Abstract
Chromosome 5, especially the 5q31-33 region, may contain one or more loci to control total serum IgE as well as asthma and bronchial hyperresponsiveness. To investigate the regions related with IgE level in chromosome 5, we performed a case-control association study on 105 high-IgE-level and 85 normal-IgE-level asthmatic children using 43 microsatellite markers that span the whole chromosome 5 with 5 cM intervals. One of microsatellite marker, D5S2011, had significantly different allele frequency between the two asthmatic groups. E allele (143 bp) of the D5S2011 marker was more frequent in high-IgE asthmatics. CD14 is the candidate gene of atopy and asthma and is distant from D5S2011 by about 1 Mb. We analyzed the SNP genotypes in the CD14 gene region alone and in combination with microsatellite marker D5S2011. The CD14/−2984 polymorphism but not the CD14/−159 is associated with IgE level in Taiwanese asthmatic children. The CD14/−159 allele was observed only to be associated with IgE level when −159T was part of a haplotype containing a D5S2011 E allele. The combination analysis using SNP and STRP markers provided a novel method for increasing detection power in candidate gene association studies.
Similar content being viewed by others
Introduction
Associating phenotypes with genotypes is the fundamental aim of genetics. Linkage analysis and association analysis are the two principal experimental strategies commonly used by researchers to identify genetic changes underlying disease etiology. It is likely that linkage analysis yielded success in the Mendelian diseases because they are characterized by a string correlation between the genotype and phenotype of a single gene. However, this type of correlation may not exist in most common diseases, which often skip generations without a clear inheritance pattern. Risch and Merikangas (1996) and Altshuler et al. (2000) promoted the concept of case-control studies using single nucleotide polymorphisms (SNPs) in candidate genes. However, this strategy lacks a number of advantages that the linkage strategy may bring to the dissection of complex traits.
In general, common diseases are diagnosed on the basis of consensus criteria that are often rooted in clinical experience but may not reflect the biology of the disease. Although, subclassification of the phenotypes may provide more precise clinical definition, it does not enhance the level of correlation between genotype and phenotype. Indeed, different subphenotypes within a family may represent the variable expressiveness of a particular gene, in which case fractionation will only decrease power. Although the genealogic approach may represent a more useful method for dissection of complex traits, it may only apply in isolated populations, such as in Iceland (Gulcher et al. 2001).
Genetic association studies by the candidate gene approach are more popular than a genome-wide scan due to genotyping cost and feasibility in statistical analysis. Unfortunately, related literature is teeming with reports of association studys that either cannot be replicated or for which corroboration by linkage cannot be established. (Terwillger and Weiss 1998; Gambaro et al. 2000; Weiss and Terwilliger 2000). Explanations for this lack of reproducibility include poor study design, incorrect assumptions on the underlying genetic architecture, and overinterpretation of data (Cardon and Bell 2001). Another important issue is the degree to which populations differ in terms of genetic susceptibility and linkage disequilibrium (LD) structure and therefore the need to sample from different population groups in disease studies. Owing to historical isolation, major racial/ethnic groups differ in allele frequencies (Stephens et al. 2001; Calafell et al. 1998). These differences are highly significant, especially for less common alleles; often, a particular allele was found in one population at modest frequency and missing from others (Stephens et al. 2001).
The association between total serum IgE levels and CD14/−159 genotype reached statistical significance only among non-Hispanic white children (Baldini et al. 1999). The result seems to be consistent with reports from the Collaborative Study of the Genetics of Asthma, which indicated that linkage between markers in chromosome 5q and asthma was present among non-Hispanic whites but not Hispanic subjects. Similar studies were applied to many racial/ethnic groups, such as subjects in the Czech (Buckova et al. 2003), Dutch, (Koppelman et al. 2001) and Hong Kong Chinese (Leung et al. 2003) populations, which showed association between IgE level and CD14/−159 polymorphism, but not in German (Sengler et al. 2003) and Polish (Lis et al. 2001) populations. Not surprisingly, these inconsistent data suggest the CD14/−159 polymorphism may only be one of many mechanisms involved in IgE regulation.
