Abstract
Graves’ disease (GD) was believed to be a polygenic disease. Several chromosomal regions were linked to GD, and the 5q31 chromosome regions containing several interleukin genes cluster were worth observing. In this study, IL4, IL13, IRF1 and UGRP1 genes were sequenced, and 5, 3, 7 and 7 polymorphisms respectively were discovered. Then an extended association study for the attracting polymorphisms was performed with 146 sporadic Graves’ patients, 142 unrelated controls and the 54 multiplex Graves’ families. However, the genotype and allele frequency distribution of these polymorphisms had similar distribution between the Graves’ patients and unrelated controls, and transmission disequilibrium tests indicated that none of them showed dominant transmission from heterozygous parents to the affected offsprings. Comparison of the clinical variables of the Graves’ patients indicated that the onset ages of the patients carrying TT at IRF1 6477 T/G locus were younger than those having variant allele (TG, GG); the difference was of statistical significance (P=0.005, Pc=0.020). Our association study revealed that, IL4, IL13, IRF1 and UGRP1 genes in chromosomal 5q31 regions might not confer susceptibility to Chinese GD. But those individuals who were TT homozygous at IRF1 6477 T/G locus seemed to be attacked by GD much earlier than others.
Similar content being viewed by others
Introduction
Graves’ disease (GD) is an organ-specific autoimmune thyroid disease, classically characterized by the presence of thyroid specific reactive T lymphocytes and thyroid specific autoantibodies and typical clinical manifestations (Ban and Tomer 2003; Weetman 2003). There has been very strong evidence that genetic factors contribute to the etiology of GD. Evidence for the role of genetic factors was indicated in one study by an increased relative risk of more than 15 in siblings (Vyse and Todd 1996) compared with the general population (λs), and in another study by an increased concordance rate in monozygotic, compared with dizygotic twin (Brix et al. 2001). The lack of a clear pattern of inheritance has suggested that multiple genes were involved in influencing the autoimmune events in GD (Burek et al. 1982; Tomer and Davies 2003).
Genome-wide linkage studies provided a powerful tool for disease-susceptible gene identification (Tomer and Davies 2003). Our previous study mapped the susceptible gene of GD in chromosomal 5q31 regions using 54 Chinese Han multiple pedigrees (Jin et al. 2003). Meanwhile, Sakai and co-workers mapped susceptibility loci for Hashimoto thyroiditis (HT), another autoimmune thyroid disease, to 8q23-q24, and for AITD (autoimmune thyroid disease, i.e., GD and HT) to 5q31-q33 by multipoint linkage analysis in a collection of Japanese AITD affected sib-pairs (Sakai et al. 2001). However, Tomer et al. (1997) did not achieve the same mapping results in Caucasians. The enrolled subjects of Jin and Sakai were East Asians, with the similar genetic background, which made us suppose that East Asians may share the same susceptible gene(s) for GD.
Based on the differential cytokines secreted, helper T cells were able to be subdivided into two subgroups, namely T helper 1 (Th1) and T helper 2 (Th2) cells. Cytokines produced by Th2 cells, such as interleukin 4 (IL4), interleukin 5 (IL5), interleukin (IL10) and interleukin 13 (IL13), played a more important role in antibody production than Th1 cytokines, i.e., interferon-γ (IFNγ), interleukin 2 (IL2), interleukin 12 (IL12) etc. (Romagnani 1996). GD was believed to be a Th2-biased autoimmunity in which autoantibodies against thyrotropin receptor stimulate thyroid hormone production, and Th1/Th2 cytokines disorder was frequently seen in Graves’ individuals (Tomer and Davies 2003). In the chromosomal 5q31 regions, the immune regulatory genes, IL4, IL13 and interferon regulatory factor 1 (IRF1) genes, which were believed to have an effect in the pathogenesis of Th1/Th2 malfunction, and thyroid-specific genes—Uteroglobin-Related Protein 1 (UGRP1 or SCGB3A2) gene (Niimi et al. 2001)—were plausible candidate genes for GD. The major aim of this study was to investigate the susceptible gene(s) of GD in the chromosomal 5q31 region by performing an association study with random population and multiplex pedigrees, and to study whether certain polymorphisms loci may influence Graves’ patients’ clinical parameters or course of disease.
