Abstract
The human chromosome 15q11-q13, or mouse chromosome 7C, is an imprinting domain controlled by bipartite imprinting centers (ICs): Prader-Willi syndrome (PWS)-IC and Angelman syndrome (AS)-IC. PWS-IC functions to maintain the paternal epigenotype on the paternal chromosome in somatic cells, while AS-IC plays a role in the establishment of the maternal epigenetic mark at PWS-IC in the female germline or early embryos. Several alternative exons and promoters of Snurf–Snrpn (SNRPN upstream reading frame–small nuclear ribonucleoprotein polypeptide N) are expressed as “IC transcripts”. Previous studies have shown that IC-transcript expression is restricted to the brain. We studied expression of the mouse IC-transcript in tissues including brain and oocytes as well as in cultured neurons and glia cells by RT-PCR and by in situ hybridization (ISH) in oocytes. The IC transcript was strongly expressed in brain (especially in neurons) and ovary (especially in oocytes and granulosa cells), while no expression was found in other tissues. This was confirmed by quantitative analysis and ISH. Expression levels in the brain were 7-fold higher compared to those in ovaries. ISH signals were observed in oocytes and granulosa cells of the secondary and developing follicles. These findings, together with previous data, suggest that the IC transcript may be associated with the establishment of PWS-IC methylation on the maternal chromosome as an AS-IC cis-acting element.
Similar content being viewed by others
Introduction
Genomic imprinting refers to the differential expression of genes according to their parental origin. There is evidence from human pedigrees for coordinate regulation of genomic imprinting in the Prader-Willi syndrome (PWS)/Angelman syndrome (AS) critical region at 15q11-q13 through a bipartite imprinting center (IC) extending several megabases in the imprinting domain (Nicholls and Knepper 2001). Similarly, the mouse 7C region—an imprinting domain homologous to the human AS/PWS domain—is also under the control of an IC (Nicholls and Knepper 2001). The human bipartite IC (PWS-IC and AS-IC) was defined by mapping of microdeletions in familial cases of PWS and AS, respectively (Ohta et al. 1999; Buiting et al. 1999). The human PWS-IC, which corresponds to the shortest region of overlap (SRO) (Fig. 1a) for deletions among such PWS patients, spans less than 4.3 kb and includes exon 1 of the SNURF–SNRPN (SNRPN upstream reading frame–small nuclear ribonucleoprotein polypeptide N) gene (Ohta et al. 1999). The mouse PWS-SRO ortholog is also located at the upstream region of Snurf–Snrpn (Yang et al. 1998; Fig. 1b), and functions to maintain the paternal epigenotype in the mouse PWS/AS imprinting domain in somatic cells (Bielinska et al. 2000). This interpretation is supported by evidence from a PWS patient who was mosaic for an imprinting mutation, and by analysis of mice chimeric for a deletion of PWS-SRO, which causes an imprinting defect (Bielinska et al. 2000). On the other hand, the human AS-IC (or AS-SRO) functions to establish a maternal epigenetic mark at the PWS-SRO in the female germline or in early embryonic cells as a cis-acting element (Kantor et al. 2004; Haruta et al. 2005; Fig. 2). The human SNURF–SNRPN and mouse Snurf–Snrpn are imprinted and expressed only from the paternal allele in somatic cells, and their expression is thought to be controlled with DNA methylation at the SNURF–SNRPN/Snurf–Snrpn promoter region (PWS-SRO) (Glenn et al. 1996; Shemer et al. 1997). Previous expression analyses of human SNURF–SNRPN and mouse Snurf–Snrpn demonstrated that monoallelic expression appeared from the four-cell post-fertilization stage onwards, while oocytes and early embryonic cells before the four-cell stage show biallelic expression (Huntriss et al. 1998; Szabo and Mann 1995a, b).
