Abstract
Four new 12,8-Eudesmanolides (1−4) and one known compound 5 named 13-Hydroxy-3,7(11)-eudesmadien-12,8-olide, were isolated from a mangrove rhizosphere-derived fungus Eutypella sp. 1–15. Their structures with absolute stereochemistry were determined by the comprehensive spectroscopic data, experimental and calculated ECD analysis. Compound 1 exhibited potent anticancer activity against JEKO-1 and HepG2 with IC50 values of 8.4 and 28.5 μM, respectively. Additionally, compound 1 also showed moderate antimicrobial activity.
Similar content being viewed by others
Main
Eudesmanolides, a group of well-known natural sesquiterpenoids, have diverse bioactivities, including antituberculosis,1 anticancer2 and anti-inflammatory activity.3 These compounds have been classified into two subgroups: 12,6-eudesmanolides such as ludovicin A4 and 12,8-eudesmanolides such as alantolactone.5, 6, 7 As part of our continuous screening for bioactive metabolite from microbial sources, a fungal strain Eutypella sp. 1–15 isolated from the mangrove rhizosphere was selected for its excellent cytotoxic and antimicrobial activity, previous chemical investigations of this strain afforded series of cytochalasans,8, 9, 10 a famous group of cytotoxic agent. We now have reinvestigated the strain for other bioactive constituents. In this study, we describe the fermentation, isolation, structure elucidation of four new 12,8-Eudesmanolides (1–4) along with one reported compound (5) 13-Hydroxy-3,7(11)-eudesmadien-12,8-olide,11 as well as their cytotoxic and antibacterial activity.
The strain Eutypella sp. 1-15, isolated from the soil of mangrove rhizosphere in Jimei, Fujian Province, China, was identified as Eutypella sp.10 according to its ITS sequence. The strain was inoculated on rice medium containing rice 1000 g, NaCl 100 g in 1.0 l water and cultured at 28 °C for 27 days. The final 2 l fermented culture were extracted with mixed solvent of EtOAc-MeOH-AcOH (80:15:5, v/v/v) to obtain a crude extract. The crude extract was partitioned between EtOAc and H2O, and subsquently EtOAc extract was partitioned with MeOH and petroleum ether to afford the MeOH extract. Then, the MeOH extract was subjected to medium-pressure reverse-phase liquid chromatography, Sephadex LH-20, and finally on silica column to afford compounds 1–5 (Figure 1), respectively.
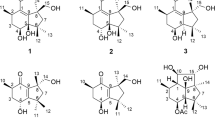
Compound 1 was obtained as colorless oil with [α]D20+10.4 (c 0.87, MeOH), UV (MeOH) λmax (log ɛ) 200 (3.47), 281 (2.78) nm, and IR (KBr) νmax=3420, 2927, 1762, 1435, 1001, 790 cm−1. The molecular formula was deduced as C15H18O3 according to HR-ESI-MS at m/z=247.1329 [M+H]+ (calcd 247.1329). The 1H, 13C NMR and HSQC spectra (Acetone-d6) of 1 display signals for two methyls, four methylenes, three methines, and six non-protonated carbons (Table 1). The HMBC correlations from H3-14 (δH 1.05) to C-1 (δC 34.9), C-5 (δC 45.2), C-9 (δC 120.7), C-10 (δC 34.6) and H3-15 (δH 1.74) to C-3 (δC 128.1), C-4 (δC 132.9), C-5, as well as 1H-1H COSY correlations between H2-1 (δH 1.70) and H2-2 (δH 2.17, 2.10), H2-2 and H-3 (δH 5.43) resulted in a six-membered ring (Figure 2a). HMBC correlations from H2-6 (δH 3.35, 2.53) to C-4, C-5, C-8 (δC 148.4), C-10, C-11 (δC 123.7) and 1H-1H COSY correlations between H-5 (δH 2.54) and H2-6 joined previously deduced fragments to form a 6/6 AB ring junction system, which was further supported by the HMBC correlations from H2-13 (δH 4.41) to C-7 (δC 150.8), C-11, C-12 (δC 169.2). Moreover, the downfield C-7 (δC 150.8) and C-8 (δC 148.4), upfield C-11 (δC 123.7) and C-9 (δC 120.7) as well as a carbonyl signal C-12 (δC 169.2) suggested a α,β-unsaturated γ-lactone (Figure 2a). Finally, plus the oxygenated methylene C-13 ((δC 54.2), H2-13 (δH 4.41)), the planar structure of 1 was elucidated as shown in Figure 1 named 13-Hydroxy-3,8,7(11)-eudesmatrien-12,8-olide. The stereochemistry of 1 was assigned as discussion below.