Subjects and methods
Subjects
We chose 48 (or 24) probands in the 54 Chinese Graves’ pedigrees previously used in genome-wide scan of Jin et al. (2003). We first performed SNP discovery with these 48 (or 24) probands and 48 (or 24) unrelated controls. Then we further performed an extended association study with 146 sporadic Graves’ patients and 142 unrelated controls. The control subjects without personal history of autoimmune or allergic diseases were clinically and bio-chemically euthyroid, and were negative for thyroid peroxidase antibodies. The diagnosis of GD was based on documented clinical and biochemical evidence of hyperthyroidism, diffused goiter and the presence of at least one of the following: positive TSH receptor antibody tests, diffusely-increased 131I (iodine-131) uptake in the thyroid gland, or presence of exophthalmos. For this study, all other pedigree members, whether thyroid autoantibody positive or negative, were classified as unaffected. The goiter size of the thyroid was graded according to the following criteria: grade 0, goiter absent; grade 1, goiter palpable but invisible with the neck in a normal position; grade 2, goiter visible with neck in a normal position; and grade 3, large goiter perceptible at a distance. For diagnostic accuracy, all individuals classified as affected were interviewed and examined by experienced clinicians. After informed consent, blood samples were collected from all participants for DNA preparation as well as biochemical measurements. The Ethical Committee of the Chinese National Human Genome Center at Shanghai approved this project for the involvement of human subjects.
DNA extracting and PCR amplifying
Genomic DNA was isolated from peripheral white blood cells using the DNA purification kit (Qiagen, Germany) or standard phenol–chloroform protocol. Based on the published sequence of IL4, IL13, IRF1 and UGRP1 genes, the primers overlapping all the exons, exon–intron adjacent areas, 3′UTR segments and more than 1,000 bp promoter regions were designed with the help of primer software v3.0 (http://frodo.wi.mit.edu/cgi-bin/primer3/primer3_www.cgi) (Rozen and Skaletsky 2000).
Polymerase chain reactions were performed in a final volume of 15 μl, containing 20–30 ng genomic DNA, 0.05 μmol of each primer, 200 μM of dNTP, 10 mM Tris–HCl (pH 8.3), 50 mM KCl, 1.5 mM MgCl2 and 1.5 U Taq DNA polymerase (Qiagen, Germany). Each PCR was performed with a 2-min hot-start procedure in GeneAmp System 9700 (PE Applied Biosystems), and amplification process was carried out using 35 cycles of denaturation at 94°C for 40 s, with annealing at 62°C for 40 s, for the first turn of cycles with 0.5°C decrease/cycle, followed by the second turn of cycle 94°C for 40 s, with annealing at 56°C for 40 s, and elongation at 72°C for 40 s, followed by a final elongation at 72°C for 10 min.
The PCR products were subjected to electrophoresis in a 1.5% agarose gel to verify the sizes. After standard procedures of purification (Wizard PCR Preps DNA Purification Resin, Cat# A7181, Promega) and extraction with phenol chloroform and ethanol, all the PCR products were subjected to sequencing reactions in the 3730 DNA auto-sequencer (BigDye Terminator Ready Reaction Kits, ABI prism 3730, PE Applied Biosystems). The sequencing data were then analyzed with Phredphrap-Polyphred-Consed software (Nickerson et al. 1997), by at least two experienced technicians separately.
Statistical analysis
Alleles and genotypes frequencies of SNPs were compared between Graves’ patients and healthy individuals, using the chi-square test or the Fisher exact probability test when appropriate. The Bonferroni method with a correction factor equal to four was used to correct the P-values in the comparisons of clinical and genetic parameters among patients stratified by the presence of the different alleles. Differences in distribution of age of disease onset were analyzed by the Kruskal–Wallis test or the Mann–Whitney test (a nonparametric test). The comparison of the other clinical variables of Graves’ patients carrying certain genotype was analyzed by chi-square test or the Fisher exact probability test when appropriate. The haploblock structures of IRF1 and UGRP1 genes were constructed using the software Haploview (Barrett et al. 2005), which had the TDT analysis implanted. A P (Pc) value <0.05 was considered significant.
