Abstract
Glucose-6-phosphate dehydrogenase (G6PD) deficiency is highly prevalent in Southeast Asians. G6PD mutations are associated with specific ethnic groups in Southeast Asia. Mon is a minority ethnic group in Myanmar, which speaks Monic, a distinct language of Mon-Khmer classification. We studied G6PD mutations in Mon and Burmese males of southern Myanmar who migrated to Thailand in Samutsakhon province. G6PD deficiency was identified in 19 (12%) of 162 Mon males and 17 (10%) of 178 Burmese males, and then assayed for G6PD mutations. Among 19 G6PD-deficient Mons, 12 were G6PD Mahidol; one case each was G6PD Jammu (871G > A; nt 1311C), G6PD Kaiping (1388G > A), G6PD Mediterranean (563C > T), a novel mutation 94(C > G); and three remain unidentified. Among 17 G6PD-deficient Burmese, 12 were G6PD Mahidol; one each was G6PD Coimbra (592C > T), G6PD Kerala-Kalyan (949G > A), and G6PD Valladolid (406C > T); and two remain unidentified. G6PD Mahidol (487G > A) is the most common mutation among Mons and Burmese. All G6PD deficient Mon and Burmese, except for a person with G6PD Valladolid, shared the same haplotype nt93T, nt1311C. Despite a similar language root with Cambodian’s Khmer language, our study suggests that Mon people share a common ancestry with Burmese rather than Cambodians.
Similar content being viewed by others
Introduction
Glucose-6-phosphate dehydrogenase (G6PD) (MIM# 305900) is an X-linked enzyme that catalyzes the production of nicotinamide adenine dinucleotide phosphate (NADPH) in the pentose phosphate pathway (PPP) of the red cell (Poggi et al. 1990). G6PD deficiency is the most common enzyme disorder in humans (WHO 1989). This disorder causes many clinical manifestations, including neonatal jaundice, acute or chronic hemolytic anemia, neonatal hyperbilirubinemia, and favism (Beutler 1994). The prevalence of G6PD deficiency is high in the Southeast Asian population where malaria has historically been endemic (Nuchprayoon et al. 2002; Iwai et al. 2001).
The G6PD gene consists of 13 exons distributed over approximately 18 kb on the distal long arm of the X chromosome (Xq28) (Martini et al. 1986). To date, at least 442 G6PD variants have been described by biochemical characterization (Xu et al. 1995), and 130 G6PD mutations by molecular technique (Hamel et al. 2002). G6PD mutations in Southeast Asians are specific to certain ethnic groups. G6PD Viangchan (871G > A; Val291Met) is the most common mutation among Thais (Nuchprayoon et al. 2002), Laotians (Iwai et al. 2001), and Cambodians (Louicharoen et al. 2005; Matsuoka et al. 2005), whereas G6PD Mahidol (487G > A; Gly163Ser) is the most common mutation among Burmese in Myanmar (Matsuoka et al. 2004).
Mon is a minority tribe in southern Myanmar. Mon people speak Monic, an Austro-Asiatic language similar to Thais and Cambodians, whereas Burmese speak Burmese (Bamar), a Tibeto-Burman language. We report here a study of G6PD deficiency and mutations in Mon and Burmese in southern Myanmar and demonstrate that their G6PD-deficient mutations are similar despite their distinct language and cultural roots.
Materials and methods
Subjects
Blood samples were collected from 162 Mon and 178 Burmese male migrant foreign laborers in Samutsakhon province, Thailand, as part of a health screening program before receiving a work permit in Thailand. Interviews of participants included their self-reported ethnic classification and place of birth. Blood samples were divided into two parts, 3 ml in acid citrate dextrose (ACD) for G6PD activity assay and 2 ml in ethylenediaminetetraacetate (EDTA) for molecular characterization, and were stored at 4°C until used. Samples that were found to be G6PD deficient were tested further for mutation analysis and haplotype analysis. Samples that were G6PD normal were randomly selected for haplotype analysis.
G6PD activity assay
G6PD activity assay was performed within 7 days according to our previous study, which was based on the World Health Organization (WHO) recommended standard test (Betke et al. 1967).