The human (a) and mouse (b) alternative transcripts expressed from the upstream region of the SNURF–SNRPN /snurf–Snrpn (SNRPN upstream reading frame–small nuclear ribonucleoprotein polypeptide N) gene. The imprinting center (IC) transcripts are expressed from multiple promoters (arrows) and include several exons. The human Angelman syndrome shortest region of overlap (AS-SRO) (AS-IC) is located upstream of SNURF–SNRPN and includes one exon (U5) (adapted from Dittrich et al. 1996), while the mouse AS-SRO has not yet been identified
Putative function of AS-IC (AS-SRO) and Prader-Willi syndrome (PWS)-IC (PWS-SRO) in somatic cells and oocytes as proposed by Nicholls and Knepper (2001). In somatic cells, a cis-acting element (arrow) from the paternal PWS-SRO maintains the paternal epigenotype in the AS/PWS imprinting domain, while the maternal PWS-SRO is methylated (shaded ovals). In oocytes, a cis-acting element (arrow) from the AS-SRO works to establish the maternal mark at the PWS-SRO
Several alternative exons upstream of SNURF–SNRPN/Snurf–Snrpn have been identified in humans (Dittrich et al. 1996) and mice (Bressler et al. 2001). In humans, alternative exons located at the upstream region of the SNRPN promoter lie around the AS-SRO (Fig. 1a), and one AS patient with an imprinting defect has a mutation at the splice site for the IC transcript (Farber et al. 1999). Several mouse IC transcripts with alternative exon(s) expressed from upstream alternative promoters of Snurf–Snrpn are also imprinted as a paternally expressed transcript, expression of which is restricted to the brain (Bressler et al. 2001). There is no similarity between the IC-transcript nucleotide sequences of human and mouse. However, the expression patterns and gene structure of the human and mouse IC transcripts are conserved. These upstream alternative exons may play an important role in the establishment of the maternal primary mark in the imprinting domain. Therefore, a cis-acting factor from the AS-SRO should be released in the female germline or the maternal chromosome in early embryos.
In this report, we aimed to examine expression of mouse IC transcripts in the germline to further assess their role in genomic imprinting of the 7C region.
Materials and methods
Reverse transcription-polymerase chain reaction
Total RNA was extracted from 3-week-old mouse tissues including brain, heart, liver, skeletal muscle, testis, kidney, and ovary using the RNeasy kit (Qiagen, Tokyo, Japan), according to the manufacturer’s recommendations. The RNA (2 μg) was used for cDNA synthesis using the SuperScript First-Strand synthesis System (Gibco BRL, Tokyo, Japan) by random hexamer priming. Reverse transcription-polymerase chain reaction (RT-PCR) was performed with primers for the mouse Snurf–Snrpn exons U1, 1 and 3. PCR conditions were as follows: initial denaturation at 94°C for 2 min, followed by 30 cycles at 94°C for 30 s, 65°C for 30 s, 72°C for 30 s, and final extension at 72°C for 10 min. The primers used for the IC-transcript expression have been published previously (Bressler et al. 2001). After visualization on a 2% agarose gel, PCR products were cloned in the cloning vector pCR2.1-TOPO (Invitrogen, Tokyo, Japan). Plasmid DNA was extracted using FastPlasmid Mini kit (Eppendorf, Hamburg, Germany). Sequencing was performed on ABI 3100 sequencer (Applied Biosystems, Foster City, CA) using Big Dye Terminator (Applied Biosystems).
Quantitative analysis of the IC transcript was performed by real-time PCR on the ABI PRISM 7900HT sequence detection system (Applied Biosystems) using SYBR Premix Ex Taq (Perfect Real Time) (Takara Bio, Shiga, Japan). Relative quantification of IC-transcript expression was performed using the standard curve method as described in user bulletin #2 (Applied Biosystems). Gapdh primers were used to make the standard curve and each experiment was performed in triplicate at least three times.