In this study, we report that the CD14/−2984 polymorphism but not the CD14/−159 polymorphism is associated with IgE level in Taiwanese asthmatic children. Our results indicated that CD14/−159 was only associated with IgE level when −159T was part of a haplotype containing a specific D5S2011 allele (D5S2011, 143 bp) and not the other D5S2011 alleles. These unexpected findings suggest that identifying subject subgroup of genetic homogeneity in the small chromosome region could enhance statistical power for genetic association study. Our approach using combination analysis of SNP and STRP markers may provide a new strategy for association studies.
Material and methods
Clinical sample collection
Asthmatic patients and age-matched controls were divided by their serum IgE levels into high IgE group (IgE value= OR >1,000 IU/ml), and normal IgE group (IgE value= OR < 200 IU/ml) for an association study for candidate genes responsible for serum IgE production. A total of 190 subjects that included 105 high-IgE-level and 85 low-IgE-level asthmatic children were collected during 1988–2001 at the National Cheng-Kung University Hospital, Tainan, Taiwan, a regional referral center for patients with asthma and other airway obstruction diseases. Patients who had symptomatic asthma without current asthma exacerbation were referred to this hospital and were studied for a standardized, complete evaluation at the outpatient clinic. At the time of initial testing, all subjects had asthma symptoms, were hyperresponsive to histamine (PC20 forced expiratory volume in 1 s [FEV1] ≥32 mg of histamine/ml, 30-s method), and were <16 years old.
This study was approved by the Medical Ethics Committee of the National Cheng-Kung University Hospital. All participants or their guardians signed the written informed-consent documents.
Clinical evaluation
Our study population consisted of asthmatic children aged between 1 and 16 years old, and the study protocol was approved by Ethical and Clinical Trial Committee of the National Cheng-Kung University Hospital. All participants or their guardian, after being well-informed of the study protocol, signed consent forms; answered a modified British Medical Society respiratory questionnaire that is exactly the same as the European Community Respiratory Health Survey (ERCHS), which has the similar validity as ISAAC (Burney et al.1994; Pearce et al. 2000); and answered additional questions pertinent to the diagnosis and assessment of asthma. Pulmonary function was tested using standard methods that included spirometry before and after the administration of two puffs of inhaled salbutamol (200 μg/puff). The conventional definition of asthma includes: (A) history of wheezing and shortness of breath during or without concurrent respiratory infections; (B) chronic cough for more than 1 month and being diagnosed by a physician as the presence of wheezing episode(s); and (C) bronchodilator test has confirmed the positive response of increased 15% of FEV1. Other evaluations included skin-prick tests for responsiveness to six common aeroallergens, a differential blood count (including total eosinophil count), and measures of total serum IgE, as well as IgE specific to house dust and mixed pollens using the Unicap system (Pharmacia, Diagnostic, Sweden). A positive skin test was defined as the presence of ≤1 reaction with a wheal diameter ≤5 mm. Total serum IgE was measured by solid-phase immunoassay (Pharmacia IgE EIA; Pharmacia Diagnostics). The confirmed asthmatic children were further divided by their serum IgE levels into the high IgE group (105 subjects) that had high IgE serum concentrations (>1,000 IU/ml) and were sensitized to at lease one aeroallergen, and the normal IgE group (85 subjects) with IgE serum levels within normal range (≤200 IU/ml) with or without detectable allergen sensitization.
DNA preparation
Genomic DNAs were extracted from 190 asthmatic Taiwanese children. DNA was isolated from blood samples using a QIAamp DNA Blood kit (QIAGEN) according to the manufacturer’s instructions. The isolated genomic DNA was quality check by agarose gel electrophoresis analysis, quantified spectrophotometrically, and stored at −80°C.
