1, whereas those in the top 25% had about 7-fold higher Cmax of 26.7 ng ml−1.
To identify genetic factors that were involved in serum level of tamsulosin hydrochloride, we performed GWAS. We genotyped 182 individuals using Illumina 610 HumanHap BeadChip. SNPs with minor allele frequency <1% were excluded for further analysis, considering the lower statistical power. After stringent quality control filtering, we obtained the genotyping result of 481 678 SNPs in 182 patients.
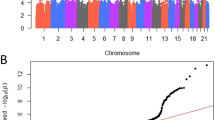
Then, we assess the association of these SNPs with Cmax using a linear regression model. A quantile–quantile plot of P-values showed that the genomic inflation factor lambda was 1000, implying a low possibility of false-positive associations due to population substructure (Figure 2). The results of association analysis revealed that SNPs rs16902947, rs7779057, rs35681285 and rs2122469 showed P-value <1.0 × 10−6 (Figure 3 and Table 1). Although these SNPs did not reach the genome-wide significance threshold after Bonferroni correction, the Bonferroni method is considered to be too conservative because many SNPs are in some degrees of linkage disequilibrium. Because all SNPs showed false discovery rate q-value <0.1, our findings would have a low possibility of false-positive associations (Table 1). Among four SNPs, SNPs rs16902947 and rs35681285 are located within genomic regions, including RAN-binding protein 3-like (RANBP3L) and isoprenoid synthase domain containing (ISPD), respectively, whereas SNPs rs7779057 and rs2122469 are located within intergenic regions far from any annotated genes.
Quantile–quantile plot of P-values in the GWAS for Cmax of tamusulosin. The horizontal axis indicates the expected −log10 (P-values). The vertical axis indicates the observed −log10 (P-values). The gray line represents y=x. The λ indicates the inflation factor of the test statistics. A full color version of this figure is available at the Journal of Human Genetics journal online.
Then, we also evaluated Cmax level stratified by SNP genotype. Cmax levels were correlated with the number of G allele at SNP rs16902947 (median 10.4, 22.6 and 65.1 ng ml−1 for AA, AG and GG genotypes, respectively, Figure 4a). Median Cmax in the patients with CC genotypes at SNP rs7779057 (10.2 ng ml−1) was less than those among TC carriers (21.5 ng ml−1, Figure 4b). Median Cmax was also lower in the patients with TT genotypes than TC genotypes at rs35681285 (10.3 and 25.0 ng ml−1, Figure 4c), and in the patients with CC genotypes than TC genotypes at rs2122469 (10.3 and 29.4 ng ml−1, Figure 4d). To assess the combined impact of these four SNPs, we defined Cmax prediction index as the number of alleles associated with higher Cmax level (G at rs16902947, T at rs7779057, C at rs35681285 and T at rs2122469). Patients were classified into three groups based on Cmax prediction index (0, 1 and ⩾2). As shown in Figure 5, Cmax prediction index was significantly associated with Cmax values (P=4.5 × 10−6, Kruskal–Wallis analysis), indicating the possible roles of these four variants in pharmacokinetics of tamsulosin hydrochloride.
Association between genotype and Cmax of tamusulosin. Box plot showing maximum tamusulosin serum concentration (Cmax) according to genotype with rs16902947 (a), rs7779057 (b), rs35681285 (c) and rs2122469 (d). The horizontal lines indicate the median values, and the boxes cover 25–75 percentiles. Points outside them show up as outliers.
CYP2D6 variations were shown to be associated with various drug metabolism.16 In addition, treatment with CYP2D6 or CYP3A4 inhibitor, such as cimetidine hydrochloride, was shown to increase serum tamusulosin concentration,17, 18 indicating the important roles of CYP2D6 and CYP3A4 in tamsulosin metabolism. In our GWAS, we analyzed five SNPs in CYP2D6 and CYP3A4 loci and found suggestive association between SNP rs28371759 (P=0.044) and Cmax (Table 2), although the association was not statistically significant after correction for multiple testing. CYP2D6 was also involved in the metabolism of tamoxifen, and its genetic variations were shown to be associated with treatment outcome of adjuvant tamoxifen therapy for estrogen receptor-positive breast cancer patients.19, 20, 21, 22 However, in our previous GWAS of adjuvant tamoxifen therapy for breast cancer patients, SNPs in CYP2D6 locus did not significantly associate with treatment response,23 probably due to the complicated haplotype structure of this gene locus. To further elucidate the role of CYP2D6 and CYP3A4 variations, comprehensive genotyping of CYP polymorphisms would be necessary.
