Abstract
Factors predisposing to the phenotypic features of higher total cholesterol (T-Cho) have not been clearly defined. Here we report an association between a C/T single nucleotide polymorphism at IVS17+8 in the inter-alpha-trypsin inhibitor heavy chain 4 gene (ITIH4) and plasma total cholesterol levels in 351 adult individuals from an east-central area of Japan. Age and gender-adjusted levels of plasma T-Cho, LDL-cholesterol, triglyceride, and HDL-cholesterol were analyzed. When we separate the subjects into two genotypic groups regarding this single nucleotide polymorphism (SNP), those who lack the T-allele had significantly higher plasma T-Cho levels than the others who bear T-allele (mean 252.3 mg/dl versus 241.7 mg/dl; p=0.009). Of the 309 individuals without the T-allele, approximately 90% presented with hypercholesterolemia, whereas only 10% were hypercholesterolemic among 42 individuals with the T-allele (p <0.0001). These data suggest that genetic variation at ITIH4 locus is one of the likely candidate determinants for plasma cholesterol metabolisms.
Similar content being viewed by others
Introduction
Accumulating evidences derived from clinical, epidemiological, and experimental studies suggest that lipid and lipoprotein concentrations in plasma reflect the influence of complex genetic loci, at least in part (Zannis and Breslow 1985); defects are known only for some classical types of hyperlipoproteinemia that affect members of families following Mendelian traits. However, the genetic mechanisms responsible for most types of familial dyslipoproteinemia appear to be complex, not monogenic.
Hyperlipoproteinemia is one of the most important predictors of cardiovascular diseases determined by genetic risk factors as well as environmental factors (Hegele 2001; Cullen et al. 1997). Clarification of the genetic risk factors is essential for diagnosis, prevention, and early effective treatment of hyperlipidemias. In addition, if the relevant genes were identified, the pathogenesis of these diseases would be explained by the variation of those genes or adjacent genes. Several gene polymorphisms have previously been associated with plasma lipoprotein abnormalities (reviewed in Goldstein et al. 1995). However, those are only a part of the all determinants, obviously.
ITIH4 is a glycoprotein mainly expressed in the liver tissue (Nishimura H et al. 1995). It belongs to the inter-α-trypsin inhibitor family of serine protease inhibitors (ITIH1, 2, 3, and 4), which have diverse functions as antiapoptotic and matrix-stabilizing molecules important throughout the development. In addition, ITIH4 gene locates in chromosomal region 3p21.1-p22 were recently described as one of the candidate quantitative trait locus (QTL) for dyslipidemias (Yuan et al. 2000). Expression in the liver, the role as a protease inhibitor, and the location in the candidate QTL region constituted rationales for selecting this gene as a candidate to study the correlation between genetic variation and lipoprotein profile.
In the course of serial investigations of population genetics for hyperlipoproteinemia in a cohort of an area located in east-central Japan, we recognized a correlation between the plasma total cholesterol (T-Cho) levels and a variation of the inter-alpha-trypsin inhibitor heavy chain 4 (ITIH4) gene. Here we focused on the analysis of the potential effect of genetic variation in ITIH4, investigating the correlation of plasma lipoprotein profile with the genetic variation of this gene.
Subjects and methods
Subjects
Subjects were obtained from participants of a cohort study that was originally carried out concurrently with health-check screening in an area in east-central Japan, as described in detail previously (Fujita et al. 2003). In brief, 22,228 participants were initially screened, and hypercholesterolemic individuals whose LDL-cholesterol (LDL-C) levels were higher than 140 mg/dl (LDL-C ≥140 mg/dl) were selected. To the present study, 351 individuals were recruited who gave their written informed consent prior to the study, which was approved by the Institutional Review Boards of the Research Consortium. Physical and clinical profiles of these subjects (ages, male to female ratios, body mass indices, and plasma lipoprotein and lipid levels) are indicated in Table 1. None of the selected participants had medical complications or was undergoing treatment for conditions known to affect plasma lipoprotein levels, such as pituitary disease, hypothyroidism or hyperthyroidism, diabetes mellitus, liver disease, and renal disease. None was receiving antihyperlipidemic therapy. The selected participants visited lipid clinics for detailed examination of their lipoprotein profile and other clinical parameters. Blood was collected after 12–16 h of fasting. Genomic DNA was extracted, as previously described (Yoshida et al. 2002).