Oocyte and cumulus cell collections
Metaphase II oocytes and cumulus cells were collected from 10-week-old female mice that had been superovulated by injection of 5 IU pregnant mares’ serum gonadotropin (Teikoku Zouki, Japan), followed by injection of 5 IU human chorionic gonadotropin (hCG, Teikoku Zouki) 47 h later. At 20 h after hCG administration, MII oocytes with cumulus cells were recovered from the oviducts, and the cumulus cells were dispersed with 1 mg/ml hyaluronidase (Sigma, St Louis, MO). Oocytes were picked up using a mouth-controlled drawn-out glass pipette and passed through five to six dishes of HTF medium to wash out surrounding somatic cells. Cumulus cell-free, non-fragmented, and “healthy” looking oocytes were chosen for RT-PCR analysis. Cumulus cells with and without oocytes obtained by the same procedure were also used for the analysis. Total RNA was then extracted from the collected oocytes and cumulus cells using an RNeasy Micro kit (Qiagen) according to the manufacturer’s instructions.
In situ hybridization
After dissection, one ovary from the same mouse whose tissues had been used for RNA extraction was fixed overnight in 4% paraformaldehyde (PFA) in PBS (120 mM NaCl, 2.7 mM KCl, and 10 mM phosphate buffer, pH 7.4), and embedded in paraffin. Sections (5 μm thick) were mounted on silane-coated glass slides. To estimate the amount of RNA retained in the paraffin sections, methyl green-pyronin Y staining was performed before in situ hybridization (ISH), according to Shutle et al. (1992). ISH was performed using digoxigenin-labeled oligonucleotide probes as previously described (Koji and Brenner 1993; Hishikawa et al. 1999). Briefly, the sections were warmed at 60°C for 30 min, deparaffinized with toluene and rehydrated through decreasing concentrations of ethanol. They were treated with 0.2 N hydrochloric acid at room temperature for 20 min and digested with 10 μg/ml proteinase K at 37°C for 15 min. After post-fixation in 4% PFA in PBS for 5 min at room temperature, the sections were immersed in 2 mg/ml glycine in PBS twice, 15 min each at room temperature. The sections were then kept in 30% deionized formamide in 4× SCC [1× SCC: 0.15M NaCl, 0.015M sodium citrate (pH 7.0)] until hybridization. Hybridization was performed at 42°C overnight with 1 μg/ml digoxigenin-labeled antisense oligo-DNA complementary to Snurf–Snrpn exon U2 dissolved in hybridization buffer containing 10 mM Tris–HCl (pH 7.4), 0.6 M NaCl, 1 mM EDTA, 1× Denhardt’s solution, 250 μg yeast tRNA, 125 μg/ml salmon testis DNA, 10% dextran sulfate, and 30% deionized formamide. Post-hybridization washing was performed five times at 37°C with 2× SCC/50% formamide for 1 h. Sections were reacted with horseradish peroxidase (HRP)-conjugated sheep antidigoxigenin antibody (Roche, Indianapolis, IN), and the HRP sites were visualized with DAB, H2O2, Co2+, and Ni2+. To confirm the specificity of the signals obtained, several control experiments were simultaneously performed. Duplicate serial sections were used in each experiment, and the sense probe was applied as a negative control. To evaluate the levels of hybridizable RNAs in the sections, 28S rRNA probe was used as a positive control (Yoshii et al. 1995) in each experiment. In some sections, competition experiments were performed with the antisense probe in the presence of 100-fold excess of either the antisense or sense non-labeled oligo-DNA to verify the sequence specificity of the signal obtained as described previously (Koji and Brenner 1993). Positive cells were evaluated based on the density using an image analyzer (DAB system, Carl Zeiss, Thornwood, NY).
IC-transcript probes (5′–3′) used were as follows: sense probe for Snrpn exon U2, GTGCAGCAGGTCCTGCTGAGCCAAAGATGCCTGTCACATCCACCC; antisense probe for exon U2, GGGTGGTTGTGACAGGCATCTTTGGCTCAGCAGGACCTGCTGCAC.