Results
SNP discovery
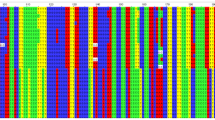
When screening polymorphisms with small subjects, we found 5, 3, 7 and 7 SNPs whose minor allele frequency was more than 5% in the IL4, IL13, IRF1 and UGRP1 genes respectively. All of these polymorphisms were in Hardy–Weinberg equilibrium (data not shown). Their description, location and allele frequency were shown in Table 1. In the IL4 gene, the −590 T/C and −34 T/C polymorphisms were in complete linkage disequilibrium (LD), and the latter three polymorphisms, i.e., 8187 G/A, 8375 G/A and 8412 G/A, were in complete LD also. In the IL13 gene, 2044 G/A and 2052 G/A polymorphisms were in complete LD. The SNPs’ LD relationships in the IRF1 gene and the UGRP1 gene were also shown below (Fig. 1).
Haploblock structure construction
Since there were seven SNPs in both the IRF1 and UGRP1 genes, we genotyped all these SNPs with the second set of samples (146 sporadic patients and 142 unrelated controls, 54 multiplex Graves’ familes with 322 individuals) and then constructed the haploblock structure for the two genes with the software Haploview.
As shown in Fig. 1, the latter five SNPs in IRF1 gene formed a haploblock. In this haploblock, there were three main haplotypes (Table 2), and 6421 G/A and 6477 T/G served the tagSNPs for these block structure. Whilst in the UGRP1 gene the latter six SNPs formed a haploblock, four main haplotypes were found in this block structure, and −626 CA/G, −112 G/A, 271 T/– could be considered as the tagSNPs. The distribution of these haplotypes in random populations is shown in Table 2, and no statistical significance was found.
Haploblock structure of the polymorphisms found in IRF1 and UGRP1 genes. The extent of LD (r2) between SNPs is shown by the color of the box, where black boxes indicate r2 =1, white ones showed r2 =0, and shades of grey mean 0< r2 <1. a Haploblock structure for the IRF1 genes; the seven polymorphisms were marked as SNP I–VII in order, and SNP III (6421 G/A) and SNP V (6477 T/G) were identified as the tagSNPs. b Haploblock structure for the UGRP1 genes; the seven polymorphisms were marked as SNP I–VII in order, and SNP II (−626 CA/G), SNP III (−112 G/A) and SNP V (271 T/–) were identified as the tagSNPs
Extended association study
The association of the IL4 –590 T/C, IL13 –1112 C/T polymorphisms with several autoimmune diseases has been extensively analyzed in many studies, but no definite conclusion has yet been formed; in the meantime, 6421 G/A and 6477 T/G polymorphisms served the tagSNPs in the haploblock for IRF1 genes , and the UGRP1 gene was a quite novel gene which was identified by Niimi in 2001. Considering the above evidence, we performed an extended association study covering these attracting SNPs, i.e., IL4 −590 T/C (rs2243250), IL13 −1112 C/T (rs1800925), IRF1 6421 G/A (rs2070728), IRF1 6477 T/G (rs2070729) and all the seven polymorphisms of UGRP1 genes. As shown in Table 3, IL4 −590 T/C, IL13 −1112 C/T, IRF1 6421 G/A, IRF1 6477 T/G did not demonstrate any statistically-significant difference between the Graves’ patients (sporadic Graves’ patients and probands of Graves’ pedigrees) and unrelated controls.
Transmission disequilibrium tests
We also performed the TDT analysis for the SNPs in IRF1 and UGRP1 genes. We genotyped all the 322 members of the 54 multiplex Graves’ families. However, transmission disequilibrium test analysis demonstrated that none of these SNPs in the IRF1 and UGRP1 genes showed dominant transmission to the affected offspring from heterozygous parents, and all the P-values were greater than 0.05 (Table 4).
Polymorphisms affecting the clinical variables for the Graves’ patients
We further compared the clinical variables for the Graves’ patients carrying different genotypes at IL4 −590 T/C, IL13 −1112 C/T, IRF1 6421 G/A, IRF1 6477 T/G, UGRP1 −626 CA/G, UGRP1 −112 G/A and UGRP1 271 T/– loci. As shown in Table 5, the onset age for those Graves’ patients who were TT genotype at IRF1 6477 T/G locus was 37.73±11.69 years, while the onset ages for the other patients were 42.16±13.75 (TG) and 43.24±15.20 (GG) years respectively, which reached statistical difference (P=0.005, Pc=0.020). In addition, pairwise comparison using the Mann–Whitney U-test gave the following P-values; P=0.014 (“TG” vs “TT”), P=0.002 (“GG” vs “TT”), and P=0.273 (“GG” vs “TG”) respectively. Meanwhile, the other loci seemed to show no statistical influence on the patients’ clinical variables, such as age of onset, gender, goiter before treatment and ophthalmopathy.
