Abstract
Interleukin (IL)-13, which is essential for IgE synthesis, mediates its effects by binding with a receptor composed of IL-4Rα and IL-13Rα1. We investigated the effects of IL-13 and IL-13Rα1 polymorphisms in Korean children with asthma, and whether these have been associated with IgE production. We enrolled 358 atopic asthmatic, 111 non-atopic asthmatic, and 146 non-atopic healthy children. IL-13 and IL-13Rα1 genotypes were identified using the PCR-RFLP method. There was an association between the asthma susceptibility and homozygosity for risk allele of IL-13 G+2044A. In children with atopic asthma, risk alleles in IL-13 (A−1512C and C−1112T) and IL-13Rα1 (A+1398G) showed increased total IgE (P=0.012, 0.015 and 0.017, respectively). Three-loci haplotype analysis for IL-13 showed that the haplotype composed of −1512C, −1112T and +2044A was associated with higher total IgE than other tested haplotypes in children with atopic asthma (P=0.003). The gene–gene interaction between risk alleles of each IL-13 promoter polymorphism and IL-13Rα1 polymorphism was associated with higher total IgE in children with atopic asthma (P=0.002, 0.010). These findings indicate that the IL-13 G+2044A is associated with asthma development and the IL-13 and IL-13Rα1 polymorphisms may interact to enhance IgE production.
Similar content being viewed by others
Introduction
Atopy, a genetically determined hypersensitivity to common environmental allergens, is associated with increased generation of IgE (Kay 2001). Asthma is associated with atopy and with IgE-mediated inflammation of the airways, which occurs via production of interleukin (IL)-4 and IL-13, both of which have been implicated in the pathogenesis of asthma in multiple human and animal studies (Rankin et al. 1996; Zhu et al. 1999; Nakamura et al. 1996; Doucet et al. 1998).
IL-13 has been noted as a critical effector in the induction and maintenance of IgE production and IgE-mediated allergic airway responses (van der Pouw Kraan et al. 1998; Wills-Karp and Chiaramonte 2003; Cohn et al. 2004). However, analyses of the association between asthma phenotype and specific polymorphisms in the IL-13 gene have produced varied results. In a Dutch population, significant associations were reported between the IL-13/−1112T allele and asthma (Howard et al. 2001; van der Pouw Kraan et al. 1999), as well as bronchial hyperreactivity (BHR) (Howard et al. 2001), skin test responsiveness (Howard et al. 2001), altered regulation of IL-13 production (van der Pouw Kraan et al. 1999), and increased binding of nuclear proteins to this region (van der Pouw Kraan et al. 1999). In German populations, the IL-13/+2044A (Arg130Gln) variant showed a significant association with high IgE levels (Graves et al. 2000), whereas this allele was found to be associated with asthma, but not IgE levels in British and Japanese populations (Heinzmann et al. 2000). Thus, IL-13 polymorphisms may have varied and population-specific effects.
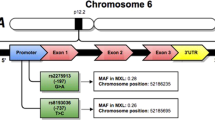
IL-13 mediates its effect via a receptor composed of a heterodimer that includes IL-4Rα and IL-13Rα1 (Miloux et al. 1997). IL-13Rα1 is widely expressed and has been identified on nearly every cell, except human T cells (Graber et al. 1998; Akaiwa et al. 2001). The gene encoding IL-13Rα1 has been mapped to the X chromosome, and in vitro expression studies of IL-13Rα1 revealed specific binding to IL-13 (Miloux et al. 1997; Hilton et al. 1996; Guo et al. 1997). By itself, IL-13Rα1 binds IL-13 with low affinity; when paired with IL-4Rα, the heterodimer binds IL-13 with high affinity and forms a functional IL-13 receptor that is capable of signal transduction (Miloux et al. 1997). Polymorphisms in these receptor molecules contribute to the genetic influences on asthma susceptibility and atopy. For example, asthma susceptibility was significantly increased in Korean children by gene–gene interaction between IL-4 T−590C and IL-4Rα Gln551Arg alleles (Lee et al. 2004) and between IL-13 C−1112T and IL-4Rα Ser478Pro alleles in Dutch population (Howard et al. 2002). Furthermore, a noncoding polymorphism in IL-13Rα1 (A+1398G) was associated with increased IgE levels in British population (Heinzmann et al. 2000). These findings highlight the importance of studying gene–gene interactions in the complex biological pathways of IL-4/IL-13 and their receptors. However, no previous study has examined gene–gene interactions involving IL-13 and IL-13Rα1. Here, we examined the polymorphisms in IL-13 and IL-13Rα1, and assessed their independent and combined effects on the development of asthma or the elevation of total serum IgE in the Korean population.