Microsatellite genotyping
Genotyping was performed using part of ABI PRISM linkage mapping sets HD-5, which were to provide coverage of chromosome 5 at 5 cM average resolution. Each marker set included a fluorescence-labeled forward primer and a tailing reverse primer. The PCR amplifications were carried out according to the manufacturer’s instructions. PCR products were separated on ABI 3700 DNA analyzers. The use of GeneScan 500 LIZ as the internal size standard assisted polymorphic fragment length calling and allowed more accurate allele calling and unambiguous comparison of data across experimental conditions. Genotypes were initially scored using Genescan and Genotyper (ABI) software and were then verified independently by three individuals without prior knowledge of phenotype.
SNP Genotyping
DNA fragment flanking the genomic region of the CD14 gene was amplified by an ABI 2700 PCR machine using two pairs of forward and reverse primers. The sequence and related information of primers are listed at Table 1. The fragments of PCR product were sequenced by ABI 3700 automatic sequencer according the manufacturer’s protocol. Sequence data were analyzed by PolyPhred software to identify the potentially candidate SNPs. The potential SNPs were manually checked to ensure the presence of a true SNP and the allele of each individual. Three independent manual confirmations were performed for all sequence data, and only those confirmed data were subjected to the subsequent statistical analysis.
Statistical analysis
To evaluate the association between STR markers and asthmatics, χ2 tests were sought with significant level estimated by Monte-Carlo simulation (Sham and Curtis 1995). The goal was to find out if the allele frequencies or the genotype frequencies of STR markers are different between IgE-high and IgE-normal asthmatics. For finding out the relationship between IgE level and SNPs in CD14, association evaluation and haplotype analysis were performed by χ2 tests and Fisher’s exact tests. The haplotype analysis included allele frequency and genotype frequency. All statistical analysis was performed by SAS. The haplotypes were constructed by the Phase program downloaded from the Web site (http://www.stat.washington.edu/stephens/software.htmll).
Results
D5S2011 E alleles were associated with IgE level
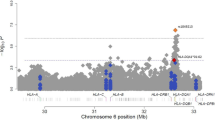
Forty-three STR markers of chromosome 5 were genotyped and analyzed. Shown in Fig. 1 is the result from the screening for loci associated with the difference between high-IgE and normal-IgE asthmatics. The highest peak was located at marker D5S2011. Allele frequencies of D5S2011 in two groups are significantly different (Table 2). The 143 bp allele, herein after defined as E allele, and genotype containing E allele, was statistically significantly more frequent in the high IgE group than the normal IgE group (Table 3). CD14, one of candidate genes for regulating IgE, was near the marker D5S2011. The physical distance between CD14 and D5S2011 was about 1.1 MB (ref: NCBI human map view, build34 version3).
Results of chromosome 5 screen for loci associated with asthmatics with the high-IgE and normal-IgE groups. Tested loci (STR markers) are arranged by number from p terminus to q terminus in chromosome 5. The x axis is the STR marker’s genetic location on chromosome 5 from p terminus to q terminus. The y axis is a natural logarithm of the χ2 test by Monte-Carlo simulation p value. y=3 is almost equal to p=0.05
CD14/−2984 polymorphisms were associated with IgE level
Five SNPs selected for this study were evenly distributed within the 5-kb region from upstream 3 kb to downstream near exon in CD14 the gene region. Although the SNPs at positions −2984 and −159 had a similar allele frequency, only the SNP at −2984 showed significant difference between the two patient groups (Table 4). The other three SNPs, −2738, −2451, and 1442. also had similar allele frequencies, but none of them showed any difference between groups. The absolute value of D′ of each two of five SNPs were all greater than 0.92 (data not shown). This result suggested that the regions studied were in one LD block.
Haplotypes that contained the D5S2011 E allele were associated with IgE level
Haplotypes of five SNPs tested in this study were reconstructed by the Phase program. The frequencies of haplotypes constructed by SNPs data alone did not bear significant difference between the two groups (data not shown). To enhance the analysis power, we further subgrouped the data for alleles of the D5S2011 locus into E and non-E categories and combined them with the SNP data to construct the haplotype. The E–TCCTG haplotype and related genotype were more frequent in the high-IgE group than the normal-IgE group and reached a statistically significant difference (Table 5).