G6PD mutations analysis
Genomic DNA was extracted using a DNA blood mini kit (QIAGEN, Germany) according to manufacturer’s instruction. To identify G6PD mutations, we initially screened for G6PD Mahidol (487G > A), which was previously found to be the most common mutation in the Burmese population (Iwai et al. 2001; Matsuoka et al. 2004). G6PD-deficient DNA samples whose mutation remained unknown were assayed for eight common mutations that were previously reported from Southeast Asians: G6PD Viangchan (871G > A), G6PD Canton (1376G > T), G6PD Union (1360C > T), G6PD Kaiping (1388G > A), G6PD Chinese-5 (1024C > T), G6PD Gaohe (95A > G), G6PD Chinese-4 (392G > T), and G6PD Coimbra (592C > T) (Nuchprayoon et al. 2002; Tang et al. 1992; Saha et al. 1994; Huang et al. 1996; Ainoon et al. 1999, 2002). For G6PD mutation assays, the target gene was amplified using polymerase chain reaction restriction fragment length polymorphism (PCR-RFLP) technique with an appropriate primer set (Table 1) (Nuchprayoon et al. 2002; Huang et al. 1996). The typical PCR reaction was carried out in a 10-μl reaction containing 1× PCR buffer, 0.5 U of Taq polymerase (Fermentas), 20 ng of each primer, 1.5 mM MgCl2, 200 μM of each dNTPs, and approximately 50 ng DNA template. After incubation at 94°C for 5 min, amplification was carried out for 35 cycles with the following temperature cycling parameters: 94°C for 0.45 min, 56°C for 0.45 min, 72°C for 0.45 min, and final extension for 7 min at 72°C. Five microliters of PCR product was digested with 1 U of an appropriate restriction enzyme digestion set (Huang et al. 1996) according to manufacturer’s protocols (New England Biolabs). The digestion was incubated for 2–4 h, subjected to electrophoresis on 6% acrylamide gel, and then stained with ethidium bromide.
For all unknown mutation samples, PCR direct sequencing was performed for each exon with primers in Table 1 (Nuchprayoon et al. 2002; Huang et al. 1996; Tang et al. 1992; Ninokata et al. 2006). For haplotype analysis, PCR-RFLP technique was performed for detecting nt1311C > T polymorphism in exon 11 and nt93T > C polymorphism in intron 11 (Vulliamy et al. 1991; Beutler et al. 1991). Cases with G6PD Vianchan (871G > A, 1311T) were distinguished from G6PD Jammu (871G > A, 1311C) by the presence of nt1311C > T.
Results
G6PD activity in 162 healthy Mon males and 178 Burmese males showed a normal distribution. The average G6PD activity was 6.34 ± 2.89 IU/g Hb [mean ± standard deviation (SD)] in Mon and 6.56 ± 2.69 IU/g Hb in Burmese. G6PD activity less than 1.5 IU/g Hb was classified as G6PD deficiency. Ethnic group, origin of subject, and number of G6PD-deficient subjects are shown in Table 2.
Nineteen (12% of 162) Mon males were G6PD deficient and assayed for mutation. Using PCR-RFLP assays, 12 (63% of 19) were G6PD Mahidol, and one each was G6PD Jammu (871G > A; nt 1311C) and G6PD Kaiping (1388G > A). We also screened for G6PD Canton (1376G > T), G6PD Union (1360C > T), G6PD Chinese-5 (1024C > T), G6PD Gaohe (95A > G), G6PD Chinese-4 (392G > T), and G6PD Coimbra (592C > T) but could not identify any of these mutations in G6PD-deficient Mons. To identify whether mutation 871G > A was G6PD Viangchan (1311C > T) or G6PD Jammu (1311C), we used a PCR-RFLP technique with restriction enzyme BclI and identified nt 1311C in the sample with 871G > A, establishing a case of G6PD Jammu in a Mon subject.
DNA sequencing from all coding exons of G6PD genes of the five remaining G6PD-deficient Mons revealed a case of G6PD Mediterranean (563C > T), and a novel mutation 94(C > G) in a Mon individual from Pha-un province in Myanmar (Fig. 1). The G6PD activity of 94(C > G) was not detectable (Table 3). The mutations of three G6PD-deficient Mons remained unidentified.
Seventeen (10% of 178) Burmese males were G6PD deficient. Twelve (71%) G6PD-deficient Burmese were G6PD Mahidol (487G > A) and one (6%) was G6PD Coimbra (592C > T). DNA sequencing from all coding exons of G6PD genes of the four remaining G6PD-deficient samples identified G6PD Kerala-Kalyan (949G > A) in one case and G6PD Valladolid (406C > T) in another case, whereas two remained unidentified.