Results
Expression of the IC transcript in the ovary
Mouse IC-transcript expression has been reported to be restricted to the brain (Bressler et al. 2001). To assess whether the IC transcript is expressed in other tissues including the ovary, we used a panel of mouse tissues—brain, ovary and testis, liver, heart, kidney, and skeletal muscle—for RNA extraction and expression analysis by RT-PCR. As described previously, the IC transcript was strongly expressed in the brain, while no expression was observed in other tissues examined except the ovary (Fig. 3a). However, Snurf–Snrpn was expressed in all tissues examined. Sequence analysis of PCR products showed that the IC-transcript exon U1 was contiguous to Snurf–Snrpn exon 2, skipping exon 1 (data not shown), as reported previously (Bressler et al. 2001). Quantitative analysis of IC-transcript expression in these tissues by real-time PCR revealed that the IC transcript was expressed in the brain and in the ovary (Fig. 3b), while no expression was found in other tissues. Expression in the brain was 7-fold higher than that in the ovary. The Snurf–Snrpn showed the highest expression in the brain followed by heart and ovary (Fig. 3c)
a Expression patterns of the mouse IC transcript by RT-PCR analysis using a primer set for U1-exon 3. The IC transcript is expressed only in the brain and ovary among the various tissues indicated, while Snurf–Snrpn (primer set for exons 1 and 2), as a positive control, is expressed in all tissues studied. Gapdh was used as an internal control. b Quantitative analysis of the IC-transcript expression by real-time PCR showing that the IC transcript is expressed in the brain and the ovary, and that expression in the brain is 7-fold higher than in the ovary. c Snurf–Snrpn is expressed in various tissues with the highest levels in the brain. Relative quantities were normalized to Gapdh expression in each sample
Cell-specific IC-transcript expression
Quantitative analysis of IC-transcript expression at the cell level was performed with real-time PCR in cultured neurons, glia cells, and fibroblasts obtained as described previously (Yamasaki et al. 2003), as well as in uncultured oocytes and granulosa cells collected as described above. The IC transcript was expressed in neurons, oocytes, and oocytes with granulosa cells (Fig. 4). Expression in neurons was 7-fold higher than in oocytes with granulosa cells, this result being concordant with the quantitative analysis of the IC transcript in brain and ovarian tissues (Fig. 3). Expression in oocytes was only around one-third of that in oocytes with granulosa cells (Fig. 4b), implying that the IC transcript is highly expressed in granulosa cells. No expression of the IC transcript was detected in glia cells and fibroblasts.
Cell-type specific expression of the IC transcript. The IC transcript is expressed in neurons, oocytes, and oocytes with granulosa cells (a), while no expression is observed in glia cells or fibroblasts. Real-time PCR analysis of the IC-transcript expression at the cell level. The IC transcript is highly expressed in neurons, and moderately in oocytes with granulosa cells (b)
Localization of the IC transcript in the mouse ovary
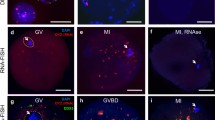
To confirm that the IC transcript is expressed from the ovarian cells, we performed non-radioactive ISH. We used an oligo-deoxynucleotide probe complementary to IC-transcript exon U2, since exon U1 (used for RT-PCR) shares sequence similarity with Snrpn exon 1. Positive ISH signals were detected in the oocyte and granulosa cells of Graafian follicles (Fig. 5a, b). Stronger signals were observed in the oocytes and granulosa cells of the secondary and developing follicles (Fig. 5e, f). To confirm the specificity of the signals obtained, we performed competition experiments using the antisense probe in the presence of an excess of non-labeled homologous oligo-DNA. The signal was notably reduced compared to the labeled antisense used alone, confirming the specificity of the signal (data not shown). As a negative control, no specific signal was observed in adjacent sections that were hybridized with the labeled sense probe (Fig. 5c, d, g, h). No specific signal was detected in the ovarian stroma.