CD14/−159 polymorphism was associated with IgE level only when subjects contain the D5S2011 E allele
Based on haplotype analysis, we could dissect the haplotypes of studied subjects into contained or noncontained E allele. When the E allele was part of the haplotype, the −159 showed different allelic frequencies between the two groups (Table 6). As Table 6 indicates, the −2984 polymorphisms were the same as −159 polymorphisms, but the other three SNPs were still not shown to have different allele frequencies between the two groups.
Allele counts of E–TCCTG, E–non-TCCTG, non-E–TCCTG, and non-E–non -TCCTG were 71, 24, 154, and 131, respectively. We assigned E as A, non-E as a, TCCTG as B, and non-TCCTG as b. Based on the equation D′= (pabpAB-paBpAb)/(pabpAB+paBpAb) (Maynard 1989), the D′ of D5S2011 and the haplotype of CD14 was 0.43.
Discussion
The data we provided suggest that the CD14/−2984 polymorphism were associated with IgE level in Taiwanese asthmatic children while the CD14/−159 polymorphism—the SNP of a previous report on non-Hispanic whites—was not associated with IgE level in our study. The D5S2011 STR markers had different allele frequencies between high-IgE and normal-IgE asthmatics, and the E allele (−143 bp) appeared to be more frequent in high-IgE subjects. Schulze et al. (2002) proposed a D′ value greater than 0.3 as the threshold of useful levels of LD for association studies. The D′ value 0.43 of linkage disequilibrium for association existing between the D5S2011 and CD14 haplotypes thus were considered a useful level. The CD14/−159 polymorphism was associated with IgE level when it was part of a haplotype containing a specific D5S2011 E allele.
Previous studies in different populations have reported associations between the CD14/−159C allele and elevated IgE levels (Baldini et al. 1999; Koppelman et al. 2001; Leung et al. 2003). However, in Hutterites, a highly inbred population, CD14/−159T was shown to be associated with atopy only when the CD14/−159T allele shared the same haplotype with the D5S642 185 bp allele (Ober et al. 2000). Furthermore, linkage but not association between CD14/−159 polymorphism and atopy had been reported (Walley et al. 2001). These conflicting results suggest that the CD14/−159 and the putative susceptibility variant are in the same LD block in some study subjects but not in all populations. Another possibility for inconsistent results from different studies is that association with IgE level should be a specific haplotype (Baldini et al. 2000; Vercelli et al. 2001a) or combination of genotypes (Vercelli et al. 2001b). That is, not only one but the combination of more than one SNPs in the CD14 promoter affects serum IgE concentration.
Theoretically, STR markers carry more power to detect LD than diallelic markers such as SNP (Ott and Rabinowitz 1997; Chapman and Wijsman 1998). The microsatellite markers, if sufficiently dense, can be used to make initial predictions about the level of short-range LD present in susceptibility regions identified by linkage study (Schulze et al. 2002). Though 5 cM resolution was not sufficiently dense for whole chromosome 5, the screen by commercially available STR markers was successful, hitting the region associated with allergy and aiding the search for the candidate genes. The D5S2011 marker, located in chromosome 5q31, is distant from the CD14 gene around 1.1 MB (ref: NCBI human map view, build34 version3). In our study subjects, for some unknown reason, linkage disequilibrium exists between the specific D5S2011 E allele and the specific haplotype of the CD14 gene region. However, the result seems to suggest multiallelic loci, such as STR or haplotype, developed long-range linkage disequilibrium, which was useful for association studies.
The case-control studies suffer in the case of locus heterogeneity and allelic heterogeneity. The number of genes involved in the pathogenesis of each common disease is large, with each gene producing only a small-to-modest effect. Finding a relatively genetic homogeneous subgroup in the outbred population should be a useful approach to avoid some mapping problem, such as lost of statistical power resulting from locus and allelic heterogeneity. The genealogy built in the outbred population has never been proved to be useful in association studies due to locus and allelic heterogeneity introduced by outbreading. The alternative approach is finding local genetically homogeneous populations base candidate gene association studies. The genetic homogeneity may exist in small chromosome regions that were passed down from the same ancestry or founder. The highly polymorphic STR markers could distill out this type of genetic homogeneity and thus further reduce the disturbance of allelic heterogeneity and locus heterogeneity for association studies.