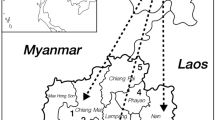
Mon and Burmese who had G6PD Mahidol came from various places of birth in Myanmar (Fig. 2; Table 2). Subjects with G6PD Mahidol had variable G6PD activities ranging from no detectable activity (8 of 24) to some residual activities. The average G6PD activity (± SD) for G6PD Mahidol was 0.43 ± 0.48 IU/g Hb (Table 3). All cases of G6PD Mahidol had haplotype 93T, 1311C. This haplotype was also the predominant haplotype in all G6PD-deficient Mons and Burmese, except for one case with G6PD Valladolid (406C > T).
Among 31 G6PD-normal Burmese randomly selected for haplotype analysis, 93T, 1311C (n = 22, 71.0%) was the most common, followed by 93C, 1311T (n = 8, 25.8%) and 93T, 1311T (n = 1, 3.2%), whereas 93C, 1311C was not identified. Among 32 G6PD-normal Mon randomly selected for haplotype analysis, only two haplotypes were found. Similar to the Burmese subjects, haplotype 93T, 1311C (n = 23, 71.9%) was more common than 93C, 1311T (n = 9, 28.1%), but the haplotype 93T, 1311T was not identified.
Discussion
Through a study for G6PD deficiency, we found the prevalence of G6PD deficiency to be high in Mon (12%) and Burmese (10%) males. Our result in Mons was higher than the previous report (6.7%; 3 of 42) (Iwai et al. 2001). The sample of Mons in our study was 3.6 times larger than that study and thus more accurate. The high prevalence of G6PD deficiency in Southeast Asians may be due to natural selection by malaria, which is endemic in Southeast Asia (Flatz et al. 1963).
There are many distinct ethnic groups in Myanmar. The eight largest groups are Burmese (Burman), Karen, Rakhine, Kachin, Kayin, Kayah, Chin, and Mon. Because of their distinctive language and cultural background and closed society until very recently, we could rely only on the self-reported ethnic classifications of the study participants.
Mon people speak Monic (Mon), which is an Austro-Asiatic, Mon-Khmer language, spoken by Mons and Cambodians (Gordon 2005). Historically, Mons were among the earliest people to settle in present-day southern Myanmar. Burmese people migrated to Myanmar around 800 AD and became the largest ethnic group in Myanmar. A Mon kingdom existed until 1757, after which it was repressed by the Burmese (Church 2003). Until now, there were few reports on genetic markers in Mon.
Burmese natives speak Burmese (Bamar), a Tibeto-Burman language. Because of their distinctive language, it has been controversial whether today’s Mon share a common ancestry with Burmese or Thai-Cambodian. We found that G6PD Mahidol was the most common mutation in Mon and Burmese males, with an allele frequency of 0.08 and 0.07, respectively. This finding confirms the results of previous reports that G6PD Mahidol is the predominant mutation in Burmese and Mon people in Myanmar (Iwai et al. 2001; Matsuoka et al. 2004). Our findings in Mon are in sharp contrast with two previous studies of Cambodians in which G6PD Viangchan was found to be the most common G6PD deficient mutation: 28 of 34 (82.4%) in one study (Louicharoen and Nuchprayoon 2005) and 46 of 47 (96.5%) in the other (Matsuoka et al. 2005) and G6PD Mahidol was not present in any of the G6PD-deficient Cambodians.
We also found that all subjects with G6PD Mahidol and almost all other Mon and Burmese mutations had haplotype 1311C and 93T. This haplotype is present in the majority of G6PD-normal Mon as well as Burmese. This is different from G6PD Viangchan in Cambodians and other populations, which have haplotype 1311T by definition, and 93T (Yan et al. 2006). Although haplotypes of Cambodians have not been reported in the literature, this evidence supports a common ancestry between Mon and Burmese rather than Cambodians.
In addition to G6PD Mahidol, we also identified a few other G6PD-deficient mutations and a novel mutation: a 94 (C > G) in a Mon individual from Pha-un province in Myanmar. The point mutation 94 (C > G), which occurs in exon 3, is predicted to change amino acid from histidine to aspartic acid at residue 32 and result in no G6PD activity in the subject, thus is a class II variant. This mutation is close to G6PD Gaohe (95A > G), which changes the same amino acid residue from histidine to arginine.
