Abstract
Cockayne syndrome is a rare autosomal recessive neurodegenerative disorder. It is considered to be a heterogeneous condition based on complementation in cell fusion studies, with two major forms, namely CS-A and CS-B. CKN1 is the gene responsible for CS-A, whose mutations disrupt the transcription-coupled repair system of the actively transcribed DNA. Mutation analysis of the CKN1 gene in eight typical CS-A Brazilian patients from six families showed a gene alteration in all of them. We found a total of five novel mutations that were absent from healthy control subjects. Six affected subjects were simple homozygotes and two affected siblings were each compound heterozygotes. While the findings extend the range of mutations in CS-A, there is no obvious genotype-phenotype correlation across the mutational spectrum.
Similar content being viewed by others
Introduction
Cockayne syndrome (CS) is a rare autosomal recessive disorder characterized by cachectic dwarfism, premature aging, mental deficiency, microcephaly, intracranial calcifications, neurological degeneration, retinal abnormalities and sensorineural hearing loss. It is considered to be a heterogeneous disorder based on complementation in cell fusion studies: sub-types have been called CS-A and CS-B (Lehmann et al. 1994). The gene that has been shown to be responsible for CS-A—CKN1, also called ERCC8—encodes a Walker domain (WD)-repeat protein (Henning et al. 1995). Mutations in the CKN1 gene disrupt the transcription-coupled repair system of the actively transcribed DNA, which is the presumed pathogenic mechanism in CS-A. We analyzed the CKN1 gene in eight typical CS-A patients from six Brazilian families.
Materials and methods
Subjects
Eight patients (four males and four females) from six families were evaluated. Two unrelated affected individuals and a pair of affected siblings were each the product of consanguineous mating (families 1, 2 and 3). Informed consent to allow genetic testing was obtained from all participants. The ages of the subjects ranged from 3 years and 8 months to 12 years (mean age of 8 years and 4 months). All patients shared several typical clinical findings (Table 1), including developmental delay, microcephaly, failure to thrive, short stature, enophthalmia, “salt-and-pepper” chorioretinitis, prominent nose, erythematous lesions in the malar and nasal regions, and flexion contractures, especially involving the knees. Neuroimaging studies in affected individuals showed marked cortical atrophy in the majority, ventriculomegaly, atrophy of the cerebellum, and basal ganglia calcifications.
DNA analysis
In each affected subject, we directly sequenced the 12 exons of the CKN1 gene plus a minimum of 50 nucleotide base pairs (bp) at each intron-exon boundary, which were amplified from genomic DNA using previously reported primers and conditions (Cao et al. 2004). Once a CKN1 mutation was identified in an affected subject, a rapid genotyping method was developed in order to screen healthy control subjects to confirm absence of the mutation from the genomes of 200 healthy control subjects. All genotype reactions were run in parallel with sequence-proven reference standards.
For the IVS4 + 1G > T mutation, we used an allele-specific genotyping method. We amplified a 257 bp fragment containing exon 4 using primers 5′-TCA TCC TAC CTG AGA CTA CTA CTT GTT and 5′- TCC TAC AAA GCA ATC TGC AA. This was followed by treatment with shrimp alkaline phosphatase (SAP) and ExoI to remove primers and unincorporated dNTPs and then followed by ddNTP extension (SnaPShot, PE Applied Biosystems, Mississauga, ON) with primer 5′ GGG ATA CAA ATA CAT TAC AA and analysis on a 3730 DNA Sequencer (PE Applied Biosystems, Mississauga, ON).
For the T104fsdelTG > 109X mutation, we amplified a 257 bp fragment containing exon 4 using primers 5′-TCA TCC TAC CTG AGA CTA CTA CTT GTT and 5′- TCC TAC AAA GCA ATC TGC AA. We then digested the amplified fragment with restriction endonuclease BsrGI, and electrophoresed the resulting fragments in 2.5% agarose gels. Digestion of the wild type allele produced two fragments with sizes 132 and 125 bp, while digestion of the mutant allele produced only a single fragment 257 bp in size.
For the T204K mutation, we used an allele-specific genotyping method. We amplified a 294 bp fragment containing exon 7 using primers 5′- TTT GGC CTC ACT TTC TTC AG and 5′- CAA TAC TTC ATT TTA AAG GAG TGA A. This was followed by treatment with SAP and ExoI to remove primers and unincorporated dNTPs and then followed by ddNTP extension (SnaPShot) with primer 5′ GAC TAT ATC TTG GCA A and analysis on a 3730 DNA Sequencer.
For the IVS7 –1G > A mutation, we amplified a 281 bp fragment containing exon 8 using primers 5′- CGA GAC CTC TGT GTG CCA TA and 5′-TGC GTT TAT TAT GTG GCT TCA. We then digested the amplified fragment with restriction enzyme DdeI, and electrophoresed the resulting fragments in 2.5% agarose gels. Digestion of the wild type allele produced two fragments with sizes 190 and 91 bp, while digestion of the mutant allele produced only a single fragment 281 bp in size.
For the D266G mutation, we used an allele-specific genotyping method. We amplified a 256 bp fragment containing exon 9 using primers 5′- CAT CAA ACA TTA AAT ATG TCC TGA A and 5′- AAA TCA AGT GTA TGT CAC AGA TCC A. This was followed by treatment with SAP and ExoI to remove primers and unincorporated dNTPs and then followed by ddNTP extension (SnaPShot) with primer 5′ CTC ACT GTT GGT ACA G and analysis on a 3730 DNA Sequencer.