Measurement of lipoproteins
Lipid and lipoprotein concentrations were measured by procedures described previously (Hattori et al. 2002); i.e., plasma cholesterol and triglyceride concentrations were assayed enzymatically, and concentrations of HDL-cholesterol (HDL-C) were determined by the MgCl2-dextran precipitation method. LDL-C concentration was calculated by subtracting HDL-C level from the fraction of both LDL-C and HDL-C, as described elsewhere (Ishii et al. 2002).
Selection criteria of single nucleotide polymorphisms (SNPs)
We focused on all SNPs that either accompanies amino acid substitution, SNPs adjacent to the exon/intron junction, and SNPs that locate in the putative promoter region in the entire ITIH4 gene. The complete list of such SNPs in Table 2 describes nucleotide variation, amino acid variation, location, reference database (JSNP and dbSNP), allele frequency, and heterozygosity in the population. We examined SNPs whose percent-heterozygosity exceeded 10% to obtain substantial statistic analytical power. Thus, out of the six SNPs listed in Table 2, IVS17+8C/T and IVS20+8A/G were selected for the present study.
Genotyping for SNP in the ITIH4 gene
PCR amplification of the flanking sequence for the polymorphism IVS17+8C/T and IVS20+8A/G at the ITIH4 locus was performed using a condition described previously (Iida et al. 2002). The surrounding sequence of the SNP as well as the primer sequences and the experimental condition were obtained from the published reports (Haga et al. 2002). SNP genotyping was performed by Invader assay (Third Wave Technologies. Madison, WI, USA) (Saito et al. 2002) using PCR products of the flanking sequence and the probes designed and synthesized by the supplier (Ohnishi et al. 2001).
Statistical analysis
Plasma levels of lipoproteins were adjusted by gender and ages of the subjects using standard data obtained from 11,994 individuals of a 2001 cohort study of the general Japanese population. Coefficients of skewness and kurtosis were calculated to test deviation from a normal distribution. Because the clinical and biochemical traits in each genotypic group did not always distribute normally, we applied a nonparametric Mann-Whitney test or an analysis of variance (ANOVA) with linear regression analysis as a post hoc test (p <0.05) to compare those traits among groups divided by a single SNP (Fujiwara et al. 2002). Fisher’s exact test was used to compare differences in the prevalence of hypercholesterolemia among the population. Chi-square tests were invoked to detect Hardy-Weinberg equilibrium.
Results
To carry out a correlation analysis of the potential effect of SNPs, IVS17+8C/T and IVS20+8A/G, 351 individuals were selected from participants for a cohort of an area initially screened by criteria that defined individuals harboring hyperlipidemic risks. Because of initial screening for the health-check assessment, basal level of each value was a little higher than that of data from the general Japanese population. However, the average differences were less than 0.5×SD in general. When the subjects were genotypically categorized into three groups for each SNP (for instance, three homozygous minor T-allele carriers, 39 heterozygous carriers and 309 homozygous C-allele carriers, for IVS17+8C/T), no deviation of genotype frequencies from Hardy-Weinberg equilibrium was observed (p=0.38, ϰ2-test).
Distribution and mean values of T-Cho, TG, LDL-C and HDL-C were analyzed among the genotypically divided groups by each SNP, and significant difference in T-Cho and HDL-C was detected among the three groups divided by IVS17+8C/T genotypes. Plasma T-Cho levels of homozygous minor T-allele carriers (n=3), heterozygous carriers (n=39), and homozygous C-allele carriers (n=309) were 200.4±39.7 mg/dl, 244.9±19.5 mg/dl, and 252.3±23.5 mg/dl respectively, indicating a codominant T-Cho lowering effect of the minor T-allele (r=0.18, p=0.0006). Similarly, plasma HDL-C levels were 31.1±6.9 mg/dl, 48.1±10.2 mg/dl, and 51.5±12.6 mg/dl, respectively, indicating a codominant HDL-C lowering effect of the minor T-allele (r=0.15, p=0.006). No statistically significant difference was detected in the plasma lipoprotein levels among the groups categorized by IVS20+8A/G, suggesting presence of a break of haplotype blocks within the gene (data not shown).