CD14/−159 T was associated with high IgE phenotype and showed same allele frequency with CD14/−2984 T after the alleles of subjects were further subgrouped by the E allele of STR marker D5S2011. It is possible that the D5S2011 E allele was simply linked to a yet-to-be-identified susceptibility locus. On the other hand, the D5S2011 E allele may represent another gene, which interacts with CD14 and co-contributes to total IgE concentration in serum. FGF1 is also distant from D5S2011 within 1 Mb (ref: NCBI human map view, build34 version3). Although we have not yet identified significant SNPs in exons or promoters associated with IgE level (data not shown), several intron SNPs of FGF1 were shown to have significant differences in allele frequency between high-IgE and normal-IgE asthmatic groups. This result suggests that D5S2011 extended the linkage disequilibrium to those intron SNPs without any functional implication.
In summary, we performed a genome screening using STR markers in different IgE-level asthmatics and confirmed that long-distance linkage disequilibrium developed by multiallelic markers is useful for association studies. Furthermore, the results provided a method for identifying subject subgroups of genetic homogeneity in the small chromosome region to enhance statistical power for genetic association studies.
References
Altshuler D, Hirschhorn JN, Klannemark M, Lindgren CM, Vohl MC, Nemesh J, Lane CR, Schaffner SF, Bolk S, Brewer C, Tuomi T et al (2000) The common PPAR-γ Pro12Ala polymorphism is associated with decreased risk of type 2 diabetes. Nat Genet 26:76–80
Baldini M, Lohman IC, Halonen M, Erickson RP, Holt PG, Martinez FD (1999) A polymorphism in the 5′ franking region of the CD14 gene is associated with circulating soluble CD14 levels and with total serum immunoglobulin E. Am J Respir Cell Mol Biol 20:976–983
Baldini M, Kabesch M, Graves PE, Erickson RP, Vercelli D, Martinez FD (2000) Detection of four novel polymorphisms in the CD14 promoter and associations of their haplotypes with total serum IgE levels. Am J Respir Crit Care Med 161:A928
Buckova D, Holla LI, Schuller M, Znojil V, Vacha J (2003) Two CD14 promoter polymorphisms and atopic phenotypes in Czech patients with IgE-mediated allergy. Allergy 58:1023–1026
Burney PG, Luczynska C, Chinn S, Jarvis D (1994) The European Community Respiratory Health Survey. Eur Respir J 5:954–960
Calafell F, Shuster A, Speed WC, Kidd JR, Kidd KK (1998) Short tandem repeat polymorphism evolution in humans. Eur J Hum Genet 6:38–49
Cardon LR, Bell JI (2001) Association study designs for complex diseases. Nat Rev Genet 2:91–99
Chapman NH, Wijsman EM (1998) Genome screens using linkage disequilibrium tests: optimal marker characteristics and feasibility. Am J Hum Genet 63:1872–1885
Gambaro G, Anglani F, D’ Angelo A (2000) Association studied of genetic polymorphisms and complex disease. Lancet 355:308–311
Gulcher JR, Kong A, Stefansson K (2001) The role of linkage studies for common diseases. Curr Opin Genet Dev 11:264–267
Koppelman GH, Reijmerink NE, Stine OC, Howard TD, Whittaker PA, Meyers DA, Postma DS, Bleecker ER (2001) Association of a promoter polymorphism of the CD14 gene and atopy. Am J Respir Crit Care Med 163:965–969
Leung TF, Tang NL, Sung YM, Li AM, Wong GW, Chan IH, Lam CW (2003) The C−159T polymorphism in the CD14 promoter is associated with serum total IgE concentration in atopic Chinese children. Pediatr Allergy Immunol 14:255–260