Results
DNA analysis
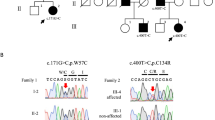
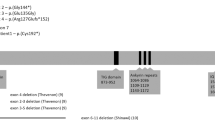
Sequence analysis revealed a total of five novel mutations in our CS-A patients (Table 1; Fig. 1). The TG dinucleotide insertion within CKN1 exon 4 codon 104, which shifted the reading frame and predicted early termination of the protein at 109 amino acids (designated T104fs delTG), was repeatedly found in our cohort: two unrelated cases were homozygous (from families 1 and 2) and both affected siblings from family 4 were compound heterozygotes for this mutation (Fig. 1a). The other mutation in the compound heterozygotes from family 4 was the CKN1 T204K missense mutation. Two affected siblings from family 3 were simple homozygotes for the CKN1 D266G missense mutations. The CKN1 mutation within the splice acceptor site at the exon 3-intron 4 boundary designated IVS4 + 1G > A, which is predicted to eliminate the splice acceptor recognition sequence, was found in a homozygote from family 5 (Fig. 1b). The CKN1 IVS7-1G > A mutation was found in one homozygous patient from family 6. We found that all novel CKN1 mutations were absent from the genomes of 200 normal healthy subjects.
Electropherograms of genomic DNA sequence of the CKN1 mutations. Panels a, b, c, d and e show the T104fsdelTG > 109X (homozygote), IVS4 + 1G > A (homozygote), T204K (heterozygote), IVS7 –G > A (homozygote) and D266G (homozygote) mutations, respectively. The position of the mutant nucleotide sequence is shown with an arrow, and in panel A the deleted nucleotides are indicated. Each panel shows nucleotide sequence (coding in upper case, non-coding in lower case), and single letter codes for amino acid residues. Sequences of the analogous regions from healthy subjects are shown above each mutant sequence for comparison
Discussion
To date, fewer than ten reported CS-A patients have been screened for mutations in the CKN1 gene. To the previously reported nonsense mutations E13X and T322X (Henning et al. 1995; Cao et al. 2004; Ridley et al. 2005), we now add T104fs delTG. The recurrence of the mutation T104fs delTG in 2 non-related patients and in a pair of siblings suggests that a founder effect may exist in Brazilian CS-A patients.
To the previously reported splicing mutation in CKN1 (Komatsu et al. 2004) we now add IVS4 + 1G > A and IVS7-1G > A. Previous findings of splicing mutations in CS-A patients, taken together with our finding of homozygosity for the altered splice donor and acceptor sequences in our cohort, indicates that abnormal splicing is an important subset of the molecular etiologies of this disorder.
To the previously reported missense mutations Q106P, A160V and A205P (Ren et al. 2003; Cao et al. 2004; Ridley et al. 2005), we now add T204K and D266G. Most of the missense mutations in CKN1, including the two new mutations reported here, occur within functional domains. For instance, Q106P occurs within the second Walker domain (WD), T204K and A205P both occur within the third WD, and D266G occurs within the fourth WD. Furthermore, the T204 residue is highly conserved in mammalian species, down to mice, while the D266 residue is conserved in both mammalian and avian species, attesting to the likely functional importance of these two residues. Interestingly, exon 4 has been repeatedly involved as the site for CS-A patient mutations, not just in our cohort but also in other reports (Ren et al. 2003; Komatsu et al. 2004).
There was no obvious difference in phenotype between our subjects with the splicing and frameshift mutations. However, the small total number of molecularly characterized affected CS-A subjects at present precludes an analysis of genotype-phenotype correlation, although our findings expand the spectrum of mutations and mutation types for this disorder.
References
Cao H, Williams C, Carter M, Hegele RA (2004) CKN1 (MIM 216400): mutations in Cockayne syndrome type A and a new common polymorphism. J Hum Genet 49:61–63
Henning KA, Li L, Iyer N, McDaniel LD, Reagan MS, Legerski R et al (1995) The Cockayne syndrome group A gene encodes a WD repeat protein that interacts with CSB protein and a subunit of RNA polymerase II TFIIH. Cell 82:555–564
Komatsu A, Suzuki S, Inagaki T, Yamashita K, Hashizume K (2004) A kindred with Cockayne syndrome caused by multiple splicing variants of the CSA gene. Am J Med Genet 128A:67–71
Lehmann AR, Bootsma D, Clarkson SG, Cleaver JE, McAlpine PJ, Tanaka K et al (1994) Nomenclature of human DNA repair genes. Mutat Res 315:41–42
Ren Y, Saijo M, Nakatsu Y, Nakai H, Yamaizumi M, Tanaka K (2003) Three novel mutations responsible for Cockayne syndrome group A. Genes Genet Syst 78:93–102
Ridley AJ, Colley J, Wynford-Thomas D, Jones (2005) Characterisation of novel mutations in Cockayne syndrome type A and xeroderma pigmentosum group C subjects. J Hum Genet 50:151–154
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Bertola, D.R., Cao, H., Albano, L.M.J. et al. Cockayne syndrome type A: novel mutations in eight typical patients. J Hum Genet 51, 701–705 (2006). https://doi.org/10.1007/s10038-006-0011-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10038-006-0011-7
Keywords
This article is cited by
-
Molecular spectrum of excision repair cross-complementation group 8 gene defects in Chinese patients with Cockayne syndrome type A
Scientific Reports (2017)
-
A possible cranio-oro-facial phenotype in Cockayne syndrome
Orphanet Journal of Rare Diseases (2013)