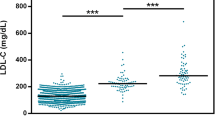
We repeatedly evaluated the genotypic effect by categorization of the subjects into two groups, those who lack the T-allele (C/C) and those who bear at least one T-allele (C/T and T/T), since the homozygous subjects carrying the minor allele was rare (n=3). By this categorization, the former subjects had significantly higher plasma T-Cho levels than the latter (mean±SD 252.3±23.5 mg/dl versus 241.7±23.8 mg/dl; p=0.009) (Fig. 1A). Similarly, the former subjects had significantly higher HDL-C levels than the latter (mean±SD 51.5±12.6 mg/dl versus 46.9±10.9 mg/dl; p=0.014).
Comparison of plasma total cholesterol (T-Cho) levels among genotypically determined groups according to the ITIH4 variation. A Adjusted plasma T-Cho levels were compared between genotypic groups with or without the ITIH4 IVS17+8 T-allele (p=0.009). Open circles represent mean values. Error bars represent the standard deviation (SD). B Distribution analysis of hypercholesterolemic patients among ITIH4 IVS17+8C/T classified-genotypes; T-allele (+)/(-). The 2×2 table was analyzed by Fisher’s exact test (p<0.0001). High T-Cho phenotypes were defined by adjusted plasma T-Cho levels above the 220 mg/dl
Since the data presented here suggested an involvement of ITIH4 locus in the expression of the hypercholesterolemic phenotype in these populations, we correlated the manifestation of the hypercholesterolemia with respect to presence or absence of T-alleles of ITIH4 IVS17+8C/T. Hypercholesterolemia was defined as plasma cholesterol level above the reference values after age and gender adjustment (T-Cho >220 mg/dl). This criterion is based on a recommended preclinical level of hyperlipidemic individuals who have to be alerted for high risk for ischemic heart disease. It classified 291 subjects as having hypercholesterolemia among the study population consisting of 351 individuals. Of the 309 individuals lacking the T-allele, 286 presented with hypercholesterolemia (93%) whereas only five did so among 42 individuals with T-allele (12%). Distribution difference was significant when tested by Fisher’s exact test (p <0.0001; Relative risk = 2.44; 95% CI: 1.80–3.30) (Fig. 1B). As to plasma HDL-C, the distribution analysis (low HDL <40 mg/dl versus high HDL ≥ 40 mg/dl) was not significantly deviated.
Correlation analysis of a population of 108 normolipidemic subjects from the same geographical area that we carried out preliminarily did not reach statistical significance (data not shown), suggesting that the ITIH4 may exert its modifying effect only in the hypercholesterolemic metabolic state. A subsequent, large-scale cohort study is obviously necessary to clarify whether or not the ITIH4 effect is also present in normolipidemic populations.
Discussion
Hypercholesterolemia is one of the important risk factors of ischemic heart disease (Matsuzawa 1995; Norioka 2000). In the present study, we showed a correlation between hypercholesterolemia and homozygosity for the C-allele of IVS17+8 C/T SNP in the ITIH4 gene. An association study revealed an elevating effect of plasma T-Cho of the homozygosity for the C-allele. The high prevalence (93%) of hypercholesterolemia (mean±SD 252.3±23.5 mg/dl) among the individuals with this genotype (C/C) is in contrast to the scarceness (12%) of hypercholesterolemia (mean±SD 244.9±19.5 mg/dl and 200.4±39.7 mg/dl) among the rest of the study subjects (C/T and T/T). Our results indicated that ITIH4 variation might modify the lipoprotein phenotype of plasma lipoprotein metabolism.
ITIH4 belongs to the inter-α-trypsin inhibitor family of serine protease inhibitors, which have diverse functions as an antiapoptotic and matrix-stabilizing molecule important throughout the development. Four genes, designated ITIH1, 2, 3 and 4, are involved in the synthesis of inter-α-trypsin inhibitor family members. All four heavy chain precursors and the resulting mature heavy chains harbor a so-called von Willebrand type-A (vWA) domain (Chan P et al. 1995, Bork P et al. 1991). Only the heavy chain 4 precursor polypeptide harbors a plasma Kallikrein-released bradykinin-like domain on its C-terminal sequence (Nishimura H et al. 1995) and a domain with some similarity to the ATP-dependent proteases (Saguchi K et al. 1995). Although the suspected link between functional changes of ITIH4 protein and plasma T-Cho metabolism was not a commonly expected one, it may have unidentified functions in regulation of cholesterol synthesis and catabolism in the liver.