Materials and methods
Subjects
The enrolled study subjects consisted of 615 Korean children who visited the Asthma and Allergy Clinics of the Asan Medical Center. They were classified into three groups (358 atopic asthmatics, 111 nonatopic asthmatics, and 146 nonatopic controls without asthma) and adjusted according to age and sex (Table 1).
Asthma phenotypes and bronchial responsiveness were determined by a physician. Following the American Thoracic Society guidelines (American Thoracic Society 1987), asthma was confirmed by a history of dyspnea and wheezing during the previous 12 months, a greater than 12% reversibility of FEV1 spontaneously or after β2-agonist inhalation, and/or a methacholine provocation test result with a PC20 less than 16 mg/ml. Atopy was defined by a positive skin prick test (wheal diameter ≥3 mm) to at least one of 27 common aeroallergens in Korea (Lee et al. 2004) or by elevated allergen-specific IgE. Non-atopy was defined by negative specific IgE in response to Dermatophagoides farinae (D.f, and Dermatophagoides pteronyssinus (D.p), and negative skin prick test to the 27 common aeroallergens. The nonatopic control subjects had no history of asthma or other allergic diseases, negative skin prick tests, normal total IgE values (≤100 IU/ml), normal lung function tests, and no airway hyperresponsiveness (PC20>16 mg/ml). The ethics committee of the Asan Medical Center Institutional Review Board approved the study, and written informed consents were obtained from the parents of all subjects.
Serum IgE, skin prick test and methacholine challenge test
The concentrations of total IgE and D.f- and D.p-specific IgE were measured by fluorescent enzyme immunoassay (AutoCAP System; Phadia, Uppsala, Sweden). Specific IgE was graded into classes from 1 to 6, according to the manufacturer’s instructions, with a specific IgE concentration ≥0.35 kIU/l scored as positive.
Skin prick testing was performed with 27 common aeroallergens (Allegopharma, Germany) in Korea (Lee et al. 2004), which were considered positive if the maximum wheal diameter was ≥3 mm.
For the methacholine challenge test, we used a dosimeter with a concentration range of 0.625–25 mg/ml, as previously described (Chai et al. 1975). FEV1 was measured after each inhalation with a spirometer (Microspiro HI298; Tokyo, Japan). Airway responsiveness was expressed as the concentration of methacholine that provoked a 20% fall in FEV1 (PC20), and airway hyperresponsiveness to methacholine was defined as PC20<16 mg/ml.
Genotyping of IL-13 and IL-13Rα1 polymorphisms
IL-13 and IL-13Rα1 polymorphisms were analyzed by polymerase chain reaction (PCR), combined with restriction fragment length polymorphism (RFLP) analysis. Fragments were amplified in 15 μl reaction mixtures containing 20 ng genomic DNA, 0.033 mM of each dNTP, 1× PCR buffer (10 mM Tris–HCl, pH 8.3, 50 mM KCl, 1.25 mM MgCl2), 5 pmol of each primer, and 0.25 units AmpliTaq Gold Taq polymerase (Applied Biosystems, Foster City, Calif.). The thermocycling conditions consisted of an initial denaturation at 95°C for 12 min, followed by 35 cycles of denaturation at 95°C for 30 s, annealing for 2 min at 65°C for IL-13 −1512A/C, 54°C for IL-13 −1112C/T, 55°C for IL-13 +2044G/A or 45°C for IL-13Rα1 +1398A/G, and extension at 72°C for 40 s, followed by a final extension step at 72°C for 5 min, using a Gene-Amp PCR System 9700 (Applied Biosystems, Norwalk, Conn.).
