Abstract
Lafora’s disease (LD) is an autosomal recessive and fatal form of progressive myoclonus epilepsy with onset in late childhood or adolescence. LD is characterised by the presence of intracellular polyglucosan inclusions, called Lafora bodies, in tissues including the brain, liver and skin. Patients have progressive neurologic deterioration, leading to death within 10 years of onset. No preventive or curative treatment is available for LD. At least three genes underlie LD, of which two have been isolated and mutations characterised: EPM2A and NHLRC1. The EPM2A gene product laforin is a protein phosphatase while the NHLRC1 gene product malin is an E3 ubiquitin ligase that ubiquitinates and promotes the degradation of laforin. Analyses of the structure and function of these gene products suggest defects in post-translational modification of proteins as the common mechanism that leads to the formation of Lafora inclusion bodies, neurodegeneration and the epileptic phenotype of LD. In this review, we summarise the available information on the genetic basis of LD, and correlate these advances with the rapidly expanding information about the mechanisms of LD gained from studies on both cell biological and animal models. Finally, we also discuss a possible mechanism to explain the locus heterogeneity observed in LD.
Similar content being viewed by others
Introduction
Lafora’s progressive myoclonus epilepsy or Lafora’s disease (LD; OMIM 254780) is an autosomal recessive, fatal disorder with pathognomonic periodic acid–Schiff-positive (PAS+) staining intracellular inclusion bodies (Van Heycop Ten Ham 1975). Besides the central nervous system, the large basophilic PAS+ inclusions, called Lafora polyglucosan bodies, are also found in the retina, heart, liver, muscle and skin (Van Heycop Ten Ham 1975; Delgado-Escueta et al. 2001). LD is classically described as symptoms starting in early adolescence as stimulus-sensitive grand mal tonic–clonic, absence, visual and myoclonic seizures. Rapidly progressive dementia, psychosis, cerebellar ataxia, dysarthria, amaurosis, mutism, muscle wasting and respiratory failure lead to death within 10 years of onset. LD, although relatively rare in the outbred populations of the United States, Canada, China and Japan, is commonly encountered in the Mediterranean basin of Spain, France and Italy, in restricted regions of central Asia, India, Pakistan, northern Africa and the Middle East, in ethnic isolates from the southern United States and Quebec, Canada, and in other parts of world where a high rate of consanguinity is practised (Delgado-Escueta et al. 2001). No preventive or curative treatment is available for LD at present.
Locus heterogeneity in LD
A gene for LD was localized to a 3 cM region on 6q24 (Serratosa et al. 1995; Sainz et al. 1997). Minassian et al. (1998), Serratosa et al. (1999) and Ganesh et al. (2000) have independently cloned the EPM2A gene and showed that LD patients were homozygous or compound heterozygotes for presumably loss-of-functions mutations. The EPM2A gene encodes a protein phosphatase named laforin and is composed of four exons (Ganesh et al. 2000). Laforin contains an N-terminal carbohydrate-binding domain (CBD) encoded mainly by exon 1, and a dual-specificity phosphatase domain (DSPD) spanning exons 3 and 4 (Fig. 1) (Minassian et al. 2000a; Ganesh et al. 2002a). Ganesh et al. (2002a) proposed two subsyndromes for LD: (1) a classical LD with adolescent onset stimulus sensitive grand mal, absence and myoclonic seizures followed by dementia and neurologic deterioration associated mainly with exon 4 mutations, and (2) an atypical LD with childhood-onset dyslexia and learning disorder followed by epilepsy and neurologic deterioration associated mainly with mutations in exon 1. Ganesh et al. (2002a) also provided clues for the distinct role of the EPM2A gene in the two subsyndromes of LD, demonstrating that mutations affecting the functions of DSPD but not the CBD, led to the classical form of LD, and mutations affecting both domains might develop into the atypical LD form. Such correlations have been confirmed by independent studies (Annesi et al. 2004).