Discussion
With a better understanding of the mechanisms underlying the heterogeneity in the response to a variety of therapeutic agents, future research will focus on the establishment of individualized treatment based on physiological variables, genetic characteristics and environmental factors. Recent advances in pharmacogenetic research revealed a number of genetic variants that affect the efficacy and safety of drugs.16, 24, 25, 26, 27, 28 For example, analysis of VKORC1 and CYP2C9 genotypes can predict warfarin sensitivity and maintenance dosage.26, 27 The new clinical trial, Clarification of Optimal Anticoagulation through Genetics, will test this approaches to determining the initial dose of warfarin in patients who are expected to need therapy for 3 months or longer.27 Variations in CYP2D6 and ABCC2 were also shown to be significantly associated with the prognosis of breast cancer patients receiving tamoxifen monotherapy.16 In addition, three ABCB1 variants were associated with higher digoxin serum concentrations in a cohort of European digoxin users.28
As tamsulosin hydrochloride is widely used for the treatment of BPH because of its better tolerability than other type of α1AR antagonists,10, 29 prediction of drug efficacy and toxicity would contribute to better treatment of BPH patients. In general, serum drug concentration is presumably correlated with the efficacy of drugs. Therefore, we conducted GWAS to elucidate the genetic factors that associate with serum tamsulosin hydrochloride concentration. Recently, GWAS have enabled us to detect genetic variations those are associated with the risk of diseases,30 laboratory test values,31 and drug adverse event32 without prior knowledge of position or function. To our knowledge, this is the first GWAS for serum drug concentration.
In our GWAS, four SNPs indicated suggestive association on Cmax of tamsulosin hydrochloride. However, these SNPs did not reach genome-wide significance based on Bonferroni correction. Although Bonferroni correction is the best-known approach, GWAS may fall short of discovering any true positive gene, particularly when it has a relatively small sample size. On the other hand, false discovery rate is another statistical method in multiple testing that offers a more liberal criterion.15 In the current analysis, four SNPs showed q-value <0.1, indicating the strong association of these SNPs with Cmax of tmasulosin.
Multicenter randomized studies have shown the correlation between α1-blocker dosage and their effects.9, 26 In addition, the mean changes in the American Urological Association symptom score were significantly greater in the 0.8 mg per day tamsulosin hydrochloride group compared with the 0.4 mg per day group.33 Therefore, higher serum tamsulosin hydrochloride concentration is very likely to improve drug efficacy. The individuals with prediction index of 0 indicate nearly one-fourth Cmax level compared with the subjects with index score of ⩾2. Thus ‘Cmax prediction index’ would be inversely related with the required dosage.
SNP rs16902947 is located at 7 kb upstream of RANBP3L, which encodes a paralog of RANBP3. RanBP3 functions as a negative regulator of transforming growth factor-beta signaling through the interaction with the R-SMAD proteins.34 In addition, transforming growth factor-beta was shown to induce uridine diphosphate glucose dehydrogenase,35 which acts as a substrate for glucuronosyl transferase and regulates glucuronidation. Because glucuronidation is a key step of tamsulosin metabolism, RanBP3L may also regulate glucuronidation and, subsequently, alter tamsulosin metabolism. SNP rs35681285 is located within an intron 5 of ISPD. Mutations of ISPD were shown to be the cause of Walker–Warburg syndrome, a rare form of autosomal-recessive congenital muscular dystrophy.36, 37 ISPD belongs to the IspD family and is required for protein O-linked mannosylation. However, there is no report that mannosylation affects the drug metabolism so far. SNPs rs7779057 and rs2122469 are at more than 40 kb away from any known gene or microRNA. Further functional analysis is necessary to elucidate the molecular mechanism, whereby these variations exert their effects on the serum tamsulosin hydrochloride concentration.