Discussion
GD is an organ-specific autoimmune disorder characterized by diffuse goiter and thyroid hormone oversecretion as a result of TSH receptor antibody stimulation. Although the etiology of GD remains unclear, it is believed to be caused by a complex interaction between genetic and environmental factors (Tomer and Davies 2003).
Recent genome-wide researches in Chinese multiplex families and Japanese affected sib-pairs have mapped the susceptible gene(s) for GD in chromosomal 5q31 regions (Sakai et al. 2001; Jin et al. 2003), but this chromosomal regions were not linked with GD in Caucasian populations (Tomer et al. 1997). This might suggest that these loci were specific to East Asian populations. This chromosomal region encoded a cluster of cytokine genes, including IL4, IRF1 and IL13, and these cytokine genes were believed to participate in maintaining the Th1/Th2 equilibrium (Ban and Tomer 2003; Weetman 2003).
IL4 is mainly produced by Th2 cells, and could cause proliferation of IgE- and IgG-secreting B cells. It also stimulates the expression of HLA class II antigens via STAT6 (signal transducer and activator of transcription-6) (Kelso 1998) and opposes the Th1 cell inflammatory response. Indeed, IL4 hes been considered the pivotal cytokine polarizing the immune response toward a Th2 cell response, and although the initial trigger for IL4 production remains unknown, genetic variations are likely to play a role (Romagnani 1996). The −590 T/C polymorphism in its promotor region appears to be functional, with increased transcriptional activity attributed to the variant allele (Rosenwasser et al. 1995). Association of this polymorphism (Rosenwasser et al. 1995; Walley and Cookson 1996; Noguchi et al. 1998) with atopy has provided evidence that IL4 is an important mediator of allergic disease. Recently, the association of IL4 −590 T/C polymorphism with GD in Caucasians has been regarded as controversial (Hunt et al. 2000; Heward et al. 2001), and requires confirmation in other populations.
IL13 is an important immunoregulatory protein produced primarily by activated Th2 cells (Chomarat and Banchereau 1998) and is involved in B cell maturation. It up-regulates CD23 and major histocompatibility complex class II expression (Briere et al. 1993) and promotes IgE isotype switching in human B cells (Punnonen et al. 1993). IL13 also downregulates macrophage activity, thereby inhibiting the production of proinflammatory cytokines and chemokines such as IL1α, IL1β, IL6, IL8 and TNFα. Thus, IL13 might be a potential candidate gene contributing to the development of GD or influencing its clinical course. Currently, the −1112 C/T polymorphism in its 5′-flanking region has been identified as having a biological function, and has been shown to regulate gene transcription (van der Pouw Kraan et al. 1999; Howard et al. 2001). In addition, the association of −1112 C/T polymorphism with GD and other atopy diseases has been widely studied (Bednarczuk et al. 2003; Hummelshoj et al. 2003; Hiromatsu et al. 2005), but the results need support from further studies.
IRF1 is a member of the IRF family of transcription factors that is involved in the regulation of genes that control cell growth, differentiation and death. In addition, IRF1 is involved in several steps of the immune response that may be pertinent to GD, including the polarization of the cytokine response in CD4+ T cells. The transcriptional activator (IRF1) plays a crucial role in the activation of IFNγ, a kind of Th1 cytokine, induced gene expression. In subjects with atopic disease, the reduction of IFNγ secretion has been reported (Jujo et al. 1992; Tang et al. 1993). In addition, IRF1 has been found to affect Th1/Th2 cytokine balance in IRF1 knockout experiments. Deficiency of the IRF1 gene in mice leads to strongly impaired Th1 responses and enhanced Th2 responses, with an increase in the antigen-specific IgE response (Lohoff et al. 1997). Hence it is possible that the IRF1 gene polymorphism might alter Th1/Th2 cytokine production and contribute to an inherited predisposition to GD in Chinese.
