Abstract
The Ainu, a minority ethnic group from the northernmost island of Japan, was investigated for DNA polymorphisms both from maternal (mitochondrial DNA) and paternal (Y chromosome) lineages extensively. Other Asian populations inhabiting North, East, and Southeast Asia were also examined for detailed phylogeographic analyses at the mtDNA sequence type as well as Y-haplogroup levels. The maternal and paternal gene pools of the Ainu contained 25 mtDNA sequence types and three Y-haplogroups, respectively. Eleven of the 25 mtDNA sequence types were unique to the Ainu and accounted for over 50% of the population, whereas 14 were widely distributed among other Asian populations. Of the 14 shared types, the most frequently shared type was found in common among the Ainu, Nivkhi in northern Sakhalin, and Koryaks in the Kamchatka Peninsula. Moreover, analysis of genetic distances calculated from the mtDNA data revealed that the Ainu seemed to be related to both the Nivkhi and other Japanese populations (such as mainland Japanese and Okinawans) at the population level. On the paternal side, the vast majority (87.5%) of the Ainu exhibited the Asian-specific YAP+ lineages (Y-haplogroups D-M55* and D-M125), which were distributed only in the Japanese Archipelago in this analysis. On the other hand, the Ainu exhibited no other Y-haplogroups (C-M8, O-M175*, and O-M122*) common in mainland Japanese and Okinawans. It is noteworthy that the rest of the Ainu gene pool was occupied by the paternal lineage (Y-haplogroup C-M217*) from North Asia including Sakhalin. Thus, the present findings suggest that the Ainu retain a certain degree of their own genetic uniqueness, while having higher genetic affinities with other regional populations in Japan and the Nivkhi among Asian populations.
Similar content being viewed by others
Introduction
The Ainu, the aboriginal inhabitants of northernmost island (Hokkaido) of the Japanese Archipelago, are ethnic minority population in Japan. They generally show unique physical characteristics such as hairiness, wavy hair, and deep-set eyes, which are very different from those of the ordinary Japanese. The ethnic origin and genetic affiliation of the Ainu, therefore, have been debated for more than 100 years in and outside Japan (reviewed by Hanihara 1983). Several lines of morphological evidence have revealed that physical features of the Ainu are closely related with those of the Neolithic Jomon people in Japan, and hence they are considered to be modern descendants of pre-agricultural aboriginal groups in the Japanese Archipelago (Yamaguchi 1982; Hanihara 1991). In addition, mitochondrial DNA (mtDNA) sequences from ancient Jomon and Ainu bones have suggested a common genetic origin of these two temporal groups (Horai et al. 1989, 1991). On the other hand, the Ainu seem to be in a unique phylogenetic position in modern Asian phylogeny at the population level, although showing smaller genetic distances from other East Asian populations among the contemporary human populations (Harihara et al. 1988; Bannai et al. 1996, 2000; Horai et al. 1996; Tajima et al. 2002). Thus, we need to expand our knowledge of how the Ainu have been genetically formed.
One way to deduce the history of human population is to examine the geographic distribution of individual genetic lineages observed in the population (Avise 2000; Underhill et al. 2001). For this purpose, a combined phylogeographic analysis of two uniparentally inherited polymorphic loci, mtDNA and non-recombining portion of the Y chromosome (NRY), may be effective (e.g., Thangaraj et al. 2003). Because mtDNA and NRY are regarded as genetically independent loci in their modes of inheritance, several evolutionary forces (such as genetic drift and natural selection) could separately operate on each locus, thereby possibly resulting in differences in the geographic variation between the two loci. By contrast, common phylogeographic features provided by the combined analysis could reflect historical events (such as human migration) rather than the evolutionary forces.
In the present study, we have examined DNA polymorphisms in the major non-coding (D-loop) region of mtDNA and NRY in the Ainu. To obtain detailed phylogeographic views at the levels of mtDNA sequence types and Y-haplogroups, we have also investigated DNA samples from other human populations in North, East, and Southeast Asia. Based on the geographic distributions of both maternal and paternal genetic variants shared by the Ainu and other Asian populations, we have intended to elucidate the genetic characteristics of the Ainu.