The tested polymorphisms of IL-13 (Graves et al. 2000) and IL-13Rα1 (Heinzmann et al. 2000) were amplified with gene-specific PCR primers and digested with specific restriction endonucleases. For −1512A/C, the primers were 5′-CAA CCG CCG CGC CAG CGC CTT CTC-3′ and 5′-CCG CTA CTT GGC CGT GTG ACC GC-3′, and the restriction enzyme was BstUI (New England BioLabs, Beverly, Mass., USA), which digested the −1512A allele into 216 and 29 bp fragments and the −1512C allele into 194, 29, and 22 bp fragments. For −1112C/T, the primers were 5′-GGA ATC CAG CAT GCC TTG TGA GG-3′ and 5′-GTC GCC TTT TCC TGC TCT TCC CGC-3′, and the restriction enzyme was BstUI, which digested the −1112C into 223 and 23 bp fragments and yielded a single 246 bp band for the −1112T allele. For +2044G/A, the primers were 5′-CTT CCG TGA GGA CTG AAT GAG ACG GTC-3′ and 5′-GCA AAT AAT GAT GCT TTC GAA GTT TCA GTG GA-3′, and the restriction enzyme was NlaIV (New England BioLabs), which digested the +2044G into 178 and 32 bp fragments and yielded a single 210 bp band for the +2044A allele. For +1398A/G, the primers were 5′-TCA GTG ATG GAG ATA ATT TA-3′ and 5′-TGA GCT GCC TGT TTA TAA AT-3′, and the restriction enzyme was MseI (New England BioLabs), which digested the +1398A allele into 85 and 45 bp fragments and yielded a single 130 bp band for the +1398G allele. The PCR products were digested with 5 U of the listed restriction enzyme at 37°C for 4 h, and separated on 3% agarose gels (FMC BioProducts, Rockland, Me., USA). To confirm the accuracy of these RFLP analyses, 20% of the subjects were randomly selected for DNA sequencing.
Statistical analysis
Clinical phenotypes, including total IgE and PC20, were analyzed as a quantitative trait. Differences of clinical phenotypes between groups were analyzed by the Kruskal–Wallis and Mann–Whitney tests. Quality variables, including sex and age, were analyzed by multiple logistic regression to determine whether there was a disproportionate presence of a given allele in affected individuals. To analyze associations between genotypes and atopic or non-atopic asthma, a dominant model was assumed because there were only small proportions of homozygotes for the risk alleles. Unconditional logistic regression analysis adjusted for age and sex was used to calculate odds ratios (ORs), their 95% confidence intervals (CIs), and P values. For the three-loci haplotype frequency analyses between the case and control groups, we used an Expectation-Maximization algorithm to model the combined IL-13 polymorphisms of each patient. Five haplotypes (ACG, CTA, ACA, CCG and CTG) were predicted, and accounted for 99% of all those seen; the other options, having frequencies smaller than 1%, were excluded because of possible genotyping errors. Chi-square tests were used to determine the difference of clinical phenotypes between the IL-13 risk haplotype (C-T-A) and the other tested haplotypes. Chi-square tests were used to determine the association among total IgE levels and the gene–gene interaction between IL-13 and IL-13Rα1 polymorphisms. Bonferroni correction was also applied to make the association more appropriate with the gene–gene interaction. All statistical analyses were performed using SAS program (Version 8e; SAS Institute, Cary, N.C.).