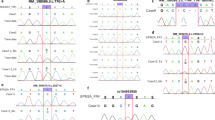
Representation of 38 known mutations in the EPM2A gene. This schematic diagram shows the domain organisation of laforin protein, the positions of various mutations found in Lafora’s disease (LD) families and their frequency (number of independent families with a given mutation). The figure also shows (bottom) the genomic organisation of the EPM2A gene and locations of the large deletions associated with LD. Mutations were tabulated from the following PubMed-indexed English articles reporting the mutations: Minassian et al. (1998, 2000a, b); Serratosa et al. (1999); Gomez-Garre et al. (2000); Ganesh et al. (2002a); Ki et al. (2003); Ianzano et al. (2004); Annesi et al. (2004); Singh et al. (2005). Amino acid positions were assigned based on the GenBank reference sequence for the EPM2A gene, NM_005670. CBD Carbohydrate-binding domain, DSPD dual-specificity phosphatase domain
Despite the clinical homogeneity in LD phenotype, with the presence of Lafora bodies in all affected individuals, multiple LD families were identified in which the phenotype does not segregate with the 6q24 region and in which no mutations in the EPM2A gene were found (Minassian et al. 1999; Serratosa et al. 1999; Ganesh et al. 2002). The simplest explanation for this genetic heterogeneity is that another gene(s) in the same metabolic pathway is/are altered in LD families not linked to 6q24. Chan et al. (2003a) mapped a second LD locus at 6p22, leading to the identification of a second LD gene named NHLRC1. NHLRC1 encodes malin, an E3 ubiquitin ligase with a RING finger domain and six NHL motifs (Fig. 2) (Chan et al. 2003b; Gentry et al. 2005). Several disease-causing mutations in NHLRC1 have been identified in LD patients from diverse ethnic backgrounds (Chan et al. 2003b; Gomez-Abad et al. 2005; Singh et al. 2005). While patients with mutations in EPM2A and NHLRC1 express similar clinical manifestations, patients with EPM2A-associated LD seem to have a more severe clinical course because NHLRC1-defective patients tend to live longer than those with EPM2A defects (Gomez-Abad et al. 2005). At least one other, still unknown, gene also causes LD; Chan et al. (2004a) used sequencing, haplotype and linkage analyses to demonstrate the presence of a third locus for LD. This has also been supported by an independent study (Singh et al. 2005).
Representation of 34 known mutations in the NHLRC1 gene. This schematic diagram shows the domain organisation of the malin protein, the positions of various mutations found in LD families and their frequency (number of independent families with a given mutation). Mutations were tabulated from the following PubMed-indexed articles in English reporting the mutations: Chan et al. (2003b); Gomez-Abad et al. (2005); Singh et al. (2005). Amino acid positions were assigned based on the GenBank reference sequence for the NHLRC1 gene, NM_198586. RING RING finger domain, NHL NHL repeats
Mutations and polymorphisms
EPM2A
To date, 38 different mutations and several polymorphisms have been described in the EPM2A gene encoding laforin phosphatase (Fig. 1). These include 16 missense mutations, 6 nonsense mutations, 6 insertion mutations of a few bases, 7 deletion mutations of a few bases and 3 large deletion mutations involving several kilobases. Except for the larger deletions, all these mutations are distributed evenly across the four known exons of the EPM2A gene (Fig. 1). Intriguingly, no splice-site mutation has been identified for LD. It is remarkable, however, that the occurrence of missense mutations in EPM2A (42%) is somewhat less when compared with observations for other genes (60–70%) and all the known missense mutations target either the CBD (seven mutations) or the DSPD (nine mutations) of laforin (Fig. 1). Given the high allelic heterogeneity observed in LD, it is likely that the majority of the mutations arise as a single event, and that only a very small proportion of mutant alleles can be predicted in certain populations. An exception to this suggestion is the common EPM2A mutation R241X in the Spanish population (Fig. 1). It has been shown that the high prevalence of the R241X mutation is due to both a founder effect and recurrent events (Gomez-Garre et al. 2000; Ganesh et al. 2002a), perhaps explaining why LD is more common in the Mediterranean basin. Recurrent deletion mutations have also been reported for the EPM2A gene, accounting for up to 8% of total mutations (Fig. 1). Three deletion mutations with different deletion breakpoints have been identified in LD patients from Arabic, Jewish and Spanish populations (Minassian et al. 1998; Serratosa et al. 1999; Minassian et al. 2000a, b; Gomez-Garre et al. 2000; Ganesh et al. 2002a). These deletions, which range in size from 30 to 80 kb, removed the first, second, or both first and second exons, of the EPM2A gene, thus creating a null allele. Interestingly, the presence of mammalian-wide interspersed repeats (MIR) in the flanking sequence of the deletion breakpoints has been identified (Minassian et al. 2000a). Thus, an unequal recombination process mediated by MIR elements is likely to be the cause for the generation of large deletion mutations in LD. Among the polymorphisms, A46P was found to be specific to the Japanese and Chinese populations (Ganesh et al. 2001a).
NHLRC1 (EPM2B)
To date, 34 mutations and several polymorphisms have been reported in the NHLRC1 gene (Fig. 2). These include 17 missense mutations, 5 nonsense mutations, 8 deletion mutations of a few bases, and 3 insertion mutations. Of the 17 missense mutations known in NHLRC1, 5 target the RING finger domain and 8 target the NHL repeat domains of the malin protein (Fig. 2). Interestingly, five missense mutations are present in the ‘linker regions’ connecting the functional domains, suggesting tight restraints on amino acid composition well beyond the predicted domains. Deletions involving a few bases of the coding regions (eight distinct mutations) constitute the second largest group of mutations in NHLRC1. This observation is intriguing, because NHLRC1 is a single exon gene, and no large deletion has so far been reported for NHLRC1. The G158 fs mutation, involving the removal of two bases in the coding region (468–469delAG), is by far the most common deletion mutation, and is the second most frequent mutation observed in NHLRC1. Missense mutation P69A, affecting the RING finger domain, is the most frequent mutation observed in NHLRC1. Both these mutations have been identified in several ethnic groups, suggesting a recurrent mutational event, and these two sites represent hot-spots for NHLRC1 mutations. Missense mutation C26S is prevalent in the French–Canadian ethnic isolates and is yet to be detected in other populations (Chan et al. 2003b). Interestingly, the chromosome 6p22 haplotype of these French-Canadian pedigrees did suggest a founder effect for the C26S mutation (Chan et al. 2003a).