Materials and methods
Subjects and DNA samples
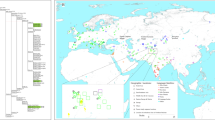
We examined 51 unrelated individuals from the Ainu for mtDNA analysis and 16 males for Y-haplogroup analysis. In addition, we included a total of 1,103 samples from 15 other Asian populations (for mtDNA analysis), and 702 males from 11 populations (for Y-haplogroup analysis) in this study. These populations were classified into three regional groups: North, East, and Southeast Asian populations (Fig. 1). The number of DNA samples from each population was as follows (the male samples analyzed in parenthesis): 110 Koryak, 35 Even, 61 (61) Buryat, 57 (21) Nivkhi, 60 (49) Northern Han, 64 Korean, 82 (82) Honshu-Japanese, 104 (104) Kyushu-Japanese, 45 (45) Okinawa-Japanese, 66 (21) Taiwan Han, 180 (223) aboriginal Taiwanese, 59 (50) Philippine (Tagalog language group), 74 (34) Thai (ordinary Thai from Chiang Mai and Khon Kaen provinces), 52 (12) Malaysian (ordinary Malay near Kuala Lumpur), and 54 Indonesian. This study was approved by the Ethics Committee of the Graduate University for Advanced Studies (Sokendai), Hayama.
Analysis of mtDNA polymorphism
Nucleotide sequences of 563-bp fragments (positions 16048–16569 followed by positions 1–41 in the reference mtDNA sequence of Anderson et al. 1981) were determined by direct sequencing of PCR-amplified products as described previously (Tajima et al. 2003). The 435 sequences from the following five populations: Ainu, Korean, Taiwan Han (Horai et al. 1996), Thai (Fucharoen et al. 2001), and aboriginal Taiwanese (Tajima et al. 2003) have been previously reported, although some sequences from Taiwan Han lack sequence information between positions 16048 and 16128. Thus, we have newly obtained 719 sequences from other populations in this study and deposited those sequences in the DDBJ/EMBL/GenBank.
On the basis of estimated number of nucleotide substitutions per site between individual sequences under the two-parameter model of nucleotide substitutions (Kimura 1980), a phylogenetic tree for the mtDNA sequences from the Ainu was constructed with the neighbor-joining (NJ) method (Saitou and Nei 1987). We also constructed an NJ tree for human populations with net nucleotide diversities (d A distances) between populations (Nei and Li 1979) and the Kimura’s two-parameter correction. The NJ tree was rooted with African population (Horai and Hayasaka 1990; Vigilant et al. 1991) as an outgroup. The reliability of the tree was analyzed by the bootstrap method (Felsenstein 1985) with 1,000 replications.
Genotyping of Y-chromosomal biallelic polymorphisms
The polymorphic sites in the present study included a set of seven previously published Y-chromosomal biallelic markers: YAP, RPS4Y 711, P27 (DYS257 108), SRY 4064, SRY 10831, M9, and M15 (Tajima et al. 2002). In addition, we analyzed six polymorphisms (M8, M55, M119, M122, M125, and M217; Underhill et al. 1997, 2000, 2001), and two newly identified polymorphisms (subsequently referred to as AS1 and AS2). PCR-RFLP (restriction fragment length polymorphism) analyses were developed to genotype the six known polymorphisms, four of which (M8, M119, M125, and M217) were amplified with PCR primers including a mismatch primer that harbors non-complementary nucleotide(s) to generate the appropriate restriction sites (information about the primer sequences and the restriction endonuclease used is available on request).
The two new T→C mutations at position 46,812 (AS1) and position 57,316 (AS2) of the RP11-418M8 clone (accession no. AC009491.3) were also typed with PCR-RFLP methods. A 245-bp fragment containing AS1 was amplified with the following PCR primers: 5′-GGGGCTCTCTGCCAAATTAT-3′ and 5′-GGTTCATGGTCTCGTTTTGG-3′, and then digested with the HpyCH4IV enzyme. A 183-bp fragment encompassing AS2 was amplified with the following primer pairs: 5′-GAGTTGGAAGCACTTTCTGTGTCATCAGTTTTCTCTGACT-3′ and 5′-ATTCCTCCATAGTCTTGTGA-3′. The underlined bases in one primer indicate non-complementary nucleotides, which create the HinfI recognition sequence for this genotyping.