In 2001, Niimi et al. identified the UGPR1 gene as the downstream gene of TTF1 (thyroid transcription factor 1) through suppressive substractive hybridization, and its expression product was mainly observed in the lung and thyroid (Niimi et al. 2001). The past several years have witnessed an increasing interest in association studies of the UGRP1 gene with asthma, another autoimmune disease whose susceptible loci has also been mapped in the 5q31 chromosomal region (Niimi et al. 2002; Heinzmann et al. 2003; Jian et al. 2003).
These positional and biological data have supported the theory that the immune regulatory genes and thyroid specific genes might be the plausible candidate genes contributing to the development of GD or influencing its clinical severity and course. However, the association studies were controversial, and have seldom been able to be repeated in different populations. In this study, we sequenced all the coding area and more than 1,000 bp promoter regions of the IL4, IL13, IRF1 and UGRP1 genes with certain probands of multiplex GD families and unrelated controls. In IL4 and IL13 genes, we chose IL4 −590 T/C and IL13 −1112 C/T, which has been shown to be associated with GD in other studies, for further analysis. Meanwhile, for IFR1 and UGRP1 genes we constructed the LD block, and IRF1 6421 G/A, IRF1 6477 T/G, UGRP1 −626 CA/G, UGRP1 −112 G/A and UGRP1 271 T/- polymorphisms can be considered as the tagSNPs for IRF1 and UGRP1 genes. Then we carried out an extended associated study for these attracting SNPs. However, these polymorphisms did not show any differential genotypic or allelic distribution between the Graves’ patients and unrelated controls. Moreover, they did not exhibit any dominant transmission from heterozygous parents to the affected offsprings. We further compared clinical variables for the Graves’ patients carrying different genotype at these above loci, and found that the onset age for those Graves’ patients who were TT genotype at IRF1 6477 T/G locus was 37.73±11.69 years, whilst the onset ages for the other patients were 42.16±13.75 (TG) and 43.24±15.20 (GG) years respectively, which represented a statistically-significant difference (P=0.005, Pc=0.020). In addition, pairwise comparison using the Mann–Whitney U-test gave the following P-values; P=0.014 (“TG” vs “TT”), P=0.002 (“GG” vs “TT”), and P=0.273 (“GG” vs “TG”) respectively.
Our association study revealed that, the plausible candidate genes IL4, IL13, IRF1 and UGRP1 gene in chromosomal 5q31 regions might not confer susceptibility to GD. However, those individuals who were TT homozygous at IRF1 6477 T/G locus seemed to be attacked by GD much earlier than others who carry the TG or GG allele.
References
Ban Y, Tomer Y (2003) The contribution of immune regulatory and thyroid specific genes to the etiology of Graves’ and Hashimoto’s diseases. Autoimmunity 36:367–379
Barrett JC, Fry B, Maller J, Daly MJ (2005) Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics 21(2):263–265
Bednarczuk T, Placha G, Jazdzewski K, Kurylowicz A, Kloza M, Makowska U, Hiromatsu Y, Nauman J (2003) Interleukin-13 gene polymorphisms in patients with Graves’ disease. Clin Endocrinol (Oxford) 59:519–525
Briere F, Bridon JM, Servet C, Rousset F, Zurawski G, Banchereau J (1993) IL-10 and IL-13 as B cell growth and differentiation factors. Nouv Rev Fr Hematol 35:233–235
Brix TH, Kyvik KO, Christensen K, Hegedus L (2001) Evidence for a major role of heredity in Graves’ disease: a population-based study of two Danish twin cohorts. J Clin Endocrinol Metab 86:930–934
Burek CL, Hoffman WH, Rose NR (1982) The presence of thyroid autoantibodies in children and adolescents with autoimmune thyroid disease and in their siblings and parents. Clin Immunol Immunopathol 25:395–404