EPM2C
Approximately 10% of LD families screened for the EPM2A and NHLRC1 genes showed no sequence variations, suggesting the presence of a third locus for LD. To rule out the potential existence of mutations in the regulatory regions of these two genes, Chan et al. (2004a) genotyped a consanguineous LD family with multiple affected children and excluded the involvement of the EPM2A and NHLRC1 loci. Thus, there appears to be at least one additional, as yet unknown, genetic locus for LD.
Biological and clinical significance of EPM2A and NHLRC1 mutations
EPM2A
Most of the EPM2A mutations are predicted to be functionally ‘null’ (nonsense and frameshift mutations) and to have lost phosphatase activity. EPM2A transcripts harbouring a premature termination codon may perhaps be unstable as a result of nonsense-mediated RNA decay (Ganesh et al. 2002a). On the other hand, LD-associated missense mutations in EPM2A must affect the structure or function of the laforin phosphatase in a manner equally as devastating, producing a ‘null’ effect. This prediction has proved true; the majority of EPM2A missense mutations found in LD patients result in a lack of phosphatase activity in vitro (Fernandez-Sanchez et al. 2003). Such an effect is not restricted to mutations located in the DSPD; loss of phosphatase activity was also shown for several CBD missense mutants (Wang et al. 2002; Fernandez-Sanchez et al. 2003). A likely explanation for this observation could be that the mutations affect proper folding of the laforin protein. Support for this notion came from transfection experiments where overexpression of missense mutants resulted in ubiquitin-positive cytoplasmic aggregates, suggesting that they were folding mutants destined for degradation (Ganesh et al. 2000, 2002a). The potential binding affinity of the CBD to glycogen, and the effects of missense mutation on this property were also tested. In addition, missense mutations were shown to affect the subcellular localization of laforin (Ganesh et al. 2002a). Importantly, missense mutations disrupt the interaction of laforin with R5 and malin, proteins that interact with laforin in vivo (Fig. 3) (Fernandez-Sanchez et al. 2003; Gentry et al. 2005). Taken together, these results make laforin one of the few disease-associated proteins for which the effects of mutations have been well characterized. These results, in addition to reinforcing the concept that LD-associated missense mutations produce ‘null’ effects, identified critical domains of laforin involved in specific physiological functions and associated clinical subtypes of LD (Ganesh et al. 2002a).
Schematic diagram showing interacting partners of laforin. Laforin interacts with HIRIP5 (Ganesh et al. 2003), EPM2AIP1 (Ianzano et al. 2003), R5 (Fernandez-Sanchez et al. 2003), malin (Gentry et al. 2005), and glycogen synthase kinase 3 (GSK3) (Lohi et al. 2005a). Laforin–laforin interaction has also been documented (Fernandez-Sanchez et al. 2003). R5 also interacts with protein phosphatase 1 (PP1) and glycogen synthase (GS) to regulate glycogen metabolism (Fernandez-Sanchez et al. 2003). An interaction between malin and GS has also been established. Malin interacts at least with four distinct E2 ubiquitin conjugating enzymes (Ubc) involved in the ubiquitination process: UbcH2, UbcH5a, UbcH5c and UbcH6 (Gentry et al. 2005). GS is regulated by GSK3, and the activity of GSK3 is shown to be modulated by laforin (Lohi et al. 2005a). Laforin and malin, therefore, appear to regulate critical steps in glycogen metabolism. The identification of novel interacting partners (HIRIP5 and EPM2AIP1) suggest a role for laforin in vital physiological processes besides glycogen metabolism. CBD Carbohydrate binding domain, DSPD dual-specificity phosphates domain, filled arrows protein–protein interactions
An interesting observation on the cellular function of laforin has come from the identification of an insertion of a ‘T’ at position 950 in exon 4 (c.950insT) of an LD patient (Ianzano et al. 2004). This mutation, which causes a frameshift starting at the 319th amino acid of laforin (Q319 fs), replaces the last 13 amino acids at the carboxyl terminal with a new sequence (Fig. 4). While the mutation did not affect the subcellular localization of the laforin mutant, the phosphatase activity of the protein was greatly reduced (Ianzano et al. 2004). Intriguingly, the effect of this mutation is limited to one of the two laforin isoforms produced by differentially spliced transcripts of the EPM2A gene (Fig. 4) (Ganesh et al. 2002b; Ianzano et al. 2004). The unique carboxyl terminal of isoform 2, generated by the alternative splicing, targets the protein to the nucleus, a distinctive feature that was not observed for laforin isoform 1 (Ganesh et al. 2002b). Laforin isoform-2 would not be affected by the Q319fs mutation because it is intronic in the transcript that encodes this protein. These results suggest that disturbances in the physiological functions of laforin isoform 1 appear to underlie the pathogenesis in LD, and that isoform 2 cannot functionally substitute for laforin isoform 1.