In situ hybridization (ISH) of Snurf–Snrpn exon U2 in sections from the mouse ovary. ISH signals appear in the oocyte and granulosa cells (a, b, e, f), but no specific signals were present when a sense probe was used (c, d, g, h). ISH-positive signals were highlighted with red color by an image analyzer (b, d, f, h). The Graffian follicle (a–d) and developing follicles (e–h) are shown. Representative ISH results were obtained from at least three different experiments
Discussion
The present RT-PCR and ISH analyses in the oocyte of the developing follicle of 3 and 10-week-old mice showed that the IC transcript was expressed from alternative exons. This was confirmed by sequencing of the RT-PCR product, which showed the presence of fused exons U1 and 2, skipping exon 1. Interestingly, a previous study demonstrated that Snurf–Snrpn is expressed from both parental alleles in early mouse embryos before the four-cell stage, and that Snurf–Snrpn monoallelic expression in human is established after the four-cell stage, as well as the regular global transcription activation (Huntriss et al. 1998). The global gene activation in early embryos takes place later than the two-cell stage in the mouse and the four-cell stage in the human (Braude et al. 1988). Prior to these stages, the embryo relies on transcription products that have been accumulated in the oocyte. Therefore, biallelic Snurf–Snrpn expression observed previously in early embryonic cells may reflect expression of both IC transcripts accumulated from the maternal oocyte and paternally expressed products from a diploid chromosome. Since Snurf–Snrpn and the IC transcript share exons 2–10, an RT-PCR primer designed to anneal downstream of exon 2 would show an expression pattern as if it was biparental.
We have observed that the alternative IC transcript is expressed only in the ovary and brain among the tissues examined. Unexpectedly, ISH analysis showed that the IC transcript is also expressed in granulosa cells, but never from other somatic cells in the ovary. It has been demonstrated that oocyte–granulosa cell communication is essential for normal growth and development of both the oocyte and the follicle, and that this communication involves paracrine signaling and exchange of small regulatory molecules through gap-junctions (Gilchrist et al. 2004). However, it is unlikely that there is a direct exchange of RNA transcripts, such as the IC transcript, between the oocyte and granulosa cells. The expression of the IC transcript we observed in granulosa cells may be important for the expression of other factors necessary for oocyte development. As the oocyte and granulosa cells have similar receptors for some signaling molecules (Gilchrist et al. 2004), it is possible that the expression of the IC transcript in granulosa cells is also concomitant to the expression in the oocyte, but that it does not have a significant biological role.
The results of our ISH and RT-PCR analyses indicate that the IC transcript in the female germline uses an alternative promoter upstream of the Snurf–Snrpn promoter (Fig. 6). In other words, the alternative promoter may be required for expression of the IC transcript in the haploid oocyte. A similar phenomenon was seen in an oocyte- and early-embryo-specific DNA methyltransferase, Dnmt1o, which was previously isolated as an alternative transcript of the Dnmt1 gene (Wilkins 2005). Since the maintenance of DNA methylation with a methyltransferase is required in the female germline, the alternative promoter is used for its expression (Wilkins 2005). In the case of Snurf–Snrpn, it is also possible that the Snurf–Snrpn protein as well as the Dnmt1 is required for oogenesis. The alternative promoter for the IC transcript might be used to express Snurf–Snrpn. However, previous analysis of a knockout mouse experiment (Yang et al. 1998) demonstrated that, unlike the Dnmt1o protein, the Snurf–Snrpn protein is not essential for normal mouse development. Therefore, the role of the IC transcript may be different from that of the alternative transcript of the Dnmt1 gene.
Alternative usage of Snurf–Snrpn promoters in the oocyte, early embryonic cells, and somatic cells. In the oocyte, the alternative IC transcript upstream of Snurf–Snrpn is expressed from the IC transcript exon U1 promoter, whereas in early embryonic cells, methylation (closed circles) at the PWS-IC is established, which may be influenced by IC-transcript expression in the oocyte. In somatic cells, Snurf–Snrpn is expressed from the exon 1 promoter only on the paternal chromosome 7
The IC transcript is highly expressed from the paternal chromosome in the brain. Paternal expression of the imprinted genes in the 7C domain is controlled by the PWS-IC in somatic cells (Nicholls and Knepper 2001). Therefore, the paternal imprinting expression of the IC transcript in the brain results from a cis-acting factor from the PWS-IC on the paternal chromosome. However, oocytes must have a different mechanism to express the IC transcript, since the PWS-IC is methylated and therefore does not function.