A few mutations were suggestive of an Indian contribution to the Mon and Burmese G6PD-deficient gene pool: G6PD Jammu (871G > A; 1311C), G6PD Mediterranean, and G6PD Kerala-Kalyan (949G > A). G6PD Jammu was previously found in an Indian (Beutler et al. 1991). G6PD Mediterranean is widely distributed in different populations in the Mediterranean regions of southern Europe, the Middle East, and India. The haplotype of our G6PD Mediterranean case was 1311C and 93T, which is similar to those with the Indian, rather than the Middle Eastern, type (Beutler and Kuhl 1990). G6PD Kerala-Kalyan (949G > A) was reported to be prevalent (24.9%) in India (Sukumar et al. 2004). None of these mutations were found in Thai, Laotians, or Cambodians, whereas G6PD Mediterranean was found in approximately 30% of G6PD-deficient Indonesians from central Java (Soemantri et al. 1995), 27% of G6PD-deficient Malaysian Malays (Ainoon et al. 2002), occasionally in Thais of the southern province of Thailand (Laosombat et al. 2005), and G6PD Kerala-Kalyan was also identified in Urak-Lawoi, a sea Gypsy population of the Andaman Sea who inhabited Phuket Island in southern Thailand (Ninokata et al. 2006). Both polymorphisms C and T are present at nt 1311 in Indians with G6PD Kerala-Kalyan (Sukumar et al. 2004). Our finding of G6PD Kerala-Kalyan in a Burmese from Yangon suggests the flow of this gene from India southward.
G6PD Kaiping is a predominant mutation among Chinese (Yan et al. 2006). This mutation is also the second most common mutation in southern Thailand (Laosombat et al. 2005). It is found in a few cases of Phuket Islanders in southern Thailand (Ninokata et al. 2006) and a few cases of Malaysian Malay (Ainoon et al. 2002). The haplotype of Chinese with G6PD Kaiping is also uniformly 1311C and 93T (Yan et al. 2006), similar to our Mon case in this study.
G6PD Coimbra (592C > T) is widely distributed across Europe and Asia but at low frequencies (Corcoran et al. 1992). G6PD Coimbra was previously reported in two G6PD-deficient Burmese (Matsuoka et al. 2004): one was from Yangon, which is similar to our case. G6PD Coimbra was also found in 3.5% of G6PD-deficient Malaysian Malay (Ainoon et al. 2002; Iwai et al. 2001) and in Flores Island, Indonesia (Kawamoto et al. 2006).
G6PD Valladolid (406C > T) has been previously identified in Spaniards (Vives-Corrons et al. 1997; Zarza et al. 1997) and Mexicans (Vaca et al. 2003). This mutation occurs in exon 5 and is predicted to code for cysteine at residue 135 instead of arginine and has reduced G6PD activity (0.64 IU/g Hb). How this mutation occurs in a G6PD-deficient Burmese subject is unclear. The mutation could have arisen independently, as there is no known strong historical linkage between Spain and Myanmar. However, a distinct haplotype seen in this case was different than in Burmese at large, suggesting a different ethnic origin of this individual, possibly during British occupation of Myanmar.
In summary, the G6PD mutations in Burmese are more heterogenous than previously suggested (Iwai et al. 2001; Matsuoka et al. 2004). Despite this heterogeneity, G6PD Mahidol is by far the most common G6PD-deficient mutation among Burmese and Mon populations. Its uniform and identical haplotype in both Mon and Burmese also confirm the common origin of these ethnic groups.