Schematic diagram of the two EPM2A isoforms, their genomic organisation and relationship with the LD mutation, Q319 fs. a Differential splicing in exon 4 results in two EPM2A isoforms. EPM2A isoform 1 encodes the 331 amino acid long laforin 1 (C-terminal sequence shown in b) that targets rough endoplasmic reticulum (Ganesh et al. 2000). EPM2A isoform 2 encodes a laforin variant (isoform 2; shown in b) with a unique C-terminal end, targeting the nucleus (Ganesh et al. 2002b). b Schematic diagram showing the difference in the amino acid sequence at the C-terminal ends of laforin isoforms 1 and 2. The location of the c.950insT mutation causing a frameshift at the 319th amino acid (Q319 fs) of laforin isoform 1 (Ianzano et al. 2004) is also shown. The effect of this mutation is restricted to laforin isoform 1 because it is intronic in the transcript that encodes laforin isoform 2 (Ianzano et al. 2004). CBD Carbohydrate binding domain, DSPD dual-specificity phosphates domain
NHLRC1
Analysis of the genotype–phenotype relationship of NHLRC1 mutations showed that nearly all mutations are predicted to result in the loss of function of the encoded malin protein (Chan et al. 2003b; Gomez-Abad et al. 2005; Singh et al. 2005). Patients with mutations in EPM2A and NHLRC1 express similar clinical manifestation, although the course of the disease is longer in patients with NHLRC1 mutations. Thus, patients with NHLRC1 mutations appear to live longer than those with EPM2A mutations (Gomez-Abad et al. 2005). One of the reasons suggested for this difference is the physiological functions of malin (Gomez-Abad et al. 2005). Malin contains the RING finger ligase domain and six NHL repeats functioning as a substrate-interacting motif (Chan et al. 2003b; Gentry et al. 2005). Malin has been shown to be a single subunit E3 ubiquitin ligase involved in the ubiquitin-mediated proteolysis cascade (Gentry et al. 2005; Lohi et al. 2005a). Additionally, malin interacts with, and ubiquitinates, laforin, leading to its degradation (Gentry et al. 2005). Missense mutations in the RING domains abolish malin’s ubiquitin ligase activity, and mutations located on the NHL motifs disrupt the interaction between malin and laforin (Gentry et al. 2005). Thus, one of the critical functions of malin is to regulate the cellular concentration of laforin protein via ubiquitin-mediated degradation, and missense mutations associated with LD disrupt this function (Gentry et al. 2005).
Disease mechanism
One of the characteristic features of LD is the presence of Lafora polyglucosan bodies. Sakai et al. (1970) and Yokoi et al. (1975) have demonstrated that Lafora bodies contain 80–93% glucose and 6% protein. Histochemical studies have suggested that the inclusions are a mixture of acid mucopolysaccharides and glycoproteins (Schwartz and Yanoff 1965). Lafora bodies stain positive for anti-ubiquitin and anti-advanced glycation end products antibodies, suggesting that these inclusions are not only rich in glucose but also contain higher levels of glycosaminoglycans (Cavanagh 1999; Ganesh et al. 2002c). Thus, Lafora bodies indicate the existence of an unknown biochemical pathway, related to glycogen metabolism, defects of which results in the accumulation of polyglucosan bodies in LD. Laforin is a dual-specificity phosphatase and, if LD is a disorder of glycogen metabolism, the presence of mutations in the EPM2A gene could be exerting its deleterious effects on one or several steps in glycogen synthesis (Ganesh et al. 2002c). Curiously, Lafora bodies are most commonly found in the organs with the highest glucose metabolism, namely brain, heart and liver, and all these tissues abundantly express laforin (Ganesh et al. 2001b, 2002c). Indeed, it has been shown recently that the CBD of laforin targets the protein to glycogen and Lafora inclusion bodies in vitro, and LD mutations in laforin could affect such an affinity (Wang et al. 2002; Ganesh et al. 2004). Similar results were also observed when a substrate-trap mutant of laforin was overexpressed in a mouse model of LD (Chan et al. 2004b). Fernandez-Sanchez et al. (2003) have demonstrated that laforin indeed interacts with R5, a regulatory subunit of protein phosphatase 1 that enhances glycogen accumulation (Fig. 3). R5 also acts as a molecular scaffold assembling PP1 with its substrate, glycogen synthase (GS) at the intracellular glycogen particles, suggesting that laforin is indeed involved in the regulation of glycogen metabolism (Fernandez-Sanchez et al. 2003). Direct evidence for the role of laforin in the glycogen metabolic pathway was provided recently by Lohi et al. (2005a). Using in vitro and in vivo approaches, the authors demonstrated that malin interacts with GS and that laforin dephosphorylates glycogen synthase kinase 3 (GSK3), the principal inhibitor of GS (Lohi et al. 2005a). Consistent with these observations, we propose here the existence of a laforin-mediated glycogen metabolic pathway regulating the generation and/or disposal of pathogenic polyglucosan inclusions (see Fig. 5).