As for the establishment of the maternal mark, there is a hypothesis that a specific trans-acting factor may bind to the AS-SRO on the unmethylated chromosome in the female germline, where some proteins, such as co-factors, are recruited for modification of chromatin structure resulting in a chromatin loop to establish maternal methylation at the PWS-IC. A similar mechanism has been described in the locus-controlling region of the beta-globin gene locus (Li et al. 2002). If this hypothesis is correct, only transcription factors and/or co-factors would function in establishment of maternal methylation at the PWS-SRO (PWS-IC), and IC-transcript expression would result from the altered chromatin structure resulting from trans-acting protein(s) bound to the AS-IC region. In this case, IC-transcript expression would not contribute to establishment of the maternal mark. However, the conserved structure of the IC transcript between human and mouse (Fig. 1), results of previous AS/PWS microdeletion analysis, and the expression patterns observed in the oocyte in our experiment suggest that the IC transcript functions as an AS-IC cis-acting element to establish methylation of the PWS-IC on the maternal chromosome.
References
Bielinska B, Blaydes SM, Buiting K, Yang T, Krajewska-Walasek M, Horsthemke B, Brannan CI (2000) De novo deletions of SNRPN exon 1 in early human and mouse embryos result in a paternal to maternal imprint switch. Nat Genet 25:74–78
Braude P, Bolton V, Moore S (1988) Human gene expression first occurs between the four- and eight-cell stages of preimplantation development. Nature 332:459–461
Bressler J, Tsai TF, Wu MY, Tsai SF, Ramirez MA, Armstrong D, Beaudet AL (2001) The SNRPN promoter is not required for genomic imprinting of the Prader-Willi/Angelman domain in mice. Nat Genet 28:232–240
Buiting K, Lich C, Cottrell S, Barnicoat A, Horsthemke B (1999) A 5-kb imprinting center deletion in a family with Angelman syndrome reduces the shortest region of deletion overlap to 880 bp. Hum Genet 105:665–666
Dittrich B, Buiting K, Korn B, Rickard S, Buxton J, Saitoh S, Nicholls RD, Poustka A, Winterpacht A, Zabel B, Horsthemke B (1996) Imprint switching on human chromosome 15 may involve alternative transcripts of the SNRPN gene. Nat Genet 14:163–170
Farber C, Dittrich B, Buiting K, Horsthemke B (1999) The chromosome 15 imprinting centre (IC) region has undergone multiple duplication events and contains an upstream exon of SNRPN that is deleted in all Angelman syndrome patients with an IC microdeletion. Hum Mol Genet 8:337–343
Gilchrist RB, Ritter LJ, Armstrong DT (2004) Oocyte–somatic cell interactions during follicle development in mammals. Anim Reprod Sci 82–83:431–446
Glenn CC, Saitoh S, Jong MT, Filbrandt MM, Surti U, Driscoll DJ, Nicholls RD (1996) Gene structure, DNA methylation, and imprinted expression of the human SNRPN gene. Am J Hum Genet 58:335–346
Haruta M, Meguro M, Sakamoto YK, Hoshiya H, Kashiwagi A, Kaneko Y, Mitsuya K, Oshimura M (2005) Narrowed abrogation of the Angelman syndrome critical interval on human chromosome 15 does not interfere with epigenotype maintenance in somatic cells. J Hum Genet 50:124–132
Hishikawa Y, Koji T, Dhar DK, Kinugasa S, Yamaguchi M, Nagasue N (1999) Metallothionein expression correlates with metastatic and proliferative potential in squamous cell carcinoma of the oesophagus. Br J Cancer 81:712–720
Huntriss J, Daniels R, Bolton V, Monk M (1998) Imprinted expression of SNRPN in human preimplantation embryos. Am J Hum Genet 63:1009–1014