Lis G, Kostyk E, Sanak M, Pietrzyk JJ (2001) Molecular studies in a population of children with bronchial asthma. I. Polymorphism in the promoter region of gene CD14. Pneumonol Alergol Pol 69:265–272
Maynard SJ (1989) Evolutionary genetics. Oxford University Press, Oxford
Ober C, Tsalenko A, Parry R, Cox NJ (2000) A second-generation genomewide screen for asthma-susceptibility alleles in a founder population. Am J Hum Genet 67:1154–1162
Ott J, Rabinowitz D (1997) The effect of marker heterozygosity on the power to detect linkage disequilibrium. Genetics 147:927–930
Pearce N, Sunyer J, Cheng S, Chinn S, Bjorksten B, Burr M, Keil U, Anderson HR, Burney P (2000) Comparison of asthma prevalence in the ISAAC and the ECRHS. ISAAC Steering Committee and the European Community Respiratory Health Survey. International Study of Asthma and Allergies in Childhood. Eur Respir J 16:420–426
Risch N, Merikangas K (1996) The future of genetic studies of complex human diseases. Science 273:1516–1517
Schulze TG, Chen YS, Akula N, Hennessy K, Badner JA, McInnis MG, DePaulo JR, Schumacher J, Cichon S, Propping P, Maier W, Rietschel M, Nothen MM, McMahon FJ (2002) Can long-range microsatellite data be used to predict short-range linkage disequilibrium? Hum Mol Genet 11:1363–1372
Sengler C, Haider A, Sommerfeld C, Lau S, Baldini M, Martinez F, Wahn U, Nickel R, German Multicenter Allergy Study Group (2003) Evaluation of the CD14 C−159T polymorphism in the German Multicenter Allergy Study cohort. Clin Exp Allergy 33:166–169
Sham PC, Curtis D (1995) Monte Carlo tests for associations between disease and alleles at highly polymorphic loci. Ann Hum Genet 59:97–105
Stephens JC, Schneider JA, Tanguay DA, Choi J, Acharya T, Stanley SE, Jiang R, Messer CJ, Chew A, Han JH et al (2001) Haplotype variation and linkage disequilibrium in 313 human genes. Science 293:489–493
Terwillger JD, Weiss KM (1998) Linkage disequilibrium mapping of complex disease: fantasy or reality? Curr Opin Biotechnol 9:578–594
Vercelli D, Baldini M, Martinez FD (2001a) The monocyte/IgE connection: may polymorphisms in the CD14 gene teach us about IgE regulation? Int Arch Allergy Immunol 124:20–24
Vercelli D, Baldini M, Stern D, Lohman IC, Halonen M, Martinez F (2001b) CD14: a bridge between innate immunity and adaptive IgE responses. J Endotoxin Res 7:45–48
Walley AJ, Wiltshire S, Ellis CM, Cookson WOCM (2001) Linkage and allelic association of chromosome 5 cytokine cluster genetic markers with atopy and asthma associated traits. Genomics 72:15–20
Weiss KM, Terwilliger JD (2000) How many diseases does it take to map a gene with SNPs?. Nat Genet 26:151–157
Acknowledgements
We thank Drs. Chunlin Su and Dennis Chen for advice, Dr. Monica Shian-Jy Bey for sample collection, and Julia Kuei-Ting Yu and Jiayu Huang for data management. This study was funded by an internal research grant at Vita Genomics, Inc.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Wang, JY., Wang, LM., Lin, C.GJ. et al. Association study using combination analysis of SNP and STRP markers: CD14 promoter polymorphism and IgE level in Taiwanese asthma children. J Hum Genet 50, 36–41 (2005). https://doi.org/10.1007/s10038-004-0215-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10038-004-0215-7
Keywords
This article is cited by
-
An Association Study of 13 SNPs from Seven Candidate Genes with Pediatric Asthma and a Preliminary Study for Genetic Testing by Multiple Variants in Taiwanese Population
Journal of Clinical Immunology (2009)
-
Association of CD14 promoter polymorphisms and soluble CD14 levels in mite allergen sensitization of children in Taiwan
Journal of Human Genetics (2006)