Proposed models to explain locus heterogeneity in LD. In each model, malin and laforin together regulate a common substrate (substrate X). Loss of either laforin or malin would result in ‘hyperactivity’ of the common substrate leading to LD. a In this model, laforin is proposed as a positive regulator for malin. b Here, malin indirectly regulates the laforin substrate. c A multi-protein complex (that includes, but is not limited to, laforin, malin, a ubiquitin conjugating enzyme) regulates the substrate. Ubc Ubiquitin conjugating enzymes, P phosphorylation, Ub ubiquitination
Besides R5, GS, GSK3 and malin, laforin is known to interact with at least two other proteins (Fig. 3). These are HIRIP5, a cytosolic protein involved in iron metabolism, and EPM2AIP1, a protein with unknown functions (Ganesh et al. 2003; Ianzano et al. 2003). These data suggest that the laforin–malin pathway is involved in vital physiological processes besides glycogen metabolism. This suggestion is interesting because the involvement of Lafora bodies in the epileptic phenotype of LD is debatable (Ganesh et al. 2002c, 2004). Targeted disruption of the Epm2a gene in mice caused formation of Lafora inclusion bodies, neurodegeneration, ataxia, myoclonus epilepsy, and impaired behavioural response (Ganesh et al. 2002c). Dying neurons characteristically exhibited swelling in the endoplasmic reticulum, Golgi networks and mitochondria in the absence of apoptotic bodies or fragmentation of DNA. Since the majority of degenerating neurons do not contain Lafora bodies, and not all cells that contain Lafora inclusions degenerate, the formation of Lafora bodies is not likely to result in neuronal cell death. The mouse model thus suggests that LD is a primary neurodegenerative disorder that may utilise a non-apoptotic mechanism of cell-death (Ganesh et al. 2002c). This notion was further supported by a report on the neuropathology of a transgenic mouse line (Chan et al. 2004b). Mice overexpressing the dominant negative laforin C266S mutant developed Lafora inclusion bodies in different tissues, including neurons, but had no signs of epileptic seizure (Chan et al. 2004b). In this regard, it is of interest to note that in adult polyglucosan body disease, polyglucosan inclusions accumulate in neurons but the patients do not develop epilepsy (Robitaille et al. 1980; Cavanagh 1999). Instead, they develop motor neuron deficit and dementia, a subset of features that are also seen in LD (Robitaille et al. 1980). Taken together, these data question a direct link between Lafora bodies and epileptic symptoms. It is likely therefore that Lafora bodies and epileptic seizures are independent consequences of defects in a common physiological pathway and that some LD symptoms could be secondary to Lafora bodies. Because LD has been traditionally classified as a ‘glycogen storage disease’, only a few reports have described the degenerative changes in the neuropile using autopsy or biopsy specimens (Van Heycop Ten Ham 1975; Busard et al. 1987). Moreover, these results were complicated by chronic administration of anti-epileptic drugs. The finding that laforin-deficient mice are associated with widespread neuronal degeneration throws new light on the evolution and pathogenesis of LD (Ganesh et al. 2002c). These observations also suggest that laforin is critical for neuronal survival and that some of the symptoms of LD are initiated by neuronal death.
