Abstract
The alpha subunit of interleukin 5 receptor (IL5RA) on chromosome 3p26-p24 is known to regulate the development and function of eosinophils. In an effort to discover additional polymorphism(s) in genes whose variant(s) have been implicated in asthma, we investigated the genetic polymorphisms in IL5RA to evaluate the gene as a potential candidate for a host genetic study of asthma. By direct DNA sequencing in 24 individuals, we identified 22 sequence variants within exons and flanking regions including a 1.5-kb promoter region of IL5RA; 10 common polymorphic sites were selected for genotyping in our asthma cohort (n=587). Two haplotype blocks were identified in a Korean population. Statistical analysis revealed that one promoter SNP, c.-5993A>G, and one ins/del polymorphism in intron 3, c.-480_482insdelGTT, showed significant association with the risk of asthma development. The genetic effects of c.-5993A>G and c.-480_482insdelGTT on asthma were more apparent among atopic subjects. Our findings suggest that polymorphisms in IL5RA might be among the genetic risk factors for asthma development, especially in atopic populations. IL5RA variant/haplotype information identified in this study will provide valuable information for strategies for the control of asthma.
Similar content being viewed by others
Introduction
Asthma is a common respiratory disease characterized by periodic obstruction of the airways and respiratory symptoms caused by acute and chronic bronchial inflammation (McFadden and Gilbert 1992). It has been well documented that the presence of atopy may precede the development of clinical asthma (Burrows et al. 1989). The development of asthma appears to be determined by an interaction between host genetic factors and a variety of environmental exposures.
IL5 is a T-cell-derived cytokine which is particularly important in the development of asthma for the terminal differentiation, activation and survival of committed eosinophil precursors (Bradley et al. 1991; Laitinen et al. 1993). IL5 stimulates the release of eosinophils into the circulation and prolongs their survival via the specific alpha subunit of interleukin 5 receptor (IL5RA on 3p26-p24) expressed on eosinophils (Isobe et al. 1992; Kotsimbos and Hamid 1997). Challenges to the airways due to allergens increase the local concentration of IL5, which correlates directly with the degree of airway eosinophilia (Sedgwick et al. 1991). In mice lacking the gene for IL5, eosinophilia does not occur after challenge by allergens (Foster et al. 1996). Moreover, the link between T-cell-derived IL5 and eosinophil release in asthmatic airways has also been supported by the demonstration that there are increased numbers of IL5RA mRNA positive cells in the bronchial biopsies of asthmatic subjects compared with controls, and that eosinophils are the predominant site of the increased IL5RA mRNA expression (Yasruel et al. 1997).
The human IL5 receptor complex is a heterodimer consisting of a unique alpha subunit predominantly expressed on eosinophils, and a more widely expressed beta subunit which is shared between the receptors for IL3 and GM-CSF (Murata et al. 1992). The alpha subunit is required for ligand-specific binding, whereas association with the beta subunit results in increased binding affinity (Boulay and Paul 1992). The alternative splicing of the IL5RA gene, which contains 14 exons, can yield several IL5RA isoforms including a membrane-anchored isoform (alpha IL5Rm) and a soluble isoform (alpha IL5Rs) (Tavernier et al. 1992).
Based on the biological properties involved in allergic inflammatory reactions as described above, it is hypothesized that IL5RA plays an important role in coordinating the release of eosinophil and IgE production against antigens leading to the development of atopy and asthma. We performed extensive screening for IL5RA by direct sequencing to detect polymorphisms and examined genetic association with the risk of asthma. Here, we present 22 genetic polymorphisms found in IL5RA and the results of the association study in a Korean asthma cohort.
Materials and methods
Subjects
Subjects were recruited from the Asthma Genome Research Center, which consists of four tertiary hospitals in Korea (Soonchunhyang University Hospital, Ajuo University Hospital, Ulsan University Hospital, and Choong-Ang University Hospital). Ethical approvals were obtained from the institutional review board of each hospital. All patients had the clinical symptoms and the physical examination results compatible with asthma. Each patient showed airway reversibility as documented by an inhalant bronchodilator-induced improvement of more than 15% of FEV1 and/or an airway hyperreactivity with less than 10 mg/ml of methacholine. Normal subjects were recruited from spouses of the patients and the general population who answered negatively to a screening questionnaire for respiratory symptoms and had FEV1 greater than 75% predicted, provocation concentration causing a fall in the FEV1 of 20% (PC20) with methacholine greater than 10 mg/ml, and normal findings on a simple chest radiogram. Total IgE and specific IgE to Dermatophagoides farinae (D.f.) and Dermatophagoides pteronyssinus (D.p.) were measured using the UniCAP system (Pharmacia Diagnostics, Sweden). Atopy was defined as having a wheal reaction to an allergen equal to or greater than that to histamine (1 mg/ml) or 3 mm in diameter and/or positive response of specific IgE to D.p. and D.f. The clinical parameters are summarized in Table 1.