Results
Geographic distribution of 25 mtDNA lineages from the Ainu
Previously, Horai et al. (1996) have observed 25 types of mtDNA sequences in 51 individuals from the Ainu people. To reveal phylogenetic relationships among the 25 sequence types from the Ainu, we constructed an unrooted NJ phylogenetic tree, as shown in Fig. 2. In the tree, we found no apparent phylogenetic clusters, suggesting the presence of diversified mtDNA sequence types in the maternal gene pool of the Ainu. When we compared mtDNA sequences of a total of 1,154 individual samples from the Ainu and 15 other Asian populations, 51.0% of the Ainu individuals exhibited 11 unique sequence types (types 5, 7, 9–13, 20, 21, 24, and 25; Table 1), which were not shared by the Ainu and any other populations studied. Thus, the Ainu maintain a certain degree of their own maternal genetic characteristics.
Unrooted neighbor-joining tree, showing the relationships of the 25 mtDNA sequence types from the Ainu. Each sequence type (in circle) with the observed number of individuals is characterized in Table 1. The area of each circle is proportional to the number of Ainu individuals, and the smallest circle denotes one individual. Open and closed circles represent the sequence types unique to the Ainu, and the shared types observed between the Ainu and other human population(s), respectively. The scale for genetic distance is shown upper left
On the other hand, the remaining 14 types for the Ainu were widely distributed among other human populations in North, East, and Southeast Asia (Table 1). Five (types 3, 4, 15, 22, and 23) of the 14 shared types were geographically confined to the Ainu and North Asian populations. Moreover, the Ainu shared seven sequence types (types 1, 2, 6, 8, 14, 16, and 19) only with other East Asian populations, and one (type 18) with Southeast Asian populations. Only one shared type (type 17) was distributed in seven human populations including the Ainu, spanning two different geographic regions (North and East Asia).
The most frequently shared type (type 4) was found in 30 individuals: three Ainu, 21 Nivkhi in northern Sakhalin, and six Koryaks in the Kamchatka Peninsula. The Ainu and Nivkhi had a total of two different types (types 3 and 4) in common, and the two types were shared by ten Ainu (19.6% of the population) and 26 Nivkhi (45.6% of the population). Four shared sequence types (types 4, 15, 22, and 23) were observed in seven Ainu (13.7% of the population) and 15 Koryaks (13.6% of the population). Therefore, at least 14 (27.5%) of the 51 Ainu individuals possess maternal genetic elements (types 3, 4, 15, 22, and 23) from North Asia, thereby implying extensive genetic contacts among the Ainu and North Asian populations inhabiting Sakhalin and the Kamchatka Peninsula.
Phylogenetic position of the Ainu in Asian populations inferred from mtDNA data
To investigate population relatedness between the Ainu and other Asian populations, we constructed a rooted NJ tree for the 16 Asian populations (Fig. 3), together with African population (Horai and Hayasaka 1990; Vigilant et al. 1991). In this tree, the Ainu and Nivkhi were neighbors (bootstrap value, 64%), although the length of branch between them (d A distance, 0.15%) was not so short, in part owing to low value of intrapopulational nucleotide diversity in the Nivkhi (data not shown). Thus, this genetic link may reflect the sharing of mtDNA sequence types between these two populations. On the other hand, based on d A distances between the Ainu and other populations, we observed the smallest value (0.035%) in comparison with Honshu-Japanese, followed by Okinawa-Japanese (0.038%) and Kyushu-Japanese (0.043%). The relatively small genetic distances between the Ainu and other regional populations in Japan (such as mainland Japanese and Okinawan) are consistent with the previous findings undertaken with other genetic markers (Nei 1995; Omoto and Saitou 1997; Bannai et al. 2000; Tajima et al. 2002). Therefore, these analyses suggest that the Ainu are genetically related to both other Japanese populations and the Nivkhi at the population level.