Compound 2 was purified as colorless solid with [α]D20+70.2 (c 1, MeOH). The 1H and 13C NMR spectra of 2 exhibited a pattern analogous to that of 1 (Table 1). The molecular formula was deduced as C15H16O3 on the basis of HR-ESI-MS at m/z=245.1172 [M+H]+ (calcd 245.1172), suggesting that 2 was the dehydrogenated derivative of 1. Further analysis of HSQC and HMBC spectra of 2 revealed the appearance of a pair of sp2 carbons at C-5 (δC 155.1) and C-6 (δC 110.6) compared to 1, which was indicated that the new generated double bond was between C-5 and C-6, therefore, 2 was determined to be 13-Hydroxy-3,5,8,7(11)-eudesmatetraen-12,8-olide (Figure 2a).
Compound 3 was obtained as yellow powder with [α]D20+162.1 (c 0.5, MeOH). The 1H, 13C and HSQC spectra of 3 revealed the absence of the methylene C-2 and the presence of a carbonyl group (δC 195.6). The downfield shift and doublet peak of H2-1 (δH 2.66, 2.55, (d, J=16.0 Hz)) indicated the methylene was replaced by a carbonyl group at C-2. The deduction was confirmed by the molecular formula of 3, C15H14O4, established by HR-ESI-MS at m/z=259.0965 [M+H]+ (calcd 259.0965). Therefore, 3 was assigned as 2-One-13-hydroxy-3,5,8,7(11)-eudesmatetraen-12,8-olide (Figure 1).
Compound 4 was obtained as colorless oil with [α]D20-40.9 (c 0.6, MeOH). The molecular formula was deduced as C15H20O4 according to HR-ESI-MS at m/z=265.1435 [M+H]+ (calcd 265.1434). The 1H and 13C NMR spectra of 4 exhibited a pattern analogous to that of 1 (Table 1). Detailed analysis of the 2D NMR spectra revealed the double bond between C-8 (δC 148.4) and C-9 (δC 120.7) in 1 was reduced. The doublet peak of H2-9 (δH 2.23, 1.46, (d, J=13.4 Hz)) and chemical shift of C-8 (δC 103.4) in 4 suggested the appearance of a hemiketal moiety at C-8 (Figure 1). Therefore, compound 4 was determined to be 8,13-Dihydroxy-3,7(11)-eudesmadien-12,8-olide. The NOESY cross-peaks at H3-14/OH-8 indicated a syn orientation between 14-Me and 8-OH.
Compound 5 was found to be 13-Hydroxy-3,7(11)-eudesmadien-12,8-olide by comparing the NMR, MS and specific rotation data ([α]D20-191.4) with the reported one ([α]D25-37.3).11 To determine the absolute configuration of these 12,8-Eudesmanolides, the calculated ECD spectra using time-dependent density functional theory (TDDFT) was applied. We found that calculated ECD spectrum of the conformers of 3 with 10S is in good accordance with the experimental spectrum (Figure 2b), also the calculated specific rotation of +531.9 further confirm the 10S conformer. Consequently, the absolute configuration of 3 was unambiguously assigned as shown in Figure 1. Considering the specific rotations of 2 and 3 were [α]D20+70.2 and [α]D20+162.1, respectively, we assume the absolute configuration of 2 was consistent with 3. As for 1, the good accordance of the calculated ECD spectrum of the conformers of 1 with 5R, 10R with the experimental ECD spectrum of 1 (Figure 2d) supported the stereochemistry of 1 as shown (Figure 1), the calculated specific rotation +101.5 also supported the conclusion. The trans orientation of 14-Me and 5-H of 1 was consistent with 5, considering the same biogenetic origin and similar ECD curves of 1, 4 and 5 (Figure 2c), we assume the absolute configuration of 1, 4 and 5 was assigned as shown in Figure 1.