Protein ubiquitination and phosphorylation have much in common, as both the processes occur rapidly and both are quickly reversed by a large set of dedicated enzymes termed deubiquitination enzymes and phosphatases, respectively. In addition, these two protein-modification events are known to cooperate in mobilising a particular pathway (Ben-Neriah 2002). Given the fact that defects in either laforin or malin lead to LD, it is logical to presume that these two proteins together regulate a critical pathway involved in neuronal function and that they act on a common substrate(s), either directly or indirectly. Laforin has been shown to be a substrate for malin (Gentry et al. 2005). The cellular function of malin could not be limited to laforin degradation, as defects in this process cannot explain the locus heterogeneity observed in LD. For example, if it were the only function of malin, then defects in malin will lead to increased levels of laforin, and, on the contrary, null mutations in EPM2A will result in the absence of laforin. We envision three alternate models to explain locus heterogeneity in LD (Fig. 5). One of the simplest models would be that laforin activates malin through dephosphorylation, and malin in turn regulates the half-life of a critical factor (Fig. 5a). Alternatively, laforin and malin may either act together as a multi-protein complex (Fig. 5c), or use an unknown protein intermediate to regulate a common substrate (Fig. 5b). In each of these models, absence of either malin or laforin would lead to an increase in the level of the ‘active’ form of the ‘critical substrate’, which may be pathogenic and result in LD. This model would also accommodate a third player in LD. For example, the third player could positively regulate malin in the model shown in Fig. 5b, or it could be one of the critical components of the laforin–malin complex of the model in Fig. 5c. In either case, loss of the third factor would result in increased levels of the active form of the ‘critical substrate’, leading to the production of LD. In each of these models, the critical step is the modification of proteins, which in turn would modify the functional state of the target protein. Transcriptional profiling in Epm2a knockout mice indeed suggests that dysregulation of genes involved in the modification of proteins is one of the major components of the pathological process (Ganesh et al. 2005). Thus, identification of the third player, and cellular substrates for laforin and malin would be important milestones in unravelling the physiological pathway defective in LD. With the availability of LD animal models for both EPM2A and NHLRC1 gene defects (Ganesh et al. 2002c; Lohi et al. 2005b), the identification of additional players in LD will aid in the development of both drugs and treatments.
References
Annesi G, Sofia V, Gambardella A, Candiano IC, Spadafora P, Annesi F, Cutuli N, De Marco EV, Civitelli D, Carrideo S, Tarantino P, Barone R, Zappia M, Quattrone A (2004) A novel exon 1 mutation in a patient with atypical Lafora progressive myoclonus epilepsy seen as childhood-onset cognitive deficit. Epilepsia 45:294–295
Ben-Neriah Y (2002) Regulatory functions of ubiquitination in the immune system. Nat Immunol 3:20–26
Busard HLSM, Renier WO, Gabreels FJM, Jasper HHJ, Slooff JL, Janssen AJM, van Haelst UJG (1987) Lafora disease: a quantitative morphological and biochemical study of the cerebral cortex. Clin Neuropathol 6:1–6
Cavanagh JB (1999) Corpora-amylacea and the family of polyglucosan diseases. Brain Res Rev 29:265–295
Chan EM, Bulman DE, Paterson AD, Turnbull J, Andermann E, Andermann F, Rouleau GA, Delgado-Escueta AV, Scherer SW, Minassian BA (2003a) Genetic mapping of a new Lafora progressive myoclonus epilepsy locus (EPM2B) on 6p22. J Med Genet 40:671–675
Chan EM, Young EJ, Ianzano L, Munteanu I, Zhao X, Christopoulos CC, Avanzini G, Elia M, Ackerley CA, Jovic NJ, Bohlega S, Andermann E, Rouleau GA, Delgado-Escueta AV, Minassian BA, Scherer SW (2003b) Mutations in NHLRC1 cause progressive myoclonus epilepsy. Nat Genet 35:125–127
Chan EM, Omer S, Ahmed M, Bridges LR, Bennett C, Scherer SW, Minassian BA (2004a) Progressive myoclonus epilepsy with polyglucosans (Lafora disease): evidence for a third locus. Neurology 63:565–567
Chan EM, Ackerley CA, Lohi H, Ianzano L, Cortez MA, Shannon P, Scherer SW, Minassian BA (2004b) Laforin preferentially binds the neurotoxic starch-like polyglucosans, which form in its absence in progressive myoclonus epilepsy. Hum Mol Genet 13:1117–1129