Results
Genotype distributions of the IL-13 and IL-13Rα1 polymorphisms
Medline searches allowed us to identify asthma development- or asthma-related phenotype-associated polymorphisms in the promoter and coding regions of the IL-13 gene on chromosome 5q31 (Howard et al. 2001; van der Pouw Kraan et al. 1999; Graves et al. 2000; Heinzmann et al. 2000; Vercelli et al. 2005). The −1512A, −1112C and +2044G IL-13 polymorphisms in control groups had allele frequencies of 0.70, 0.81 and 0.67, respectively, and were concordant with Hardy–Weinberg equilibrium (Table 2).
Only the A+1398G polymorphism in the noncoding region of IL-13Rα1 gene was relatively common (Heinzmann et al. 2000; Guo et al. 1997), with the +1398A allele having a frequency of 0.51 in control group (Table 2). Since male subjects are hemizygous at the IL-13Rα1 gene, we adjusted the odds ratio (OR) by sex. However, asthma did not differ significantly in males (data not shown).
To investigate the associations of the IL-13 and IL-13Rα1 polymorphisms with asthma susceptibility, we determined the occurrence of these genotypes in asthmatic and control groups, using subjects homozygous for the two common alleles as a reference group. We found an association between the susceptibility of asthma and homozygosity for the risk alleles of IL-13 G+2044A (OR=1.95, 95% CI=1.08–3.53; Table 3), but not with the susceptibility of atopic (OR=0.99, 95% CI=0.52–1.87) or non-atopic asthma (data not shown). Other IL-13 promoter polymorphisms and IL-13Rα1 polymorphism were not associated with the susceptibility of asthma or atopic asthma (Table 2).
Total IgE levels in asthmatics with IL-13 and IL-13Rα1 polymorphisms
We found that total IgE was significantly higher in children with atopic asthma with hetero- or homozygous for the risk alleles in the promoter region of IL-13 (A−1512C and C−1112T) and IL-13Rα1 (A+1398G) versus those with homozygous for common alleles (P=0.012, 0.015, and 0.017, Table 3). A similar result was observed in the asthmatic group (P=0.049, 0.024, and 0.034; Table 3), but not in the control group.
Three-loci haplotype analysis of IL-13 polymorphisms
There was no significant association between the haplotype with risk alleles (C-T-A) and asthma susceptibility in Korean children with asthma or atopic asthma (data not shown). However, children with atopic asthma with the C-T-A haplotype showed significantly higher levels of total IgE versus those with the other haplotypes (P=0.006; Table 4). Similarly, total IgE levels were significantly higher in children with asthma with the C-T-A haplotype versus those with the other haplotypes (P=0.019; Table 4). Conversely, this association was not observed in the control group.
Association between total IgE and the combination of IL-13 and IL-13Rα1 polymorphisms
Because each of the tested IL-13 promoter polymorphisms and IL-13Rα1 polymorphism were associated with total IgE levels, we investigated whether there might be an association between total IgE levels and a combination of the IL-13Rα1 risk allele along with one of the IL-13 risk alleles. We observed that children with atopic asthma, who had a combination of hetero- or homozygous for −1512C or −1112T in IL-13 and +1398G in IL-13Rα1, showed higher total serum IgE levels than those with atopic asthma, who had a combination of homozygous for −1512A or −1112C in IL-13 and +1398A in IL-13Rα1 (P=0.002, 0.010 after Bonferroni correction, respectively; Table 5). However, the combination of hetero- or homozygous for +2044A in IL-13 and +1398G in IL-13Rα1 tended to have higher total serum IgE levels versus those homozygous for +2044G in IL-13 and +1398G in IL-13Rα1, but not to a significant degree (P=0.099 after Bonferroni correction; Table 5).
Similarly, we investigated associations between total IgE levels and the combination of IL-13 and IL-13Rα1 risk alleles in children with asthma. Children with asthma with the combination of hetero- or homozygous for either IL-13 −1512A or −1112C and hetero- or homozygous for IL-13Rα1 +1398G showed significantly higher total IgE levels compared to those homozygous for the common alleles (P=0.012 and P=0.011 after Bonferroni correction, respectively; data not shown). Children with asthma with the combination of hetero- or homozygous for IL-13 +2044G and IL-13Rα1 +1398G tended to have higher IgE levels (Log IgE, 2.38 ± 0.58 mg/ml) versus those with the combination of homozygous for the common alleles (Log IgE, 2.29 ± 0.60 mg/ml), but not to a statistically significant degree (P=0.249 after Bonferroni correction; data not shown).