Kantor B, Kaufman Y, Makedonski K, Razin A, Shemer R (2004) Establishing the epigenetic status of the Prader-Willi/Angelman imprinting center in the gametes and embryo. Hum Mol Genet 13:2767–2779
Koji T, Brenner RM (1993) Localization of estrogen receptor messenger ribonucleic acid in rhesus monkey uterus by non radioactive in situ hybridization with digoxigenin-labeled oligodeoxynucleotides. Endocrinology 132:382–392
Li Q, Peterson KR, Fang X, Stamatoyannopoulos G (2002) Locus control regions. Blood 100:3077–3086
Nicholls RD, Knepper JL (2001) Genome organization, function, and imprinting in Prader-Willi and Angelman syndromes. Annu Rev Genomics Hum Genet 2:153–175
Ohta T, Gray TA, Rogan PK, Buiting K, Gabriel JM, Saitoh S, Muralidhar B, Bilienska B, Krajewska-Walasek M, Driscoll DJ, Horsthemke B, Butler MG, Nicholls RD (1999) Imprinting-mutation mechanisms in Prader-Willi syndrome. Am J Hum Genet 64:397–413
Shemer R, Birger Y, Riggs AD, Razin A (1997) Structure of the imprinted mouse Snrpn gene and establishment of its parental-specific methylation pattern. Proc Natl Acad Sci USA 94:10267–10272
Shutle EKW, Lyon HO, Hoyer PE (1992) Simultaneous quantification of DNA and RNA in tissue sections. A comparative analysis of the methyl green-pyronin technique with the gallocyanin chromalum and Feulgen procedures using image cytometry. Histochem J 24:305–310
Szabo PE, Mann JR (1995a) Allele-specific expression and total expression levels of imprinted genes during early mouse development: implications for imprinting mechanisms. Genes Dev 9:3097–3108
Szabo PE, Mann JR (1995b) Biallelic expression of imprinted genes in the mouse germ line: implications for erasure, establishment, and mechanisms of genomic imprinting. Genes Dev 9:1857–1868
Wilkins JF (2005) Genomic imprinting and methylation: epigenetic canalization and conflict. Trends Genet 21:356–365
Yamasaki K, Joh K, Ohta T, Masuzaki H, Ishimaru T, Mukai T, Niikawa N, Ogawa M, Wagstaff J, Kishino T (2003) Neurons but not glial cells show reciprocal imprinting of sense and antisense transcripts of Ube3a. Hum Mol Genet 12:837–847
Yang T, Adamson TE, Resnick JL, Leff S, Wevrick R, Francke U, Jenkins NA, Copeland NG, Brannan CI (1998) A mouse model for Prader-Willi syndrome imprinting-centre mutations. Nat Genet 19:25–31
Yoshii A, Koji T, Ohsawa N, Nakane PK (1995) In situ localization of ribosomal RNAs is a reliable reference for hybridizable RNA in tissue sections. J Histochem Cytochem 43:321–328
Acknowledgements
N.N. was supported in part by CREST from the Japan Science and Technology Agency. T.O. was supported in part by “Academic Frontier” Project for Private Universities: matching fund subsidy from the Ministry of Education, Culture, Sports, Science, and Technology (MEXT), 2002–2006, and a Grant-in-Aid for Scientific Research (Category C, No. 15590290) from MEXT. We also thank Ms. Yasuko Noguchi and Naoko Yanai for their technical assistance.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Mapendano, C.K., Kishino, T., Miyazaki, K. et al. Expression of the Snurf–Snrpn IC transcript in the oocyte and its putative role in the imprinting establishment of the mouse 7C imprinting domain. J Hum Genet 51, 236–243 (2006). https://doi.org/10.1007/s10038-005-0351-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10038-005-0351-8
Keywords
This article is cited by
-
Expression of SNURF–SNRPN upstream transcripts and epigenetic regulatory genes during human spermatogenesis
European Journal of Human Genetics (2009)
-
A targeted deletion upstream of Snrpn does not result in an imprinting defect
Mammalian Genome (2007)