Sequencing analysis of the human IL5RA gene
We sequenced exons and flanking regions, including a promoter region (1.5 kb), to discover single nucleotide polymorphisms (SNPs) in unregulated individual DNA samples from 24 Koreans using the ABI PRISM 3700 DNA analyzer (Applied Biosystems, Foster City, CA). Sixteen primer sets for amplification and sequencing analysis were designed based on GenBank sequences (see Electronic Supplementary Material) (ref. genome seq.: NT_022517 released on 19 February 2004). Information regarding primers is available on our website (http://www.snp-genetics.com/reference/IL5RA_add_info.doc). Sequence variants were verified by chromatograms (see Electronic Supplementary Material). In order to compare allele frequencies of IL5RA SNPs that were identified in the Korean population with other major ethnic groups, we also genotyped DNA from 50 Caucasians and 50 African-Americans obtained from the Human Genetic Cell Repository (http://locus.umdnj.edu/nigms/).
Genotyping with fluorescence polarization detection
For genotyping of polymorphic sites with our asthma study, amplifying primers and probes were designed for TaqMan (An et al. 2002; Wiltshire et al. 2003) (see Electronic Supplementary Material). Primer Express (Applied Biosystems) was used to design both the PCR primers and the MGB TaqMan probes. One allelic probe was labeled with the fluorescent 6-carboxyfluorescein dye and the other with the fluorescent VIC dye. PCRs were run in TaqMan Universal Master mix without UNG (Applied Biosystems) with PCR primer concentrations of 900 nM and TaqMan MGB-probe concentrations of 200 nM. Reactions were performed in 384-well format in a total reaction volume of 5 μl using 20 ng of genomic DNA. The plates were then placed in a thermal cycler (PE 9700, Applied Biosystems) and heated at 50°C for 2 min and 95°C for 10 min followed by 40 cycles of 95°C for 15 s and 60°C for 1 min with a final soak at 25°C. The TaqMan assay plates were transferred from the thermal cyclers to the Prism 7900HT instruments (Applied Biosystems) where the fluorescence intensity in each well of the plate was read. Fluorescence data files from each plate were analyzed by automated allele-calling software (SDS 2.1).
Statistics
We examined a widely used measure of linkage disequilibrium (LD) between all pairs of biallelic loci, Lewontin’s D′ (|D′|) (Hedrick 1987). Haplotypes were inferred using the algorithm developed by the Broad Institute; the algorithm (Haploview) searches for a spine of strong |D′| running from one marker to another. Phase probabilities of each site were calculated for each individual with this software. Genotype distribution of IL5RA SNPs and haplotype among the asthmatics and the normal subjects were analyzed with logistic regression models controlling age (continuous value), sex (male=0, female=1) and smoking status (nonsmoker=0, ex-smoker=1, smoker=2) as covariates.
Results
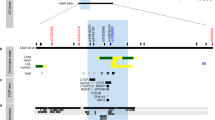
By direct DNA sequencing in 24 individuals, we identified 22 sequence variants within exons and flanking regions of IL5RA including a 1.5-kb promoter region: 13 variants in the promoter and 5′ UTR region, 3 in exons, and 6 in introns (Fig. 1a). Ten of the identified polymorphisms were selected for larger-scale genotyping based on locations, LDs, frequencies, and haplotype tagging status. The frequencies of the ten selected SNPs were 0.074 (c.-6783T>C), 0.329 (c.-6475G>A), 0.317 (c.-6173T>C), 0.493 (c.-5993G>A), 0.485 (c.-5567C>G), 0.260 (c.-5091G>A), 0.251 (c.-3643G>A), 0.191 (c.-479_481insdelTGT), 0.111 (c.82+2068T>C), and 0.348 [c.385A>G (p.I129 V)] in the Korean population (n=587, Table 1, Fig. 1).