Discussion
In this study, we found that two IL-13 promoter polymorphisms (A−1512C and C−1112T) and one IL-13Rα1 polymorphism (A+1398G) were significantly associated with increased total IgE levels in Korean children with asthma and atopic asthma. We further found that children with atopic asthma with a combination of hetero- or homozygous for each of the IL-13 polymorphism and the IL-13Rα1 polymorphism showed significantly higher total IgE levels compared to those with homozygous for the common alleles at both loci. To our knowledge, this is the first report showing that IgE phenotypes are associated with a gene–gene interaction between polymorphic variants of IL-13 and IL-13Rα1. We did not observe any association between combinations of IL-13 and IL-13Rα1 polymorphisms and other clinical phenotypes, such as eosinophil and BHR phenotypes (data not shown).
Although previous reports have shown that genetic variants in the IL-13 promoter and coding regions were involved in the pathogenesis of asthma and atopy (Howard et al. 2001; van der Pouw Kraan et al. 1999; Graves et al. 2000; Heinzmann et al. 2000; He et al. 2003; Leung et al. 2001; Arima et al. 2002), we found that G+2044A of IL-13 was associated with asthma susceptibility and that A−1512C and C−1112T of IL-13 were also significantly associated with total IgE levels in Korean children with asthma. When we investigated the effect of the risk allele haplotypes of three IL-13 gene polymorphisms (−1512C, −1112T and +2044A) in Korean children with asthma, especially with atopic asthma, we found that the C-T-A haplotype was associated with significantly higher total IgE levels compared to those found harboring all other haplotypes. The combination of the IL-13 G+2044A and IL-13Rα1 A+1398G risk alleles was associated with increased IgE levels, but not to a statistically significant degree, while the combination of either IL-13 promoter polymorphism (A−1512C and C−1112T) risk allele with +1398G of IL-13Rα1 was associated with significantly increased total IgE levels in Korean children with atopic asthma. This seems to indicate that variations in the promoter region of IL-13 affect their amount of signaling, and that there could be a gene–gene effect with the tested polymorphism in IL-13Rα1.
IL-13 mediates its effect through its receptor, a heterodimer composed of IL-4Rα and IL-13Rα1. IL-13 signaling uses the JAK-signal transducer and activator of transcription (STAT) pathway, specifically STAT6 (Takeda et al. 1996). This is thought to occur through IL-4Rα, because stimulation of the complex by both IL-4 and IL-13 results in activation of signaling intermediates characteristic of IL-4 responses, including phosphorylation of IL-4Rα, insulin receptor substrate 2 (IRS-2), JAK1, and Tyk2 (Wills-Karp 2001; Welham et al. 1995; Hershey 2003). The functional role of IL-13Rα1 and its variants remain unknown, although a previous study showed that a variant of IL-13Rα1 (+1398A) was associated with IgE levels, but not with asthma in British male subjects (Heinzmann et al. 2000). Here, we found that hetero- or homozygosity for the risk allele of IL-13Rα1 A+1398G was significantly associated with increased total IgE levels in children with asthma, especially atopic asthma. This may suggest that this non-coding polymorphism of IL-13Rα1 has a functional consequence for the binding of IL-13, or alternatively that the IL-13Rα1 A+1398G polymorphism is in linkage disequilibrium with as yet unidentified polymorphisms in the regulatory or coding regions of the gene encoding IL-13Rα1. Yet another possibility is that IL-13Rα1 could have additional but as yet unknown signaling functions that are impacted by this polymorphism.