Although no previous pharmacokinetic and pharmacodynamic studies demonstrated the correlation between Cmax and the drug efficacy, the drug dosage was shown to be correlated with the drug response,38 and drug dosage is reflected by Cmax.13 Thus, our findings would partially explain the diversity of response to tamsulosin hydrochloride treatment. Pharmacokinetics of tamsulosin was shown to be influenced by various factors, including food intake, α1-acid glycoprotein levels, hepatic and renal impairment, and comedication with CYP2A6 and/or 3A4 inhibitors.18 In addition, genetic variations in ARs might also affect the drug efficacy even if the serum drug concentrations are same. We would like to conduct the association analysis of Cmax and drug efficacy with genetic variations in the future study. For the Japanese patients, 0.2 mg of tamsulosin hydrochloride is commonly used for BPH patients as the initial dose, and the dosage is adjusted in accordance with the treatment efficacy. We hope that our study would contribute to estimate the required dosage of tamsulosin hydrochloride and, subsequently, establish the personalized medical treatment of BPH patients in the future.
References
Berry, S. J., Coffey, D. S., Walsh, P. C. & Ewing, L. L. The development of human benign prostatic hyperplasia with age. J. Urol. 132, 474 (1984).
Roehrborn, C. G. & Schwinn, D. A. α1-Adrenergic receptors and their inhibitors in lower urinary tract symptoms and benign prostatic hyperplasia. J. Urol. 171, 1029–1035 (2004).
Boesch, S. T., Corvin, S., Zhang, J., Rogatsch, H., Bartsch, G. & Klocker, H. Modulation of the differentiation status of cultured prostatic smooth muscle cells by an α1-adrenergic receptor antagonist. Prostate 39, 226–233 (1999).
Boesch, S. T., Dobler, G., Ramoner, R., Corvin, S., Thurnher, M., Bartsch, G. et al. Effects of α1-adrenoceptor antagonists on cultured prostatic smooth muscle cells. Prostate 45, 34–41 (2000).
McVary, K. T., Razzaq, A., Lee, C., Venegas, M. F., Rademaker, A. & McKenna, K. E. Growth of the rat prostate gland is facilitated by the autonomic nervous system. Biol. Reprod. 51, 99–107 (1994).
Price, D., Schwinn, D., Lomasney, J., Allen, L., Caron, M. & Lefkowitz, R. Identification, quantification, and localization of mRNA for three distinct alpha 1 adrenergic receptor subtypes in human prostate. J. Urol. 150, 546 (1993).
Lepor, H., Tang, R., Kobayashi, S., Shapiro, E., Forray, C., Wetzel, J. M. et al. Localization of the alpha 1A-adrenoceptor in the human prostate. J. Urol. 154, 2096–2099 (1995).
Fawzy, A., Braun, K., Lewis, G. P., Gaffney, M. & Ice, K. Doxazosin in the treatment of benign prostatic hyperplasia in normotensive patients: a multicenter study. J. Urol. 154, 105–109 (1995).
Lepor, H., Auerbach, S., Puras-Baez, A., Narayan, P., Soloway, M., Lowe, F. et al. A randomized, placebo-controlled multicenter study of the efficacy and safety of terazosin in the treatment of benign prostatic hyperplasia. J. Urol. 148, 1467–1474 (1992).
Foglar, R., Shibata, K., Horie, K., Hirasawa, A. & Tsujimoto, G. Use of recombinant [alpha] 1-adrenoceptors to characterize subtype selectivity of drugs for the treatment of prostatic hypertrophy. Eur. J. Pharmacol. Mol. Pharmacol. 288, 201–207 (1995).
Chapple, C., Wyndaele, J., Nordling, J., Boeminghaus, F., Ypma, A. & Abrams, P. Tamsulosin the first prostate-selective alpha 1A-adrenoceptor antagonist. A meta-analysis of two randomized, placebo-controlled, multicentre studies in patients with benign prostatic obstruction (symptomatic BPH). European Tamsulosin Study Group. Eur. Urol. 29, 155 (1996).