Gene map, haplotypes and LD coefficients in IL5RA. a Gene map and SNPs in IL5RA on chromosome 3p26-p24 (reference sequence of IL5RA mRNA: NM_000564.2). Coding exons are marked by black blocks and 5′ and 3′ UTR by white blocks. Asterisks indicate SNPs that were genotyped in the larger population. The frequencies of SNPs without larger-scale genotyping were based on sequencing data (n=24). Additional information on the 22 SNPs is available at http://www.snp-genetics.com/reference/IL5RA_add_info.doc. First base of translation site is denoted as nucleotide +1. b Linkage disequilibrium coefficient (|D′|) and LD BLOCKs among IL5RA SNPs. Triangles indicate haplotypic BLOCKs (denoted as BLOCK1 and 2). c Haplotypes of IL5RA. Haplotypes with frequency >0.03 are presented. Others denoted with superscript 1 include rare haplotypes of BLOCK1: TGTGCA, TGCGGA, TGTACG, TGCGCG, TATGCA, TATAGG, TGTAGA, TGTGGG, and CGCGGG. Others denoted with superscript 2 include rare haplotypes of BLOCK2: AinsTA, AdelTG, GdelCA, and GdelTG
Linkage disequilibrium coefficients (|D′|) and r2 among polymorphisms were also calculated (Fig. 1). Interestingly, the analysis shows that IL5RA can be parsed into two haplotype BLOCKs (1 and 2), with each block having strong LD spine. There were six and five common haplotypes in BLOCK1 and BLOCK2, respectively, and the significant differences in frequencies of SNPs and haplotypes were observed among all three ethnic groups (Fig. 1c).
Allele frequencies of each polymorphism and common haplotypes (BLOCK1-ht3, BLOCK1-ht4, BLOCK2-ht1, and BLOCK2-ht4) were compared between the patients and the normal controls using logistic regression models (Table 2). BLOCK1-ht1, BLOCK1-ht2, BLOCK1-ht5, BLOCK2-ht2, BLOCK2-ht3, and BLOCK2-ht5 were not analyzed because they were equivalent to c.-6173T>C, c.-5091G>A, c.-6783T>C, c.-3643G>A, c.->80_482insdelGTT, and c.82+2068T>C, respectively. BLOCK1-ht6 was not analyzed because the frequency was low. The G allele of c.-5993A>G showed a lower frequency in the asthma patients than in normal controls (0.48 vs 0.51, P=0.04). The protective effect of the G allele of c.-5993A>G was clearer in the atopic population (0.005–0.007, Table 3). The GTT deletion allele of c.-480_482insdelGTT in intron 3 showed higher frequency in asthma patients than in normal controls (0.21 vs 0.16, P=0.01). The susceptibility of the GTT deletion allele of c.-480_482insdelGTT was also more apparent in the atopic population (0.03–0.17, Table 3).
Discussion
The alpha subunit of interleukin 5 receptor plays a very important role in the pathophysiology of allergic rhinitis and asthma (Gorski et al. 2002). Elevated IL5RA mRNA expression was associated with significantly increased numbers of bronchial mucosal eosinophils in asthmatic patients as compared to nonasthmatic control subjects (Powell et al. 1996). Furthermore, the number of IL5RA mRNA-positive cells was significantly greater in the atopic vs nonatopic groups (Kotsimbos et al. 1996).
In this study we demonstrated that c.-5993A>G and c.-480_482insdelGTT showed significant association with the risk of asthma, especially in atopic subjects (Tables 2, 3). In further study, the promoter SNP, c.-5993A>G, was analyzed for putative transcription factor binding site using the software TFSEARCH (Searching Transcription Factor Binding Sites V1.3; putative score>0.87). As a result, the c.-5993A>G is located in the consensus sequence of cap (cAMP receptor protein) and AP-4 (activating enhancer-binding protein 4). Thus, a nucleotide change from A to G could, possibly, disrupt those transcription-factor binding sites. The c.-480_482insdelGTT in intron 3 is located in the consensus sequence (Wu and Maniatis 1993) of the 5′-end splice site of intron 3 of IL5RA (NM_000564.2). However, the change in the consensus sequence seems to induce no alternative splicing event when the splicing event is predicted using the software NNSplice version 0.9 (http://www.fruitfly.org/seq_tools/splice.html). Further functional studies for the genetic effects of promoter SNP and intronic ins/del polymorphism are needed.