The compounds 1–5 were evaluated for antimicrobial activities against Bacillus subtilis CMCC63501, Escherichia coli CMCC44103, staphylococcus aureus CMCC26003, Bacillus pumilus CMCC63202 and Micrococcus luteus CMCC28001 using a plate assay.12 Compound 1 exhibited inhibitory efficacy to Bacillus subtilis CMCC63501 and Bacillus pumilus CMCC63202 with IC50 value of 18.1 and 23.8 μM, respectively (Table 2). Compounds 2–5 did not show any activity (IC50>50 μM). Further, all isolates (1–5) were assessed for antiproliferative activities against a panel of cancer cells, including BGC823, SKGT4, MCF-7, A375, MDA-MB-231, HL-60, JEKO-1, HepG2 and Kyse450 using the MTT method.13 Compound 1 displayed growth inhibitory effect against human Mantle cell lymphoma JEKO-1 and human Hepatoma carcinoma HepG2 with IC50 values of 8.4 and 28.5 μM, respectively (Table 2). However, at a concentration of 50 μM, no significant changes in cell viability were observed in the other tested cancer cells after exposure to 1 for 48 h. Compounds 2–4 show almost no cytotoxicity to all tested cancer cells. Compound 5 show weak cytotoxicity against HepG2 with IC50 value of 48.4 μM. These data suggested that compound 1 has moderate anticancer activity. The bioactivity of these new 12,8-Eudesmanolides was mediated by an elaborate network composed of functional group, configuration and conformation of the framework.
References
Charles, C. L., Frank, R. F., Scott, G. F., Leovigildo, Q. & Nikolaus, H. F. Antimycobacterial Eudesmanolides from Inula helenium and Rudbeckia subtomentosa. Planta Med. 65, 351–355 (1999).
Yang, C., Shi, Y. P. & Jia, Z. J. Sesquiterpene lactone glycosides, eudesmanolides, and other constituents from Carpesium macrocephalum. Planta Med. 68, 626–630 (2002).
Kim, Y.H., Choo, S. J., Ryoo, I. J., Ahn, J. S. & Yoo, I. D. Eudesmanolides from taraxacum mongolicum and their inhibitory effects on the production of nitric oxide. Arch. Pharm. Res. 34, 37–41 (2011).
Goren, N., Woerdenbag, H. J. & BozokJohansson, C. Cytotoxic and antibacterial activities of sesquiterpene lactones isolated from Tanacetum praeteritum subsp praeteritum. Planta Med. 62, 419–422 (1996).
Yao, Y. et al. Alantolactone induces G1 phase arrest and apoptosis of multiple myeloma cells and overcomes bortezomib resistance. Apoptosis 20, 1122–1133 (2015).
Barrero, A.F., Rosales, A., Cuerva, J. M. & Oltra, J. E. Unified synthesis of eudesmanolides, combining biomimetic strategies with homogeneous catalysis and free-radical chemistry. Org. Lett. 5, 1935–1938 (2003).
Rezeng, C. D. et al. Alantolactone exhibited anti-herpes simplex virus 1 (HSV-1) action in vitro. Biosci. Trends 9, 420–422 (2015).
Namatame, I., Tomoda, H., Arai, M. & Omura, S. Effect of fungal metabolites cytochalasans on lipid droplet formation in mouse macrophages. J. Antibiot. 53, 19–25 (2000).
Ding, G. et al. Cytochalasans with different amino-acid origin from the plant endophytic fungus Trichoderma gamsii. J. Antibiot. 65, 143–145 (2012).
Qi, S. et al. and Sesquiterpenes from Eutypella scoparia 1-15. Nat. Prod. Commun. 10, 2027–2030 (2015).
Pittayakhajonwut, P., Usuwan, A., Intaraudom, C., Veeranondha, S. & Srikitikulchai, P. Sesquiterpene lactone 12,8-eudesmanolides from the fungus xylaria ianthinovelutina. Planta Med. 75, 1431–1435 (2009).
Li, Y. Y., Hu, Z. Y. & Shen, Y. M. Two new cyclopeptides and one new nonenolide from xylaria sp 101. Nat. Prod. Commun. 6, 1843–1846 (2011).
Huang, X. X. et al. Structure determinants of lagunamide A for anticancer activity and its molecular mechanism of mitochondrial apoptosis. Mol. Pharmaceutics 13, 3756–3763 (2016).
Acknowledgements
This work was supported by grants from the National Natural Science Foundation of China (No 81422045, U1405223 and 81661138005), the Fundamental Research Funds for the Central Universities of China (No 20720160064 and 20720160031), and Xiamen Southern Oceanographic Center (14GYY002NF02).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies the paper on The Journal of Antibiotics website
Supplementary information
Rights and permissions
About this article
Cite this article
Wang, Y., Wang, Y., Wu, Aa. et al. New 12,8-Eudesmanolides from Eutypella sp. 1–15. J Antibiot 70, 1029–1032 (2017). https://doi.org/10.1038/ja.2017.89
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ja.2017.89