Structurally, the extracellular portions of IL-13Rα1 are composed of three fibronectin type III domains, designated D1, D2, and D3 (Arima et al. 2005). Mutagenesis analyses of IL-13 and IL-13Rα have shown that mutation of Leu319 and Tyr321 in the D2 and D3 domains of IL-13Rα1, respectively, led to significant impairment of the IL-13 response (Arima et al. 2005). A truncated murine IL-13Rα1 lacking the intracellular domain was not able to mediate IL-13-induced signals or responses, supporting the possibility that IL-13Rα1 is required for signaling (Orchansky et al. 1997). Analysis of molecular interactions showed that helices A and C of human IL-13 interacted with IL-4Rα, while helix D of human IL-13 was responsible for the interaction with IL-13Rα1 (Zuegg et al. 2001). Furthermore, mutation of arginine 110 in the helix D region to a glutamine (G+2044A) resulted in enhanced IL-13 signaling (Heinzmann et al. 2000). Collectively, these studies indicate that IL-13Rα1 is likely to play a critical role not only in binding but also in signaling of IL-13.
Future work will be required to identify gene–gene and gene–environment interactions on a genome-wide level, with the aim of fully elucidating the genetic risk factors for asthma and atopy, understanding the pathogenesis of these common diseases, and perhaps devising new treatment strategies aimed at modulating IL-13 signaling via IL-4Rα/IL-13Rα1.
Abbreviations
- IL-13:
-
Interleukin-13
- IL-4Rα:
-
Interleukin-4 receptor alpha
- IL-13Rα1:
-
Interleukin-13 receptor alpha 1
- IgE:
-
Immunoglobulin E
- STAT:
-
Signal transducer and activator of transcription
- OR:
-
Odds ratio
- CI:
-
Confidence interval
- Th1:
-
T helper lymphocytes type 1
- Th2:
-
T helper lymphocytes type 2
- LD:
-
Linkage disequilibrium
References
Akaiwa M, Yu B, Umeshita-Suyama R, Terada N, Suto H, Koga T, Arima K, Matsushita S, Saito H, Ogawa H, Furue M, Hamasaki N, Ohshima K, Izuhara K (2001) Localization of human interleukin 13 receptor in non-haematopoietic cells. Cytokine 13:75–84
American Thoracic Society (1987) Standards for the diagnosis and care of patients with chronic obstructive pulmonary disease (COPD) and asthma. Am Rev Respir Dis 136:225–244
Arima K, Umeshita-Suyama R, Sakata Y, Akaiwa M, Mao XQ, Enomoto T, Dake Y, Shimazu S, Yamashita T, Sugawara N, Brodeur S, Geha R, Puri RK, Sayegh MH, Adra CN, Hamasaki N, Hopkin JM, Shirakawa T, Izuhara K (2002) Upregulation of IL-13 concentration in vivo by the IL13 variant associated with bronchial asthma. J Allergy Clin Immunol 109:980–987
Arima K, Sato K, Tanaka G, Kanaji S, Terada T, Honjo E, Kuroki R, Matsuo Y, Izuhara K (2005) Characterization of the interaction between interleukin-13 and interleukin-13 receptors. J Biol Chem 26:24915–24922
Chai H, Farr RS, Froehlich LA, Mathison DA, McLean JA, Rosenthal RR, Sheffer AL, Spector SL, Townley RG (1975) Standardization of bronchial inhalation challenge procedures. J Allergy Clin Immunol 56:323–327
Cohn L, Elias JA, Chupp GL (2004) Asthma: mechanisms of disease persistence and progression. Annu Rev Immunol 22:789–815
Doucet C, Brouty-Boye D, Pottin-Clemenceau C, Canonica GW, Jasmin C, Azzarone B (1998) Interleukin (IL)-4 and IL-13 act on human lung fibroblasts: implication in asthma. J Clin Invest 101:2129–2139