Homma, Y., Kawabe, K., Tsukamoto, T., Yamaguchi, O., Okada, K., Aso, Y. et al. Estimate criteria for efficacy of treatment in benign prostatic hyperplasia. Int. J. Urol. 3, 267–273 (1996).
Tsunoo, M. S. A., Soeishi, Y., Kobori, M. & Shimoyama, M. Phase I clinical trial of YM617, a new alpha1-adrenoceptor antagonist – second report: a single oral dose of controlled release formulation in healthy male subjects. J. Clin. Therap. Med. 6, 2529–2551 (1990).
Purcell, S., Neale, B., Todd-Brown, K., Thomas, L., Ferreira, M. A. R., Bender, D. et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 81, 559–575 (2007).
Storey, J. D. & Tibshirani, R. Statistical significance for genomewide studies. Proc. Natl Acad. Sci. USA 100, 9440 (2003).
Kiyotani, K., Mushiroda, T., Imamura, C. K., Hosono, N., Tsunoda, T., Kubo, M. et al. Significant effect of polymorphisms in CYP2D6 and ABCC2 on clinical outcomes of adjuvant tamoxifen therapy for breast cancer patients. J. Clin. Oncol. 28, 1287–1293 (2010).
Miyazawa, Y., Forrest, A., Schentag, J. J., Kamimura, H., Swarz, H. & Ito, Y. Effect of concomitant administration of cimetidine hydrochloride on the pharmacokinetic and safety profile of tamsulosin hydrochloride 0.4 mg in healthy subjects. Curr Ther Res 63, 15–26 (2002).
Franco-Salinas, G., de la Rosette, J. J. & Michel, M. C. Pharmacokinetics and pharmacodynamics of tamsulosin in its modified-release and oral controlled absorption system formulations. Clin Pharmacokinet 49, 177–188 (2010).
Kiyotani, K., Mushiroda, T., Sasa, M., Bando, Y., Sumitomo, I., Hosono, N. et al. Impact of CYP2D6*10 on recurrence-free survival in breast cancer patients receiving adjuvant tamoxifen therapy. Cancer Sci. 99, 995–999 (2008).
Kiyotani, K., Mushiroda, T., Imamura, C. K., Hosono, N., Tsunoda, T., Kubo, M. et al. Significant effect of polymorphisms in CYP2D6 and ABCC2 on clinical outcomes of adjuvant tamoxifen therapy for breast cancer patients. J. Clin. Oncol. 28, 1287–1293 (2010).
Goetz, M. P., Rae, J. M., Suman, V. J., Safgren, S. L., Ames, M. M., Visscher, D. W. et al. Pharmacogenetics of tamoxifen biotransformation is associated with clinical outcomes of efficacy and hot flashes. J. Clin. Oncol. 23, 9312–9318 (2005).
Schroth, W., Goetz, M. P., Hamann, U., Fasching, P. A., Schmidt, M., Winter, S. et al. Association between CYP2D6 polymorphisms and outcomes among women with early stage breast cancer treated with tamoxifen. JAMA 302, 1429–1436 (2009).
Kiyotani, K., Mushiroda, T., Tsunoda, T., Morizono, T., Hosono, N., Kubo, M. et al. A genome-wide association study identifies locus at 10q22 associated with clinical outcomes of adjuvant tamoxifen therapy for breast cancer patients in Japanese. Hum. Mol. Genet. 21, 1665–1672 (2012).
Drysdale, C. M., McGraw, D. W., Stack, C. B., Stephens, J. C., Judson, R. S., Nandabalan, K. et al. Complex promoter and coding region β2-adrenergic receptor haplotypes alter receptor expression and predict in vivo responsiveness. Proc. Natl Acad. Sci. USA 97, 10483 (2000).
Iyer, L., Das, S., Janisch, L., Wen, M., Ramirez, J., Karrison, T. et al. UGT1A1* 28 polymorphism as a determinant of irinotecan disposition and toxicity. Pharmacogenomics J. 2, 43–47 (2002).