The IVS17+8C/T SNP locates close to the splice junction for the donor site of exon 17 of the ITIH4 gene. Due to this polymorphic sequence near the splice junction, the ITIH4 gene may have differential splicing that results in production of distinct ITIH4 isoforms in some organs. This possibility may need to be investigated by functional studies in the future. In addition, possibilities that this SNP marker may itself be in linkage disequilibrium with other unexamined functional variants localized in its close proximity or those in nearby genes have to be tested. HapMap initiative is now in progress among the international consortium. Haplotypic knowledge of the human genome will help clarify the contribution of the ITIH4 gene in lipid profile through detailed haplotype association study in the near future. It would also be important to verify the association in geographically distinct populations and other ethnic groups in the future (Shastry 2002).
In summary, we noted an association between the variation of the inter-α-trypsin inhibitor heavy chain 4 (ITIH4 ) gene and hypercholesterolemia in 351 subjects from an area in the east-central region of Japan. Given our genetic results, we expect that the effects of multiple genes, both additive and interactive (Lalouel et al. 2001; Takada et al. 2002), will eventually prove to be responsible for many cases of common, inherited, mixed dyslipidemias.
References
Bork P, Rohde K (1991) More von Willebrand factor type A domains? Sequence similarities with malaria thrombospondin-related anonymous protein, dihydropyridine-sensitive calcium channel and inter-alpha-trypsin inhibitor. Biochem J 279:908–910
Chan P, Risler JL, Raguenez G, Salier JP (1995) The three heavy-chain precursors for the inter-alpha-inhibitor family in mouse: new members of the multicopper oxidase protein group with differential transcription in liver and brain. Biochem J 306:505–512
Cullen P, Funke H, Schulte H, Assmann G (1997) Lipoproteins and cardiovascular risk-from genetics to CHD prevention. J Atheroscler Thromb 4:51–58
Fujita Y, Ezura Y, Emi M, Ono S, Takada D, Takahashi K, Uemura K, Iino Y, Katayama Y, Bujo H, Saito Y (2003) Hypertriglyceridemia associated with amino acid variation N985Y of RP1 gene. J Hum Genet 48:305–308
Fujiwara H, Emi M, Nagai H, Nishimura T, Konishi N, Kubota Y, Ichikawa T, Takahashi S, Shuin T, Habuchi T, Ogawa O, Inoue K, Skolnick MH, Swensen J, Camp NJ, Tavtigian SV (2002) Association of common missense changes in ELAC2 (HPC2) with prostate cancer in a Japanese case-control series. J Hum Genet 47:641–648
Goldstein JL, Hobbs HH, Brown MS (1995) Familial hypercholesterolaemia. In: Scriver CT, Beaudet Al, Sly WS, Valle D (eds), The metabolic and molecular bases of inherited disease. McGraw-Hill, New York, pp1981–2030
Haga H, Yamada R, Ohnishi, Y, Nakamura Y, Tanaka T (2002) Gene-based SNP discovery as part of the Japanese Millennium Genome Project: identification of 190, 562 genetic variations in the human genome. J Hum Genet 47:605–610
Hattori H, Hirayama T, Nobe Y, Nagano M, Kujiraoka T, Egashira T, Ishii J, Tsuji M, Emi M (2002) Eight novel mutations and functional impairments of the LDL receptor in familial hypercholesterolemia in the north of Japan. J Hum Genet 47:80–87
Hegele RA (2001) Monogenic dyslipidemias: window on determinants of plasma lipoprotein metabolism. Am J Hum Genet 69:1161–1177
Iida A, Saito S, Sekine A, Mishima C, Kitamura Y, Kondo K, Harigae S, Osawa S, Nakamura Y (2002) Catalog of 77 single-nucleotide polymorphisms (SNPs) in the carbohydrate sulfotransferase 1 (CHST1) and carbohydrate sulfotransferase 3 (CHST3) genes. J Hum Genet 47:14–19