IL5 has been suggested as a specific therapeutic target for asthma therapy. In animal models, allergen challenge blockade and gene deletion have been effective in reducing airway eosinophilia and hyperresponsiveness (Mauser et al. 1995). Allergens activate mast cells and Th2 cells in the airway (Noah and Becker 2000), inducing in turn the production of mediators of inflammation and cytokines including IL4 and IL5 (Walker et al. 1991). IL5 moves to the bone marrow and causes terminal differentiation of eosinophils through the binding to IL5RA (Bochner 1997).
The effects of IL5RA genetic polymorphisms on the risk of asthma were not dramatic in the present study. Therefore, it may be argued that Bonferroni correction should be applied to the P values obtained. If Bonferroni corrections were strictly adopted, associated P values could not retain the significances [the threshold of significance would be 0.004 (14 polymorphisms)]. However, although there is a chance of type 1 error due to multiple comparisons, when considering the fact that the comparisons were not totally independent of each other due to tight LDs among SNPs/haplotype and related phenotypes, the significance of associations might be acceptable. Further biological and/or functional evidence would be needed to confirm the suggested association of IL5RA polymorphisms with the risk of asthma in this study.
In summary, we have identified 22 polymorphisms in the human IL5RA gene, and 10 common polymorphic sites were selected for genotyping in our asthma cohort. Statistical analysis revealed that c.-5993A>G and c.-480_482insdelGTT showed significant association with the risk of asthma development. The effect of c.-5993A>G and c.-480_482insdelGTT on asthma was more apparent in the atopic population. Further studies would be needed to elucidate the function of the variant, which showed significant association with atopy and asthma.
References
An P, Nelson GW, Wang L, Donfield S, Goedert JJ, Phair J, Vlahov D, Buchbinder S, Farrar W L, Modi W, O’Brien SJ, Winkler CA (2002) Modulating influence on HIV/AIDS by interacting RANTES gene variants. Proc Natl Acad Sci USA 99:10002–10007
Bochner BS (1997) Cellular adhesion and its antagonism. J Allergy Clin Immunol 100:581–585
Boulay JL, Paul WE (1992) The interleukin-4 family of lymphokines. Curr Opin Immunol 4:294–298
Bradley BL, Azzawi M, Jacobson M, Assoufi B, Collins JV, Irani AM, Schwartz LB, Durham SR, Jeffery PK, Kay AB (1991) Eosinophils, T-lymphocytes, mast cells, neutrophils, and macrophages in bronchial biopsy specimens from atopic subjects with asthma: comparison with biopsy specimens from atopic subjects without asthma and normal control subjects and relationship to bronchial hyperresponsiveness. J Allergy Clin Immunol 88:661–674
Burrows B, Martinez FD, Halonen M, Barbee RA, Cline MG (1989) Association of asthma with serum IgE levels and skin-test reactivity to allergens. N Engl J Med 320:271–277
Foster PS, Hogan SP, Ramsay AJ, Matthaei KI, Young IG (1996) Interleukin 5 deficiency abolishes eosinophilia, airways hyperreactivity, and lung damage in a mouse asthma model. J Exp Med 183:195–201
Gorski P, Wittczak T, Walusiak J, Palczynski C, Ruta U, Kuna P, Alam R (2002) Eotaxin but not MCP-3 induces eosinophil influx into nasal fluid in allergic patients. Allergy 57:519–528
Hedrick PW (1987) Gametic disequilibrium measures: proceed with caution. Genetics 117:331–341
Isobe M, Kumura Y, Murata Y, Takaki S, Tominaga A, Takatsu K, Ogita Z (1992) Localization of the gene encoding the alpha subunit of human interleukin-5 receptor (IL5RA) to chromosome region 3p24-3p26. Genomics 14:755–758
Kotsimbos AT, Hamid Q (1997) IL-5 and IL-5 receptor in asthma. Mem Inst Oswaldo Cruz 92(Suppl 2):75–91