Chomarat P, Banchereau J (1998) Interleukin-4 and interleukin-13: their similarities and discrepancies. Int Rev Immunol 17:1–52
Heinzmann A, Dietrich H, Deichmann KA (2003) Association of uteroglobulin-related protein 1 with bronchial asthma. Int Arch Allergy Immunol 131:291–295
Heward JM, Nithiyananthan R, Allahabadia A, Gibson S, Franklyn JA, Gough SC (2001) No association of an interleukin 4 gene promoter polymorphism with Graves’ disease in the United Kingdom. J Clin Endocrinol Metab 86:3861–3863
Hiromatsu Y, Fukutani T, Ichimura M, Mukai T, Kaku H, Nakayama H, Miyake I, Shoji S, Koda Y, Bednarczuk T (2005) Interleukin-13 gene polymorphisms confer the susceptibility of Japanese populations to Graves’ disease. J Clin Endocrinol Metab 90:296–301
Howard TD, Whittaker PA, Zaiman AL, Koppelman GH, Xu J, Hanley MT, Meyers DA, Postma DS, Bleecker E (2001) Identification and association of polymorphisms in the interleukin-13 gene with asthma and atopy in a Dutch population. Am J Respir Cell Mol Biol 25:377–384
Hummelshoj T, Bodtger U, Datta P, Malling HJ, Oturai A, Poulsen LK, Ryder LP, Sorensen PS, Svejgaard E, Svejgaard A (2003) Association between an interleukin-13 promoter polymorphism and atopy. Eur J Immunogenet 30:355–359
Hunt PJ, Marshall SE, Weetman AP, Bell JI, Wass JA, Welsh KI (2000) Cytokine gene polymorphisms in autoimmune thyroid disease. J Clin Endocrinol Metab 85:1984–1988
Jian Z, Nakayama J, Noguchi E, Shibasaki M, Arinami T (2003) No evidence for association between the −112G/A polymorphism of UGRP1 and childhood atopic asthma. Clin Exp Allergy 33:902–904
Jin Y, Teng W, Ben S, Xiong X, Zhang J, Xu S, Shugart YY, Jin L, Chen J, Huang W (2003) Genome-wide scan of Graves’ disease: evidence for linkage on chromosome 5q31 in Chinese Han pedigrees. J Clin Endocrinol Metab 88:1798–1803
Jujo K, Renz H, Abe J, Gelfand EW, Leung DY (1992) Decreased interferon γ and increased interleukin-4 production in atopic dermatitis promotes IgE synthesis. J Allergy Clin Immunol 90:323–331
Kelso A (1998) Cytokines: principles and prospects. Immunol Cell Biol 76:300–317
Lohoff M, Ferrick D, Mittrucker HW, Duncan GS, Bischot S, Rollinghoff M, Mak TW (1997) Interferon regulatory factor-1 is required for a T helper 1 immune response in vivo. Immunity 6:681–689
Nakao F, Ihara K, Kusuhara K, Sasaki Y, Kinukawa N, Takabayashi A, Nishima S, Hara T (2001) Association of IFN-γ and IFN regulatory factor 1 polymorphisms with childhood atopic asthma. J Allergy Clin Immunol 107:499–504
Nickerson DA, Tobe VO, Taylor SL (1997) PolyPhred: automating the detection and genotyping of single nucleotide substitutions using fluorescence-based resequencing. Nucleic Acids Res 25:2745–2751
Niimi T, Keck-Waggoner CL, Popescu NC, Zhou Y, Levitt RC, Kimura S (2001) UGRP1, a uteroglobin/Clara cell secretory protein-related protein, is a novel lung-enriched downstream target gene for the T/EBP/NKX2.1 homeodomain transcription factor. Mol Endocrinol 15:2021–2036
Niimi T, Munakata M, Keck-Waggoner CL, Popescu NC, Levitt RC, Hisada M, Kimura S (2002) A polymorphism in the human UGRP1 gene promoter that regulates transcription is associated with an increased risk of asthma. Am J Hum Genet 70:718–725
Noguchi E, Shibasaki M, Arinami T, Takeda K, Yokouchi Y, Kawashima T, Yanagi H, Matsui A, Hamaguchi H (1998) Association of asthma and the interleukin-4 promotor gene in Japanese. Clin Exp Allergy 28:449–453
van der Pouw Kraan TC, van Veen A, Boeije LC, van Tuyl SA, de Groot ER, Stapel SO, Bakker A, Verweij CL, Aarden LA, van der Zee JS (1999) An IL-13 promoter polymorphism associated with increased risk of allergic asthma. Genes Immun 1:61–65