Ishii J, Nagano M, Kujiraoka T, Ishihara M, Egashira T, Takada D, Tsuji M, Hattori H, Emi M (2002) Clinical variant of Tangier disease in Japan: mutation of the ABCA1 gene in hypoalphalipoproteinemia with corneal lipidosis. J Hum Genet 47:366–369
Lalouel JM, Rohrwasser A (2001) Development of genetic hypotheses in essential hypertension. J Hum Genet 46:299–306
Matsuzawa Y (1995) Significance of hyperglycemia in the occurrence of ischemic heart disease. J Atheroscler Thromb 2:S26–28
Nishimura H, Kakizaki I, Muta T, Sasaki N, Pu PX, Yamashita T, Nagasawa S (1995) cDNA and deduced amino acid sequence of human PK-120, a plasma kallikrein-sensitive glycoprotein. FEBS Lett 357:207–211
Norioka M, Suzuki M, Ryomoto K, Ikebuchi M, Harano Y (2000) Effect of bezafibrate treatment on the altered lipoprotein profiles in hypertriglyceridemic subjects. J Atheroscler Thromb 7:198–202
Ohnishi Y, Tanaka T, Ozaki K, Yamada R, Suzuki, H, Nakamura Y (2001) A high-throughput SNP typing system for genome-wide association studies. J Hum Genet 46:471–477
Saguchi K, Tobe T, Hashimoto K, Sano Y, Nakano Y, Miura NH, Tomita M (1995) Cloning and characterization of cDNA for inter-alpha-trypsin inhibitor family heavy chain-related protein (IHRP), a novel human plasma glycoprotein. J Biochem 117:14–18
Saito S, Iida A, Sekine A, Miura Y, Ogawa C, Kawauchi S, Higuchi S, Nakamura Y (2002) Identification of 779 genetic variations in eight genes encoding members of the ATP-binding cassette, subfamily C (ABCC/MRP/CFTR). J Hum Genet 47:147–171
Shastry B S (2002) SNP alleles in human disease and evolution. J Hum Genet 47:561–566
Takada D, Emi M, Ezura Y, Nobe Y, Kawamura K, Iino Y, Katayama Y, Xin Y, Wu LL, Shum SL, Stephenson SH, Hunt SC, Hopkins PN (2002) Interaction between the LDL-receptor gene bearing a novel mutation and a variant in the apolipoprotein A-II promoter: Molecular study in a 1135-member familial hypercholesterolemia kindred. J Hum Genet 47:656–664
Yoshida S, Harada H, Nagai H, Fukino K, Teramoto A, Emi M (2002) Head-to-head juxtaposition of Fas-associated phosphatase-1 (FAP-1) and c-Jun NH2-terminal kinase 3 (JNK3) genes: genomic structure and seven polymorphisms of the FAP-1 gene. J Hum Genet 47:614-619
Yuan B, Neuman R, Duan SH, Weber JL, Kwok PY, Saccone NL, Wu JS, Liu KY, Schonfeld G (2000) Linkage of a gene for familial hypobetalipoproteinemia to chromosome 3p21.1–22. Am J Hum Genet 66:1699–1704
Zannis VI, Breslow JL (1985) Genetic mutations affecting human lipoprotein metabolism. Adv Hum Genet 14:125–215
Acknowledgments
We thank Mitsuko Kajita, Mina Kodaira, Miho Kawagoe, Tamami Uchida, Kyoko Shimizu, Mayumi Tanaka, and Naoko Tsuruta for their expert technical assistance. This work was supported by a grant for Strategic Research from the Ministry of Education, Science, Sports and Culture of Japan; by a Research Grant for Research from the Ministry of Health and Welfare of Japan; by a Research for the Future Program Grant of The Japan Society for the Promotion of Science.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Fujita, Y., Ezura, Y., Emi, M. et al. Hypercholesterolemia associated with splice-junction variation of inter-α-trypsin inhibitor heavy chain 4 (ITIH4) gene. J Hum Genet 49, 24–28 (2004). https://doi.org/10.1007/s10038-003-0101-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10038-003-0101-8
Keywords
This article is cited by
-
Associations of cardiovascular risk with circulating peptides related to hypertensive disorders of pregnancy
Hypertension Research (2021)
-
The extracellular matrix protein ITIH5 is a novel prognostic marker in invasive node-negative breast cancer and its aberrant expression is caused by promoter hypermethylation
Oncogene (2008)
-
Association of single nucleotide polymorphisms in the promoter region of the pro-opiomelanocortin gene (POMC) with low bone mineral density in adult women
Journal of Human Genetics (2005)