References
Ainoon O, Joyce J, Boo NY, Cheong SK, Zainal ZA, Hamidah NH (1999) Glucose-6-phosphate dehydrogenase (G6PD) variants in Malaysian Chinese. Hum Mutat 14:352–359
Ainoon O, Yu YH, Amir Muhriz AL, Boo NY, Cheong SK, Hamidah NH (2002) Glucose-6-phosphate dehydrogenase (G6PD) variants in Malaysian Malays. Hum Mutat 21:101–109
Betke K, Beutler E, Brewer GH, Kirkman HN, Luzzatto L, Motulsky AG, Ramot B, Siniscalco M (1967) Standardization of procedures for the study of glucose-6-phosphate dehydrogenase. Report of a WHO scientific group. WHO Tech Rep Ser No. 366
Beutler E (1994) G6PD deficiency. Blood 84:3818–3838
Beutler E, Kuhl W (1990) The NT 1311 polymorphism of G6PD: G6PD Mediterranean mutation may have originated independently in Europe and Asia. Am J Hum Genet 47:1008–1012
Beutler E, Westwood Beryl, Kuhl W (1991) Definition of the mutations of G6PD Wayne, G6PD Viangchan, G6PD Jammu, and G6PD ‘LeJeune’. Acta Haematol 86:179–182
Church P (2003) A short history of Southeast Asia, 1st edn. Wiley, London
Corcoran CM, Calabro V, Tamagnini G, Town M, Haider B, Vulliamy TJ, Mason PJ, Luzzatto L (1992) Molecular heterogeneity underlying the G6PD Mediterranean phenotype. Hum Genet 88:688–690
Flatz G, Sringram S (1963) Malaria and glucose-6-phosphate dehydrogenase deficiency in Thailand. Lancet 14:1248–1250
Gordon RG Jr (ed) (2005) Ethnologue: languages of the world, 15th edn. SIL International, Dallas. Online version: http://www.ethnologue.com/
Hamel AR, Cabral IR, Sales TS, Costa FF, Olalla-Saad ST (2002) Molecular heterogeneity of G6PD deficiency in an Amazonian population and description of four new variants. Blood Cell Mol Dis 28:399–406
Huang CS, Hung KL, Huang MJ, Li YC, Liu TH, Tang TK (1996) Neonatal jaundice and molecular mutations in glucose-6-phosphate dehydrogenase deficient newborn infants. Am J Hematol 51:19–25
Iwai K, Hirono A, Matsuoka H, Kawamoto F, Horie T, Lin K, Tantular IS, Dachlan YP, Notopuro H, Hidayah NI, Salim AM, Fujii H, Miwa S, Ishii A (2001) Distribution of glucose-6-phosphate dehydrogenase mutations in Southeast Asia. Hum Genet 108: 445–449
Kawamoto F, Matsuoka H, Kanbe T,Tantular IS, Pusarawati S, Kerong HI, Damianus W, Mere D, Dachlan YP (2006) Further investigations of glucose-6-phosphate dehydrogenase variants in Flores Island, eastern Indonesia. J Hum Genet 51: 952–957
Laosombat V, Sattayasevana B, Janejindamai W, Viprakasit V, Shirakawa T, Nishiyama K, Matsuo M (2005) Molecular heterogeneity of glucose-6-phosphate dehydrogenase (G6PD) variants in the south of Thailand and identification of a novel variant (G6PD Songklanagarind). Blood Cell Mol Dis 34:191–196
Louicharoen C, Nuchprayoon I (2005) G6PD Viangchan (871G > A) is the most common G6PD-deficient variant in the Cambodian population. J Hum Genet 50:448–452
Martini G, Toniolo D, Vulliamy T, Luzzatto L, Dono R, Viglietto G, Paonessa G, D’Urso M, Persico MG (1986) Structure analysis of the X-linked gene encoding human glucose-6-phosphate dehydrogenase. EMBO J 5:1849–1855
Matsuoka H, Nguon C, Kanbe T, Jalloh A, Sato H, Yoshida S, Hirai M, Arai M, Socheat D, Kawamoto F (2005) Glucose-6-phosphate dehydrogenase (G6PD) mutations in Cambodia: G6PD Viangchan (871G > A) is the most common variant in the Cambodian population. J Hum Genet 50:468–472
Matsuoka H, Wang J, Hirai M, Arai M, Yoshida S, Kobayashi T, Jalloh A, Lin K, Kawamoto F (2004) Glucose-6-phosphate dehydrogenase (G6PD) mutations in Myanmar: G6PD Mahidol (487G > A) is the most common variant in the Myanmar population. J Hum Genet 49:544–547
Ninokata A, Kimura R, Samakkarn U, Settheetham-Ishida W, Ishida T (2006) Coexistence of five G6PD variants indicates ethnic complexity of Phuket islanders, Southern Thailand. J Hum Genet 51:424–428
Nuchprayoon I, Sanpavat S, Nuchprayoon S (2002) Glucose-6-phosphate dehydrogenase (G6PD) mutations in Thailand: G6PD Viangchan (871G > A) is the most common deficiency variant in the Thai population. Hum Mutat 19:185