Neighbor-joining tree for the 16 Asian populations, on the basis of d A distances estimated from the nucleotide sequences of 482-bp fragments of the D-loop region (positions 16129–16569 followed by positions 1–41 in the reference mtDNA sequence of Anderson et al. 1981) with the Kimura’s two-parameter correction (Kimura 1980). This tree is rooted with African population (Horai and Hayasaka 1990; Vigilant et al. 1991) as an outgroup. Bootstrap probabilities (over 50%) are attached to the internal branches. The scale for genetic distance is shown below the tree
Y-haplogroup analysis
Figure 4 shows evolutionary relationships among 14 Y-haplogroups defined by analyzing 15 biallelic polymorphisms on the NRY. Two newly identified SNPs (referred to as AS1 and AS2) seemed to be phylogenetically similar (or equivalent) to M175/M214 and M174, respectively, in the YCC (Y Chromosome Consortium) phylogenetic tree (The YCC 2002). Thus, we defined each of the haplogroup status according to the YCC nomenclature, although we will need to clarify phylogenetic positions of the two new markers in YCC phylogeny.
Maximum parsimonious phylogenetic tree for 14 Y-chromosome haplogroups with their frequency distributions in 12 Asian populations. The haplogroup designation in this tree conforms to the mutation-based nomenclature recommended by the Y Chromosome Consortium (The YCC 2002). The marker names on the branches of the tree represent the expected mutational events at the respective polymorphic sites
We found three Y-haplogroups (C-M217*, D-M55*, and D-M125) in 16 males from the Ainu (Fig. 4). Of these, D-M55* (81.3%) and D-M125 (6.3%) carried the M55 mutation, one of the informative markers for Asian YAP+ lineages (Underhill et al. 2000). Thus, all of the YAP+ lineages observed in the Ainu could be regarded as Asian-specific types at the haplogroup level. Moreover, these lineages were shared only with Okinawans and mainland Japanese (Honshu and Kyushu) among the Asian populations studied. The Ainu also possessed C-M217* (12.5%) belonging to haplogroup lineages defined by RPS4Y 711-T allele, whereas their paternal gene pool contained no C-M8, which was unique to other three Japanese populations (Fig. 4). A recent study has shown that C-M217* is predominantly distributed in North Asian populations such as Oroqen and Siberian Evenks (Karafet et al. 2001), and hence this haplogroup might be brought into the Ainu because of gene flow from North Asian populations. Unlike the mtDNA analysis, on the other hand, there was no shared Y-haplogroup between the Ainu and Southeast Asian populations.
Discussion
The present comparison of 1,154 mtDNA sequences from 16 Asian populations clarifies maternal genetic uniqueness of the Ainu because of the presence of 11 mtDNA sequence types (types 5, 7, 9–13, 20, 21, 24, and 25) that are specific to this ethnic group. These unique sequence types account for about half of the maternal gene pool of the Ainu, suggesting a certain degree of their genetic isolation from the surrounding human populations. Because the 11 sequence types have one polymorphism at position 16223 (16223T) in common, they are likely to be included in Eurasian super-haplogroup M at the mtDNA haplogroup level (Kivisild et al. 2002; Yao et al. 2002). It should be noted that complete mtDNA sequences of the unique lineages are needed to understand further their phylogenetic affiliations in mtDNA phylogeny.
On the other hand, the shared mtDNA types 3, 4, 15, 22, and 23 in the Ainu imply genetic contacts between the Ainu and North Asian populations. In particular, the Ainu seem to have higher genetic affinity with the Nivkhi in Sakhalin among the North Asian populations studied, as shown in Fig. 3. Analysis of human leukocyte antigen (HLA) genes and haplotypes (Tokunaga et al. 2001) has also revealed the genetic link between the Ainu and Nivkhi, both of which are geographically neighboring ethnic groups. The shared sequence types (types 3 and 4) between these two populations possess a unique combination of five transitional mutations (16126C-16189C-16231C-16266T-16519C), which is regarded as the motif sequence for mtDNA haplogroup Y1 (Kivisild et al. 2002). Recent studies have shown that this haplogroup is mostly distributed in North Asian and native Siberian populations (Schurr et al. 1999; Kivisild et al. 2002). Because of higher frequencies of the shared mtDNA types in the Nivkhi than the Ainu (Table 1), this sharing is plausibly attributed to gene flow from the Nivkhi to the Ainu.