Graber P, Gretener D, Herren S, Aubry JP, Elson G., Poudrier J, Lecoanet-Henchoz S, Alouani S, Losberger C, Bonnefoy JY, Kosco-Vilbois MH, Gauchat JF (1998) The distribution of IL-13 receptor alpha1 expression on B cells, T cells and monocytes and its regulation by IL-13 and IL-4. Eur J Immunol 28:4286–4298
Graves PE, Kabesch M, Halonen M, Holberg CJ, Baldini M, Fritzsch C, Weiland SK, Erickson RP, von Mutius E, Martinez FD (2000) A cluster of seven tightly linked polymorphisms in the IL-13 gene is associated with total serum IgE levels in three populations of white children. J Allergy Clin Immunol 105:506–513
Guo J, Apiou F, Mellerin MP, Lebeau B, Jacques Y, Minvielle S (1997) Chromosome mapping and expression of the human interleukin-13 receptor. Genomics 42:141–145
He JQ, Chan-Yeung M, Becker AB, Dimich-Ward H, Ferguson AC, Manfreda J, Watson WT, Sandford AJ (2003) Genetic variants of the IL13 and IL4 genes and atopic diseases in at-risk children. Genes Immun 4:385–389
Heinzmann A, Mao XQ, Akaiwa M, Kreomer RT, Gao P-S, Ohshima K, Umeshita R, Abe Y, Braun S, Yamashita T, Roberts MH, Sugimoto R, Arima K, Arinobu Y, Yu B, Kruse S, Enomoto T, Dake Y, Kawai M, Shimazu S, Sasaki S, Adra CN, Kitaichi M, Inoue H, Yamauchi K, Tomichi N, Kurimoto F, Harnasaki N, Hopkin JM, Izuhara K, Shirakawa T, Deichmann KA (2000) Genetic variants of IL-13 signaling and human asthma and atopy. Hum Mol Genet 9:549–559
Hershey GK (2003) IL-13 receptors and signaling pathways: an evolving web. J Allergy Clin Immunol 111:677–690
Hilton DJ, Zhang JG, Metcalf D, Alexander WS, Nicola NA, Willson TA (1996) Cloning and characterization of a binding subunit of the interleukin 13 receptor that is also a component of the interleukin 4 receptor. Proc Natl Acad Sci USA 93:497–501
Howard TD, Whittaker PA, Zaiman AL, Koppelman GH, Xu J, Hanley MT, Meyers DA, Postma DS, Bleecker ER (2001) Identification and association of polymorphisms in the interleukin-13 gene with asthma and atopy in a Dutch population. Am J Respir Cell Mol Biol 25:377–384
Howard TD, Koppelman GH, Xu J, Zheng SL, Postma DS, Meyers DA, Bleecker ER (2002) Gene–gene interaction in asthma: IL4RA and IL13 in a Dutch population with asthma. Am J Hum Genet 70:230–236
Kay AB (2001) Allergy and allergic diseases. N Engl J Med 344:30–37
Lee SG, Kim BS, Kim JH, Lee SY, Choi SO, Shim JY, Hong TJ, Hong SJ (2004) Gene–gene interaction between interleukin-4 and interleukin-4 receptor α in Korean children with asthma. Clin Exp Allergy 34:1202–1208
Leung TF, Tang NLS, Chan HIS, Li AM, Ha G, Lam CWK (2001) A polymorphism in the coding region of interleukin-13 gene is associated with atopy but not asthma in Chinese children. Clin Exp Allergy 31:1515–1521
Miloux B, Laurent P, Bonnin O, Lupker J, Caput D, Vita N, Ferrara P (1997) Cloning of the human IL-13R alpha1 chain and reconstitution with the IL4R alpha of a functional IL-4/IL-13 receptor complex. FEBS Lett 401:163–166
Nakamura Y, Azuma M, Okano Y, Sano T, Takahashi T, Ohmoto Y, Sone S (1996) Upregulatory effects of interleukin-4 and interleukin-13 but not interleukin-10 on granulocyte/macrophage colony-stimulating factor production by human bronchial epithelial cells. Am J Respir Cell Mol Biol 15:680–687