Mushiroda, T., Ohnishi, Y., Saito, S., Takahashi, A., Kikuchi, Y., Shimomura, H. et al. Association of VKORC1 and CYP2C9 polymorphisms with warfarin dose requirements in Japanese patients. J. Hum. Genet. 51, 249–253 (2006).
Klein, T., Altman, R., Eriksson, N., Gage, B., Kimmel, S., Lee, M. et al. Estimation of the warfarin dose with clinical and pharmacogenetic data. N Engl J Med 360, 753 (2009).
Aarnoudse, A. J. L. H. J., Dieleman, J. P., Visser, L. E., Arp, P. P., Van Der Heiden, I. P., Van Schaik, R. H. N. et al. Common ATP-binding cassette B1 variants are associated with increased digoxin serum concentration. Pharmacogenetics Genomics 18, 299 (2008).
Wilt, T. J., MacDonald, R. & Nelson, D. Tamsulosin for treating lower urinary tract symptoms compatible with benign prostatic obstruction: a systematic review of efficacy and adverse effects. J. Urol. 167, 177–183 (2002).
Wellcome Trust Case Control Consortium. Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature 447, 661–678 (2007).
Okada, Y., Takahashi, A., Ohmiya, H., Kumasaka, N., Kamatani, Y., Hosono, N. et al. Genome-wide association study for C-reactive protein levels identified pleiotropic associations in the IL6 locus. Hum. Mol. Genet. 20, 1224–1231 (2011).
Ozeki, T., Mushiroda, T., Yowang, A., Takahashi, A., Kubo, M., Shirakata, Y. et al. Genome-wide association study identifies HLA-A* 3101 allele as a genetic risk factor for carbamazepine-induced cutaneous adverse drug reactions in Japanese population. Hum. Mol. Genet. 20, 1034–1041 (2011).
Narayan, P., Tewari, A. A second phase III multicenter placebo controlled study of 2 dosages of modified release tamsulosin in patients with symptoms of benign prostatic hyperplasia. J. Urol. 160, 1701–1706 (1998).
Dai, F., Lin, X., Chang, C. & Feng, X. H. Nuclear export of Smad2 and Smad3 by RanBP3 facilitates termination of TGF-[beta] signaling. Dev. Cell 16, 345–357 (2009).
Bontemps, Y., Vuillermoz, B., Antonicelli, F., Perreau, C., Danan, J. L., Maquart, F. X. et al. Specific protein-1 is a universal regulator of UDP-glucose dehydrogenase expression. J. Biol. Chem. 278, 21566 (2003).
Willer, T., Lee, H., Lommel, M., Yoshida-Moriguchi, T., de Bernabe, D. B. V., Venzke, D. et al. ISPD loss-of-function mutations disrupt dystroglycan O-mannosylation and cause Walker-Warburg syndrome. Nat. Genet. 44, 575–580 (2012).
Roscioli, T., Kamsteeg, E. J., Buysse, K., Maystadt, I., van Reeuwijk, J., van den Elzen, C. et al. Mutations in ISPD cause Walker–Warburg syndrome and defective glycosylation of [alpha]-dystroglycan. Nat. Genet. 44, 581–585 (2012).
Abrams, P., Speakman, M., Stott, M., Arkell, D. & Pocock, R. A dose-ranging study of the efficacy and safety of tamsulosin, the first prostate-selective alpha 1A adrenoceptor antagonist, in patients with benign prostatic obstruction. Br. J. Urol. 80, 587–596 (1997).
Acknowledgements
The trial was led by LOTUS Co., Ltd. (Former: Order-Made Souyaku Co., Ltd.) and sponsored by the Astellas Pharma Inc.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Rights and permissions
About this article
Cite this article
Takata, R., Matsuda, K., Sugimura, J. et al. Impact of four loci on serum tamsulosin hydrochloride concentration. J Hum Genet 58, 21–26 (2013). https://doi.org/10.1038/jhg.2012.126
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/jhg.2012.126
Keywords
This article is cited by
-
Association of gene coding variation and resting metabolic rate in a multi-ethnic sample of children and adults
BMC Obesity (2017)
-
Differential Response to Medical Therapy for Male Lower Urinary Tract Symptoms
Current Bladder Dysfunction Reports (2015)