In view of the shared mtDNA types, furthermore, we also detect possible involvement of other East Asians including other regional Japanese (types 1, 2, 6, 8, 14, 16, and 19) and Southeast Asians (type 18) in the formation of the Ainu gene pool. This must be partly due to the sharing of genetic history (such as common ancestry and genetic interactions between populations), although it is not easy to determine what is the major cause of such genetic involvement. In any case, North, East, and Southeast Asian populations may have separately contributed to the maternal gene pool of the Ainu, suggesting maternal multiple origins of the Ainu.
On the paternal side, we observe no Y-haplogroups specific to the Ainu, but predominant distribution of Asian-specific YAP+ lineages (Y-haplogroups D-M55* and D-M125) among the Ainu males. The Asian-specific YAP+ lineages (Y-haplogroup D) are infrequently distributed throughout Asia except for Japan, Tibet (Underhill et al. 2000), and the Andaman islands of the Indian Ocean (Thangaraj et al. 2003). Therefore, this unusual distribution is one of the major paternal genetic characteristics of the Ainu. Moreover, the Ainu and other regional Japanese populations possess these paternal lineages (D-M55* and D-M125) in common. These paternal lineages (Y-haplogroup D) are considered to have contributed to the formation of the Neolithic Jomon males or their ancestral groups in prehistoric Japan (Hammer and Horai 1995; Tajima et al. 2002), although the migration route of the YAP+ lineages into Asia is still puzzling (Jin and Su 2000; Underhill et al. 2001). Thus, all the modern Japanese populations including the Ainu may genetically share varying degrees of the Jomon (or pre-Jomon) ancestry.
On the other hand, the present study reveals paternal genetic differences between the Ainu and other Japanese, because three Y-haplogroups (C-M8, O-M175*, and O-M122*) common to the three Japanese populations are absent in the Ainu. This observation reflects the unique paternal genetic background of the Ainu and/or their independent genetic history. Notably, Y-haplogroup O-M122* (belonging to haplotype lineages defined by M9-G allele) is thought to be the genetic trace of recent range expansions from China (Su et al. 2000; Underhill et al. 2001). Thus, the lack of O-M122* in the Ainu implies that subsequent dispersals of this haplogroup were overlaid on earlier paternal lineages (such as Y-haplogroup D) in the Japanese Archipelago. This agrees with the view that the Ainu are considered to be the modern descendants of the aboriginal people of the Neolithic Jomon Age.
It is also important to note that the Nivkhi exhibit no Asian-specific YAP+ lineages (Y-haplogroup D) that are frequent in the Ainu males (87.5% of the population), whereas one Y-haplogroup (C-M217*) is shared by the two ethnic groups. Taking into consideration the northern distribution of C-M217* in Asia (Karafet et al. 2001), these findings suggest no apparent paternal gene flow from the Ainu to the Nivkhi. This is compatible with the above-mentioned maternal view that the genetic communication between these two populations may have been unidirectional to the Ainu. One explanation is that human migrations to northern Japan including Hokkaido from North Asia throughout prehistoric and historic times (Omoto and Saitou 1997; and references therein) might have brought the northern genetic elements into the Ainu.
In conclusion, the present combined phylogeographic analyses both for mtDNA sequence types and Y-haplogroups provide new insights into the genetic backgrounds of the Ainu. Unique maternal and paternal genetic features of the Ainu clearly represent their own genetic characteristics, although the Ainu have some degree of genetic affinity with the geographically neighboring populations: other regional populations in Japan and the Nivkhi in Sakhalin. Thus, this study elucidates the maternal and paternal genetic compositions of the Ainu, whereas it is still uncertain when the respective genetic lineages were brought into the Ainu. Further genetic studies for dating mtDNA- and Y-haplogroups in Asia should be necessary to infer their temporal origins. These extensive analyses will also help us to have a firm grasp of the genetic relationship between the Neolithic Jomon people and the contemporary Ainu.