Kotsimbos TC, al Ghamdi K, Small P, Frenkiel S, Hamid QA (1996) Upregulation of Th-2 cytokine receptors in atopy- and nonatopy-associated chronic sinusitis. J Otolaryngol 25:317–321
Laitinen LA, Laitinen A, Haahtela T, Vilkka V, Spur BW, Lee TH (1993) Leukotriene E4 and granulocytic infiltration into asthmatic airways. Lancet 341:989–990
Mauser PJ, Pitman AM, Fernandez X, Foran SK, Adams GK III, Kreutner W, Egan RW, Chapman RW (1995) Effects of an antibody to interleukin-5 in a monkey model of asthma. Am J Respir Crit Care Med 152:467–472
McFadden ER Jr, Gilbert IA (1992) Asthma. N Engl J Med 327:1928–1937
Murata Y, Takaki S, Migita M, Kikuchi Y, Tominaga A, Takatsu K (1992) Molecular cloning and expression of the human interleukin 5 receptor. J Exp Med 175:341–351
Noah TL, Becker S (2000) Chemokines in nasal secretions of normal adults experimentally infected with respiratory syncytial virus. Clin Immunol 97:43–49
Powell N, Humbert M, Durham SR, Assoufi B, Kay AB, Corrigan CJ (1996) Increased expression of mRNA encoding RANTES and MCP-3 in the bronchial mucosa in atopic asthma. Eur Respir J 9:2454–2460
Schaid DJ, Rowland CM, Tines DE, Jacobson RM, Poland GA (2002) Score tests for association between traits and haplotypes when linkage phase is ambiguous. Am J Hum Genet 70:425–434
Sedgwick JB, Calhoun WJ, Gleich GJ, Kita H, Abrams JS, Schwartz LB, Volovitz B, Ben-Yaakov M, Busse WW (1991) Immediate and late airway response of allergic rhinitis patients to segmental antigen challenge. Characterization of eosinophil and mast cell mediators. Am Rev Respir Dis 144:1274–1281
Tavernier J, Tuypens T, Plaetinck G, Verhee A, Fiers W, Devos R (1992) Molecular basis of the membrane-anchored and two soluble isoforms of the human interleukin 5 receptor alpha subunit. Proc Natl Acad Sci USA 89:7041–7045
Walker C, Kaegi MK, Braun P, Blaser K (1991) Activated T cells and eosinophilia in bronchoalveolar lavages from subjects with asthma correlated with disease severity. J Allergy Clin Immunol 88:935–942
Wiltshire T, Pletcher MT, Batalov S, Barnes SW, Tarantino LM, Cooke MP, Wu H, Smylie K, Santrosyan A, Copeland NG, Jenkins NA, Kalush F, Mural RJ, Glynne RJ, Kay SA, Adams MD, Fletcher CF (2003) Genome-wide single-nucleotide polymorphism analysis defines haplotype patterns in mouse. Proc Natl Acad Sci USA 100:3380–3385
Wu JY, Maniatis T (1993) Specific interactions between proteins implicated in splice site selection and regulated alternative splicing. Cell 75:1061–1070
Yasruel Z, Humbert M, Kotsimbos TC, Ploysongsang Y, Minshall E, Durham SR, Pfister R, Menz G, Tavernier J, Kay A B, Hamid Q (1997) Membrane-bound and soluble alpha IL-5 receptor mRNA in the bronchial mucosa of atopic and nonatopic asthmatics. Am J Respir Crit Care Med 155:1413–1418
Acknowledgements
This work was supported by grant number M1-0302-00-0073 of the National Research Lab Program as part of the National Research and Development Program of the Ministry of Science and Technology of Korea.
Author information
Authors and Affiliations
Corresponding author
Electronic Supplementary Material
Rights and permissions
About this article
Cite this article
Cheong, H.S., Kim, L.H., Park, B.L. et al. Association analysis of interleukin 5 receptor alpha subunit (IL5RA) polymorphisms and asthma. J Hum Genet 50, 628–634 (2005). https://doi.org/10.1007/s10038-005-0304-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10038-005-0304-2
Keywords
This article is cited by
-
A genetic effect of IL-5 receptor α polymorphism in patients with aspirin-exacerbated respiratory disease
Experimental & Molecular Medicine (2013)
-
Interaction of C1GALT1–IL5RA on the susceptibility to IgA nephropathy in Southern Han Chinese
Journal of Human Genetics (2013)
-
The severity of FIP1L1–PDGFRA-positive chronic eosinophilic leukaemia is associated with polymorphic variation at the IL5RA locus
Leukemia (2007)