Delgado-Escueta AV, Ganesh S, Yamakawa K (2001) Advances in the genetics of progressive myoclonus epilepsy. Am J Med Genet 106:129–138
Fernandez-Sanchez ME, Criado-Garcia O, Heath KE, Garcia-Fojeda B, Medrano-Fernandez I, Gomez-Garre P, Sanz P, Serratosa JM, Rodriguez de Cordoba S (2003) Laforin, the dual-phosphatase responsible for Lafora disease, interacts with R5 (PTG), a regulatory subunit of protein phosphatase-1 that enhances glycogen accumulation. Hum Mol Genet 12:3161–3171
Ganesh S, Agarwala KL, Ueda K, Akagi T, Shoda K, Usui T, Hashikawa T, Osada H, Delgado-Escueta AV, Yamakawa K (2000) Laforin, defective in the progressive myoclonus epilepsy of Lafora type, is a dual specificity phosphatase associated with polyribosomes. Hum Mol Genet 9:2251–2261
Ganesh S, Shoda K, Amano K, Uchiyama A, Kumuda S, Moriyama N, Hirose S, Yamakawa K (2001a) Mutation screening for Japanese Lafora’s disease patients: identification of novel sequence variants in the coding and upstream regulatory regions of the EPM2A gene. Mol Cell Probes 15:281–289
Ganesh S, Agarwala KL, Amano K, Suzuki T, Delgado-Escueta AV, Yamakawa K (2001b) Regional and developmental expression of Epm2a gene and its evolutionary conservation. Biochem Biophys Res Commun 283:1046–1053
Ganesh S, Delgado-Escueta AV, Suzuki T, Francheschetti S, Riggio C, Avanzini G, Rabinowicz A, Bohlega S, Bailey J, Alonso ME, Rasmussen A, Ochoa A, Prado AJ, Medina MT, Yamakawa K (2002a) Genotype–phenotype correlations for EPM2A mutations in Lafora’s progressive myoclonus epilepsy: exon 1 mutations associate with an early onset cognitive deficit subphenotype. Hum Mol Genet 11:1263–1271
Ganesh S, Suzuki T, Yamakawa K (2002b) Alternative splicing modulates subcellular localization of laforin. Biochem Biophys Res Commun 1291:1134–1137
Ganesh S, Delgado-Escueta AV, Avila MR, Sakamoto T, Machado-Salas J, Hoshii Y, Akagi T, Suzuki T, Amano K, Agarwala KL, Hasegawa Y, Ishihara T, Hashikawa T, Itohara S, Cornford EM, Niki H, Yamakawa K (2002c) Targeted disruption of Epm2a gene causes formation of Lafora inclusion bodies, neurodegeneration, ataxia, myoclonus epilepsy and impaired behavioral response in mice. Hum Mol Genet 11:1251–1262
Ganesh S, Tsurutani N, Suzuki T, Ueda K, Agarwala KL, Osada H, Delgado-Escueta AV, Yamakawa K (2003) The Lafora disease gene product laforin interacts with HIRIP5, a phylogenetically conserved protein containing a NifU-like domain. Hum Mol Genet 12:2359–2368
Ganesh S, Tsurutani N, Suzuki T, Hoshii Y, Ishihara T, Deldago-Escueta AV, Yamakawa K (2004) The carbohydrate-binding domain of Lafora disease protein targets Lafora polyglucosan bodies. Biochem Biophys Res Commun 31:1101–1109
Ganesh S, Tsurutani N, Amano K, Mittal S, Uchikawa C, Delgado-Escueta AV, Yamakawa K (2005) Transcriptional profiling of a mouse model for Lafora disease reveals dysregulation of genes involved in the expression and modification of proteins. Neurosci Lett 387:62–67
Gentry MS, Worby CA, Dixon JE (2005) Insights into Lafora disease: malin is an E3 ubiquitin ligase that ubiquitinates and promotes the degradation of laforin. Proc Natl Acad Sci USA 102:8501–8506
Gomez-Abad C, Gomez-Garre P, Gutierrez-Delicado E, Saygi S, Michelucci R, Tassinari CA, Rodriguez de Cordoba S, Serratosa JM (2005) Lafora disease due to EPM2B mutations: a clinical and genetic study. Neurology 64:982–986
Gomez-Garre P, Sanz Y, Rodriguez De Cordoba SR, Serratosa JM (2000) Mutational spectrum of the EPM2A gene in progressive myoclonus epilepsy of Lafora: high degree of allelic heterogeneity and prevalence of deletions. Eur J Hum Genet 8:946–954
Ianzano L, Zhao XC, Minassian BA, Scherer SW (2003) Identification of a novel protein interacting with laforin, the EPM2a progressive myoclonus epilepsy gene product. Genomics 81:579–587
Ianzano L, Young EJ, Zhao XC, Chan EM, Rodriguez MT, Torrado MV, Scherer SW, Minassian BA (2004) Loss of function of the cytoplasmic isoform of the protein laforin (EPM2A) causes Lafora progressive myoclonus epilepsy. Hum Mutat 23:170–176
Ki CS, Kong SY, Seo DW, Hong SB, Kim HJ, Kim JW (2003)Two novel mutations in the EPM2A gene in a Korean patient with Lafora’s progressive myoclonus epilepsy. J Hum Genet 48:51–54
Lohi H, Ianzano L, Zhao XC, Chan EM, Turnbull J, Scherer SW, Ackerley CA, Minassian BA (2005a) Novel glycogen synthase kinase 3 and ubiquitination pathways in progressive myoclonus epilepsy. Hum Mol Genet 14:2727–2736