References
Anderson S, Bankier AT, Barrell BG, de Bruijn MH, Coulson AR, Drouin J, Eperon IC, Nierlich DP, Roe BA, Sanger F, Schreier PH, Smith AJ, Staden R, Young IG (1981) Sequence and organization of the human mitochondrial genome. Nature 290:457–465
Avise JC (2000) Phylogeography. Harvard University Press, Cambridge
Bannai M, Tokunaga K, Imanishi T, Harihara S, Fujisawa K, Juji T, Omoto K (1996) HLA class II alleles in Ainu living in Hidaka District, Hokkaido, northern Japan. Am J Phys Anthropol 101:1–9
Bannai M, Ohashi J, Harihara S, Takahashi Y, Juji T, Omoto K, Tokunaga K (2000) Analysis of HLA genes and haplotypes in Ainu (from Hokkaido, northern Japan) supports the premise that they descent from Upper Paleolithic populations of East Asia. Tissue Antigens 55:128–139
Felsenstein J (1985) Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39:783–791
Fucharoen G, Fucharoen S, Horai S (2001) Mitochondrial DNA polymorphisms in Thailand. J Hum Genet 46:115–125
Hammer MF, Horai S (1995) Y chromosomal DNA variation and the peopling of Japan. Am J Hum Genet 56:951–962
Hanihara K (1983) Ainu and Ryukyus (Okinawa Islanders). In: Kondo S (ed) Recent progress of natural sciences in Japan, vol 8. Science Council of Japan, Tokyo, pp 25–30
Hanihara K (1991) Dual structure model for the population history of the Japanese. Jpn Rev 2:1–33
Harihara S, Saitou N, Hirai M, Gojobori T, Park KS, Misawa S, Ellepola SB, Ishida T, Omoto K (1988) Mitochondrial DNA polymorphism among five Asian populations. Am J Hum Genet 43:134–143
Horai S, Hayasaka K (1990) Intraspecific nucleotide sequence differences in the major noncoding region of human mitochondrial DNA. Am J Hum Genet 46:828–842
Horai S, Hayasaka K, Murayama K, Wate N, Koike H, Nakai N (1989) DNA amplification from ancient human skeletal remains and their sequence analysis. Proc Japan Acad 65(Ser. B):229–233
Horai S, Kondo R, Murayama K, Hayashi S, Koike H, Nakai N (1991) Phylogenetic affiliation of ancient and contemporary humans inferred from mitochondrial DNA. Philos Trans R Soc Lond B Biol Sci 333:409–417
Horai S, Murayama K, Hayasaka K, Matsubayashi S, Hattori Y, Fucharoen G, Harihara S, Park KS, Omoto K, Pan IH (1996) mtDNA polymorphism in East Asian populations, with special reference to the peopling of Japan. Am J Hum Genet 59:579–590
Jin L, Su B (2000) Natives or immigrants: modern human origin in east Asia. Nat Rev Genet 1:126–133
Karafet T, Xu L, Du R, Wang W, Feng S, Wells RS, Redd AJ, Zegura SL, Hammer MF (2001) Paternal population history of East Asia: sources, patterns, and microevolutionary processes. Am J Hum Genet 69:615–628
Kimura M (1980) A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J Mol Evol 16:111–120
Kivisild T, Tolk HV, Parik J, Wang Y, Papiha SS, Bandelt HJ, Villems R (2002) The emerging limbs and twigs of the East Asian mtDNA tree. Mol Biol Evol 19:1737–1751
Nei M (1995) The origins of human populations: genetic, linguistic, and archeological data. In: Brenner S, Hanihara K (eds) The origin and past of modern humans as viewed from DNA. World Scientific, Singapore, pp 71–91
Nei M, Li WH (1979) Mathematical model for studying genetic variation in terms of restriction endonucleases. Proc Natl Acad Sci USA 76:5269–5273
Omoto K, Saitou N (1997) Genetic origins of the Japanese: a partial support for the dual structure hypothesis. Am J Phys Anthropol 102:437–446
Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4:406–425
Schurr TG, Sukernik RI, Starikovskaya YB, Wallace DC (1999) Mitochondrial DNA variation in Koryaks and Itel’men: population replacement in the Okhotsk Sea-Bering Sea region during the Neolithic. Am J Phys Anthropol 108:1–39
Su B, Xiao C, Deka R, Seielstad MT, Kangwanpong D, Xiao J, Lu D, Underhill P, Cavalli-Sforza L, Chakraborty R, Jin L (2000) Y chromosome haplotypes reveal prehistorical migrations to the Himalayas. Hum Genet 107:582–590