Punnonen J, Aversa G, Cocks BG, McKenzie AN, Menon S, Zurawski G, de Waal Malefyt R, de Vries JE (1993) Interleukin 13 induces interleukin 4-independent IgG4 and IgE synthesis and CD23 expression by human B cells. Proc Natl Acad Sci USA 90:3730–3734
Romagnani S (1996) Th1 and Th2 in human diseases. Clin Immunol Immunopathol 80:225–235
Rosenwasser LJ, Klemm DJ, Dresback JK, Inamura H, Mascali JJ, Klinnert M, Borish L (1995) Promotor polymorphisms in the chromosome 5 gene cluster in asthma and atopy. Clin Exp Allergy 25(Suppl 2):74–78
Rozen S, Skaletsky H (2000) Primer3 on the WWW for general users and for biologist programmers. Methods Mol Biol 132:365–386
Sakai K, Shirasawa S, Ishikawa N, Ito K, Tamai H, Kuma K, Akamizu T, Tanimura M, Furugaki K, Yamamoto K, Sasazuki T (2001) Identification of susceptibility loci for autoimmune thyroid disease to 5q31-q33 and Hashimoto’s thyroiditis to 8q23-q24 by multipoint affected sib-pair linkage analysis in Japanese. Hum Mol Genet 10:1379–1386
Seegers D, Borm ME, van Belzen MJ, Mulder CJ, Bailing J, Crusius JB, Meijer JW, Wijmenga C, Pena AS, Bouma G (2003) IL12B and IRF1 gene polymorphisms and susceptibility to celiac disease. Eur J Immunogenet 30:421–425
Tang M, Kemp A, Varigos G (1993) IL-4 and interferon-γ production in children with atopic disease. Clin Exp Immunol 92:120–124
Tomer Y, Davies TF (2003) Searching for the autoimmune thyroid disease susceptibility genes: from gene mapping to gene function. Endocr Rev 24:694–717
Tomer Y, Barbesino G, Keddache M, Greenberg DA, Davies TF (1997) Mapping of a major susceptibility locus for Graves’ disease (GD-1) to chromosome 14q31. J Clin Endocrinol Metab 82:1645–1648
Vandenbroeck K, Hardt C, Louage J, Fiten P, Jackel S, Ronsse I, Epplen JT, Grimaldi LM, Olsson T, Marrosu MG, Billiqu A, Opdenakker G (2000) Lack of association between the interferon regulatory factor-1 (IRF1) locus at 5q31.1 and multiple sclerosis in Germany, northern Italy, Sardinia and Sweden. Genes Immun 1:290–292
Vyse TJ, Todd JA (1996) Genetic analysis of autoimmune disease. Cell 85:311–318
Walley AJ, Cookson WOCM (1996) Investigation of an interleukin-4 promoter polymorphism for associations with asthma and atopy. J Med Genet 33:689–692
Weetman AP (2003) Autoimmune thyroid disease: propagation and progression. Eur J Endocrinol 148:1–9
Author information
Authors and Affiliations
Corresponding author
Additional information
Huang Wei and Teng Weiping equally contributed to this work
Rights and permissions
About this article
Cite this article
Yang, Y., Lingling, S., Ying, J. et al. Association study between the IL4, IL13, IRF1 and UGRP1 genes in chromosomal 5q31 region and Chinese Graves’ disease. J Hum Genet 50, 574–582 (2005). https://doi.org/10.1007/s10038-005-0297-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10038-005-0297-x
Keywords
This article is cited by
-
Genetic Associations of Interleukin-related Genes with Graves’ Ophthalmopathy: a Systematic Review and Meta-analysis
Scientific Reports (2015)
-
The association of SCGB3A2 polymorphisms with the risk of Graves’ disease: a meta-analysis
Endocrine (2014)
-
Variation in Uteroglobin-Related Protein 1 (UGRP1) gene is associated with Allergic Rhinitis in Singapore Chinese
BMC Medical Genetics (2011)
-
The −112G > A polymorphism of the secretoglobin 3A2 (SCGB3A2) gene encoding uteroglobin-related protein 1 (UGRP1) increases risk for the development of Graves’ disease in subsets of patients with elevated levels of immunoglobulin E
Journal of Applied Genetics (2011)
-
Association analysis of polymorphisms in IL-3, IL-4, IL-5, IL-9, and IL-13 with Graves’ disease
Journal of Endocrinological Investigation (2010)