Lohi H, Young EJ, Fitzmaurice SN, Rusbridge C, Chan EM, Vervoort M, Turnbull J, Zhao XC, Ianzano L, Paterson AD, Sutter NB, Ostrander EA, Andre C, Shelton GD, Ackerley CA, Scherer SW, Minassian BA (2005b) Expanded repeat in canine epilepsy. Science 307:81
Minassian BA, Lee JR, Herbrick JA, Huizenga J, Soder S, Mungall AJ, Dunham I, Gardner R, Fong CY, Carpenter S, Jardim L, Satishchandra P, Andermann E, Snead OC III, Lopes-Cendes I, Tsui LC, Delgado-Escueta AV, Rouleau GA, Scherer SW (1998) Mutations in a gene encoding a novel protein tyrosine phosphatase cause progressive myoclonus epilepsy. Nat Genet 20:171–174
Minassian BA, Ianzano L, Meloche M, Andermann E, Rouleau GA, Delgado-Escueta AV, Scherer SW (2000a) Mutation spectrum and predicted function of laforin in Lafora’s progressive myoclonus epilepsy. Neurology 55:331–333
Minassian BA, Ianzano L, Delgado-Escueta AV, Scherer SW (2000b) Identification of new and common mutations in the EPM2A gene in Lafora disease. Neurology 54:488–490
Robitaille Y, Carpenter S, Karpati G, DiMauro SD (1980) A distinct form of adult polyglucosan body disease with massive involvement of central and peripheral neuronal processes and astrocytes: a report of four cases and a review of the occurrence of polyglucosan bodies in other conditions such as Lafora’s disease and normal ageing. Brain 103:315–336
Sainz J, Minassian BA, Serratosa JM, Gee MN, Sakamoto LM, Iranmanesh R, Bohlega S, Baumann RJ, Ryan S, Sparkes RS, Delgado-Escueta AV (1997) Lafora progressive myoclonus epilepsy: narrowing the chromosome 6q24 locus by recombinations and homozygosities. Am J Hum Genet 61:1205–1205
Sakai M, Austin J, Witmer F, Trueb L (1970) Studies in myoclonus epilepsy (Lafora body form) II polyglucosans in the systemic deposits of myoclonus epilepsy and in corpora amylacea. Neurology 20:160–176
Schwartz GA, Yanoff (1965) Lafora bodies, corpora-amylacea, and Lewy bodies: morphological and histochemical study. Arch Neurobiol 28:800–818
Serratosa JM, Delgado-Escueta AV, Posada I, Shih S, Drury I, Berciano J, Zabala JA, Antunez MC, Sparkes RS (1995) The gene for progressive myoclonus epilepsy of the Lafora type maps to chromosome 6q. Hum Mol Genet 4:1657–1663
Serratosa JM, Gomez-Garre P, Gallardo ME, Anta B, de Bernabe DB, Lindhout D, Augustijn PB, Tassinari CA, Malafosse RM, Topcu M, Grid D, Dravet C, Berkovic SF, de Cordoba SR (1999) A novel protein tyrosine phosphatase gene is mutated in progressive myoclonus epilepsy of the Lafora type (EPM2). Hum Mol Genet 8:345–352
Singh S, Suzuki T, Uchiyama A, Kumada S, Moriyama N, Hirose S, Takahashi Y, Inoue Y, Kimura K, Sawaishi S, Yamakawa K, Ganesh S (2005) Mutations in the NHLRC1 gene are the common cause for Lafora disease in the Japanese population. J Hum Genet 50:347–352
Van Heycop Ten Ham MW (1975) Lafora disease, a form of progressive myoclonus epilepsy. In: Vinken PJ, Bruyn GW (eds) The epilepsies. Handbook of clinical neurology 15. North-Holland, Amsterdam, pp 382–422
Wang J, Stuckey JA, Wishart MJ, Dixon JE (2002) A unique carbohydrate binding domain targets the Lafora disease phosphatase to glycogen. J Biol Chem 277:2377–2380
Yokoi S, Nakayama H, Negishi T (1975) Biochemical studies on tissues from a patient with Lafora disease. Clin Chim Acta 62:415–423
Acknowledgements
Financial support from the Department of Science and Technology (New Delhi) and the Council of Scientific and Industrial Research (New Delhi) is gratefully acknowledged.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Ganesh, S., Puri, R., Singh, S. et al. Recent advances in the molecular basis of Lafora’s progressive myoclonus epilepsy. J Hum Genet 51, 1–8 (2006). https://doi.org/10.1007/s10038-005-0321-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10038-005-0321-1
Keywords
This article is cited by
-
Lafora disease: from genotype to phenotype
Journal of Genetics (2018)
-
Loss of malin, but not laforin, results in compromised autophagic flux and proteasomal dysfunction in cells exposed to heat shock
Cell Stress and Chaperones (2017)
-
Glycogen metabolism has a key role in the cancer microenvironment and provides new targets for cancer therapy
Journal of Molecular Medicine (2016)
-
Interdependence of laforin and malin proteins for their stability and functions could underlie the molecular basis of locus heterogeneity in Lafora disease
Journal of Biosciences (2015)
-
Expression, purification and characterization of soluble red rooster laforin as a fusion protein in Escherichia coli
BMC Biochemistry (2014)