Orchansky PL, Ayres SD, Hilton DJ, Schrader JW (1997) An interleukin (IL)-13 receptor lacking the cytoplasmic domain fails to transducer IL-13-induced signals and inhibits responses to IL-4. J Biol Chem 272:22940–22947
Rankin JA, Picarella DE, Geba GP, Temann UA, Prasad B, DiCosmo B, Tarallo A, Stripp B, Whitsett J, Flavell RA (1996) Phenotypic and physiologic characterization of transgenic mice expressing interleukin 4 in the lung: lymphocytic and eosinophilic inflammation without airway hyperreactivity. Proc Natl Acad Sci USA 93:7821–7825
Takeda K, Kamanaka M, Tanaka T, Kishimoto T, Akira S (1996) Impaired IL-13-mediated functions of macrophages in STAT6-deficient mice. J Immunol 157:3220–3222
van der Pouw Kraan TC, van der Zee JS, Boeije LC, De Groot ER, Stapel SO, Aarden LA (1998) The role of IL-13 in IgE synthesis by allergic asthma patients. Clin Exp Immunol 111:129–135
van der Pouw Kraan TC, van Veen A, Boeije LC, van Tuyl SA, de Groot ER, Stapel SO, Bakker A, Verweij CL, Aarden LA, van der Zee JS (1999) An IL-13 polymorphism associated with increased risk of allergic asthma. Genes Immunity 1:61–65
Vercelli D (2005) Genetic regulation of IgE responses: achilles and the tortoise. J Allergy Clin Immunol 116:60–64
Welham MJ, Learnmonth L, Bone H, Schrader JW (1995) Interleukin-13 signal transduction in lymphohemopoietic cells. Similarities and differences in signal transduction with interleukin-4 and insulin. J Biol Chem 270:12286–12296
Wills-Karp M (2001) IL-12/IL-13 axis in allergic asthma. J Allergy Clin Immunol 107:9–18
Wills-Karp M, Chiaramonte M (2003) Interleukin-13 in asthma. Curr Opin Pulm Med 9:21–27
Zhu Z, Homer RJ, Wang Z, Chen Q, Geba GP, Wang J, Zhang Y, Elias JA (1999) Pulmonary expression of interleukin-13 causes inflammation, mucus hypersecretion, subepithelial fibrosis, physiologic abnormalities, and eotaxin production. J Clin Invest 103:779–788
Zuegg J, Webb DC, Foster PS, Casarotto MG (2001) Structural model of human IL-13 defines the spatial interactions with the IL-13Rα/IL-4Rα receptor. Immunol Cell Biol 79:332–339
Acknowledgments
We thank all the study participants. We are also grateful to nurse Hee-Jung Seo (Asthma and Allergy Clinic, Asan Medical Center, Seoul, Korea) for her work in patient recruitment.
Author information
Authors and Affiliations
Corresponding author
Additional information
Hyo-Bin Kim, and Yong-Chul Lee contributed equally to this work. This work was supported by grants from the Asan Institute for Life Science (2005-091) and the National Research Laboratory Program, Korea Science and Engineering Foundation and by Korea Research Foundation Grants funded by the Korean Government (MOEHRD, Basic Research Promotion Fund) (KRF-2005-201-E00014), Republic of Korea.
Rights and permissions
About this article
Cite this article
Kim, HB., Lee, YC., Lee, SY. et al. Gene–gene interaction between IL-13 and IL-13Rα1 is associated with total IgE in Korean children with atopic asthma. J Hum Genet 51, 1055–1062 (2006). https://doi.org/10.1007/s10038-006-0061-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10038-006-0061-x
Keywords
This article is cited by
-
Gene-gene interaction between tuberculosis candidate genes in a South African population
Mammalian Genome (2011)
-
Lack of association between interleukin-13 gene polymorphisms (−1055 C/T and +2044 G/A) in Iranian patients with lung cancer
Molecular Biology Reports (2009)