Poggi V, Town M, Foulkes NS, Luzzatto L (1990) Identification of a single base change in a new human mutant glucose-6-phosphate dehydrogenase gene by polymerase-chain-reaction amplification of the entire coding region from genomic DNA. Biochem J 271:157–160
Saha S, Saha N, Tay JS, Jeyaseelan K, Basair JB, Chew SE (1994) Molecular characterization of red cell glucose-6-phosphate dehydrogenase deficiency in Singapore Chinese. Am J Hematol 47:273–277
Soemantri AG, Saha S, Saha N, Tay JSH (1995) Molecular variants of red cell glucose-6-phosphate dehydrogenase deficiency in Central Java, Indonesia. Hum Hered 45:346–350
Sukumar S, Mukherjee MB, Colah RB, Mohanty D (2004) Molecular basis of G6PD deficiency in India. Blood Cells Mol Dis 33:141–145
Tang TK, Huang CS, Huang MJ, Tam KB, Yeh CH, Tang CJ (1992) Diverse point mutations result in glucose-6-phosphate dehydrogenase (G6PD) polymorphism in Taiwan. Blood 79:2135–2140
Vaca G, Arambula E, Monsalvo A, Medina C, Nunez C, Sandoval L, Lopez-Guido B (2003) Glucose-6-phosphate dehydrogenase (G-6-PD) mutations in Mexico: four new G-6-PD variants. Blood Cells Mol Dis 31:112–120
Vives-Corrons JL, Zarza R, Aymerrich M, Bioxadera J, Carrera A, Colomer D (1997) Molecular analysis of glucose-6-phosphate dehydrogenase deficiency in Spain. Sangre (Barc) 42:391–398
Vulliamy TJ, Othman A, Town M, Nathwani A, Falusi AG, Mason PJ, Luzzatto L (1991) Polymorphic sites in the African population detected by sequence analysis of the glucose-6-phosphate dehydrogenase gene outline the evolution of the variants A and A. Proc Natl Acad Sci USA 88:8568–8571
WHO Working Group (1989) Glucose-6-phosphate dehydrogenase deficiency. WHO Bull OMS 67:601–611
Xu W, Westwood B, Bartsocas CS, Malcorra-Azpiazu JJ, Indrak K, Beutler E (1995) Glucose-6-phosphate dehydrogenase mutations and haplotypes in various ethnic groups. Blood 85:257–263
Yan T, Cai R, Mo O, DongLin Z, Ouyan H, Huang L, Zhao M, Huang F, Li L, Liang X, Xu X (2006) Incidence and complete molecular characterization of glucose-6-phosphate dehydrogenase deficiency in the Guangxi Zhuang autonomous region of southern China: description of four novel mutations. Haematologica 91:1321–1328
Zarza R, Pujades A, Rovira A, Saavedra R, Fernandez J, Aymerich M, Vives Corrons JL (1997) Two new mutations of the glucose-6-phosphate dehydrogenase (G6PD) gene associated with haemolytic anaemia: clinical, biochemical and molecular relationships. Br J Haematol 98:578–582
Acknowledgments
We thank Dr. Molee Wanichsuwan for his assistance in collecting blood samples at Samutsakhon Hospital. This research was supported by The Royal Golden Jubilee Ph.D. program of the Thailand Research Fund. We thank Allen Noble, PA, for editorial review of the manuscript.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Nuchprayoon, I., Louicharoen, C. & Charoenvej, W. Glucose-6-phosphate dehydrogenase mutations in Mon and Burmese of southern Myanmar. J Hum Genet 53, 48–54 (2008). https://doi.org/10.1007/s10038-007-0217-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10038-007-0217-3
Keywords
This article is cited by
-
Prevalence of G6PD deficiency and diagnostic accuracy of a G6PD point-of-care test among a population at risk of malaria in Myanmar
Malaria Journal (2023)
-
Effect of neonatal reticulocytosis on glucose 6-phosphate dehydrogenase (G6PD) activity and G6PD deficiency detection: a cross-sectional study
BMC Pediatrics (2022)
-
Haematological profile of malaria patients with G6PD and PKLR variants (erythrocytic enzymopathies): a cross-sectional study in Thailand
Malaria Journal (2022)
-
Molecular characterization of G6PD mutations reveals the high frequency of G6PD Aures in the Lao Theung population
Malaria Journal (2021)
-
Distribution of G6PD deficiency genotypes among Southeast Asian populations
Tropical Medicine and Health (2021)