Tajima A, Pan IH, Fucharoen G, Fucharoen S, Matsuo M, Tokunaga K, Juji T, Hayami M, Omoto K, Horai S (2002) Three major lineages of Asian Y chromosomes: implications for the peopling of east and southeast Asia. Hum Genet 110:80–88
Tajima A, Sun CS, Pan IH, Ishida T, Saitou N, Horai S (2003) Mitochondrial DNA polymorphisms in nine aboriginal groups of Taiwan: implications for the population history of aboriginal Taiwanese. Hum Genet 113:24–33
Thangaraj K, Singh L, Reddy AG, Rao VR, Sehgal SC, Underhill PA, Pierson M, Frame IG, Hagelberg E (2003) Genetic affinities of the Andaman islanders, a vanishing human population. Curr Biol 13:86–93
The Y Chromosome Consortium (2002) A nomenclature system for the tree of human Y-chromosomal binary haplogroups. Genome Res 12:339–348
Tokunaga K, Ohashi J, Bannai M, Juji T (2001) Genetic link between Asians and native Americans: evidence from HLA genes and haplotypes. Hum Immunol 62:1001–1008
Underhill PA, Jin L, Lin AA, Mehdi SQ, Jenkins T, Vollrath D, Davis RW, Cavalli-Sforza LL, Oefner PJ (1997) Detection of numerous Y chromosome biallelic polymorphisms by denaturing high-performance liquid chromatography. Genome Res 7:996–1005
Underhill PA, Shen P, Lin AA, Jin L, Passarino G, Yang WH, Kauffman E, Bonné-Tamir B, Bertranpetit J, Francalacci P, Ibrahim M, Jenkins T, Kidd JR, Mehdi SQ, Seielstad MT, Wells RS, Piazza A, Davis RW, Feldman MW, Cavalli-Sforza LL, Oefner PJ (2000) Y chromosome sequence variation and the history of human populations. Nat Genet 26:358–361
Underhill PA, Passarino G, Lin AA, Shen P, Lahr MM, Foley RA, Oefner PJ, Cavalli-Sforza LL (2001) The phylogeography of Y chromosome binary haplotypes and the origins of modern human populations. Ann Hum Genet 65:43–62
Vigilant L, Stoneking M, Harpending H, Hawkes K, Wilson AC (1991) African populations and the evolution of human mitochondrial DNA. Science 253:1503–1507
Yamaguchi B (1982) A review of the osteological characteristics of the Jomon population in prehistoric Japan. J Anthrop Soc Nippon 90:77–90
Yao YG, Kong QP, Bandelt HJ, Kivisild T, Zhang YP (2002) Phylogeographic differentiation of mitochondrial DNA in Han Chinese. Am J Hum Genet 70:635–651
Acknowledgements
We thank Dr. P.A. Underhill for helpful comments on our newly discovered Y-markers. This work was supported in part by Grants-in-Aid from the Ministry of Education, Culture, Sports, Science and Technology of Japan to S. Horai.
Author information
Authors and Affiliations
Corresponding author
Additional information
The nucleotide sequence data reported are available in the DDBJ/EMBL/GenBank databases under the accession numbers AB093862–AB094026 and AB122153–AB122706
Rights and permissions
About this article
Cite this article
Tajima, A., Hayami, M., Tokunaga, K. et al. Genetic origins of the Ainu inferred from combined DNA analyses of maternal and paternal lineages. J Hum Genet 49, 187–193 (2004). https://doi.org/10.1007/s10038-004-0131-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10038-004-0131-x
Keywords
This article is cited by
-
Origin of ethnic groups, linguistic families, and civilizations in China viewed from the Y chromosome
Molecular Genetics and Genomics (2021)
-
A partial nuclear genome of the Jomons who lived 3000 years ago in Fukushima, Japan
Journal of Human Genetics (2017)
-
Distribution of HLA haplotypes across Japanese Archipelago: similarity, difference and admixture
Journal of Human Genetics (2015)
-
Latitude has more significant impact on prevalence of multiple sclerosis than ultraviolet level or sunshine duration in Japanese population
Neurological Sciences (2015)
-
Mitochondrial DNA haplogroups and homogeneity in the Korean population
Genes & Genomics (2014)