Abstract
The advent of modern antibiotics contributed enormously to the dramatic extension of human lifespan since their discovery by virtue of their lethal and selective action against pathogenic microbes. And yet despite our powerful arsenal of weapons against these pathogens, the war against them has not been won. And it may never be. Drug resistance is still menacing the society with many lives being lost due to deadly infections caused by continuously evolving strains spread beyond our means to eradicate them or prevent their spreading. Herein, the emergence and evolution of antibiotics is briefly reviewed, and a select number of total syntheses of naturally occurring antibiotics from the authors’ laboratories are highlighted. The article concludes with a strong endorsement of the current efforts to intensify our fight against these dangerous pathogens with the hope that, this time, these initiatives will be sufficiently focused and serious enough so as to achieve our set goals of, at least, being prepared and ahead of them as part of our drive to improve humanity’s healthcare and wellbeing.
Similar content being viewed by others
Historical perspective
The origin of the term ‘antibiotic’ can be traced back to the word antibiose first used as an antonym to symbiosis by Paul Vuillemin1 in his 1890 publication to describe the antagonistic action between different microorganisms (for example, fungi vs bacteria; bacteria vs protozoa).2, 3 Later, the word antibiotic was used to describe naturally occurring secondary metabolites produced by bacteria and fungi possessing either growth inhibitory (bacteriostatic) or killing (bactericidal) activities against bacteria or fungi. Today the term has a broader meaning, in one sense to include designed molecules and a narrower definition, in another and with the terms antibacterial or antifungal, to designate their specific actions against bacteria and fungi, respectively, but not viruses.2 The term antiviral is reserved for the latter. In the Middle Ages, the miasma (or miasmatic, from the Greek μίασμα, meaning pollution, fowl air or contaminated vapors) theory was used to explain the causes of various diseases. In 1546 the Italian scholar Girolamo Fracastoro (Hieronymus Fracastorius) proposed transmittable, imperceptible seed-like particles (seminaria morbi) as causative agents for the outbreak of certain diseases.4 It took several centuries to prove that bacteria are one example of such ‘particles’. In the meantime, the Hungarian physician Ignaz Semmelweis, also known as the ‘savior of mothers’, recognizing the importance of hygiene in hospitals suggested and enforced hand washing with chlorinated lime solutions before patient examinations, while the Scottish surgeon Sir Joseph Lister treated surgery wounds with phenol (carbolic acid) solutions to avoid infections. In the 1860s the French physician Casimir Davaine demonstrated that blood injections from animals infected with anthrax to healthy animals caused infection, providing further clues for the cause of disease. It was also in the second half of the nineteenth century that the French chemist and bacteriologist Louis Pasteur contributed decisively to our understanding of the underlying causes of infectious diseases through his experimental studies with bacteria that culminated in his germ theory of disease. The subsequent contributions of the German physician Robert Koch also proved transformative. His discoveries, including the so-called four ‘Koch postulates’ establishing a causative relationship between microbes and disease (predicated on work of Jakob Henle,5 and together with Friedrich Loeffler), propelled bacteriology toward its modern era.6, 7, 8 In 1882 Koch, together with Bernhard Fischer and Georg Gaffky, isolated the Mycobacterium tuberculosis and in 1884 the Vibrio cholerae (a species first described in 1854 by Italian anatomist and pathologist Filippo Pacini),9 the causative strains that lead to the corresponding diseases. These milestone discoveries earned Koch one of the first Nobel Prizes in Physiology or Medicine (1905).
The first antibiotic to be discovered from nature was mycophenolic acid (1, Figure 1). Reported by the Italian physician and microbiologist Bartolomeo Gosio in 1893, this antibiotic was isolated from Penicillium glaucum (P. brevicompactum) as a crystalline solid while he was studying pellagra.10, 11 At that time it was shown that mycophenolic acid inhibits the growth of Bacillus anthracis, and later that it also possess antiviral, antifungal, antitumor and anti-psoriasis properties.12 This seminal discovery remained unnoticed (probably due to its publication in Italian) until mycophenolic acid was rediscovered in 1913 in the United States.13 Its structure, however, remained unknown until 1952,14 while its total synthesis was achieved in 1969.15, 16 It was not until 1995 that the 2-(morpholin-4-yl)ethyl ester of mycophenolic acid (a prodrug of 1) was approved by the US Food and Drug Administration, although not as an antibacterial drug but rather as an immunosuppressant to prevent transplant rejection (through a mechanism involving the inhibition of DNA biosynthesis).17
The first decade of the twentieth century witnessed the emergence of the first man-made antibacterial agent, arsphenamine (Salvarsan, 2, Figure 1). Synthesized by chemist Alfred Bertheim in the laboratories of Paul Ehrlich and approved in 1910 as a drug,18 the story of Salvarsan began when the immunologist Ehrlich was introduced to cell staining by his cousin Carl Weigert, who demonstrated that aniline dyes can be used for this purpose in histology.19, 20, 21, 22 Ehrlich was fascinated by the fact that selective staining of different cell types, including bacteria, was possible, and this led him to contemplate the idea of chemotherapia specifica and to propose his famous concept of the Zauberkugel (magic bullet) that continues to inspire biologists and chemists alike today. Starting in 1906, Ehrlich and his group started to synthesize and test compounds derived from arsanilic acid (atoxyl) against bacterial strains. In 1909, a year after Ehrlich shared the Nobel Prize in Physiology or Medicine with Ilya Mechnikov for their work on immunology, he and his assistant Sahachirō Hata found that compound 606 was effective against bacterial strains of Spirochaetes, and against protozoa Trypanosoma, findings that led to clinical trials, and eventual marketing of the drug for the treatment of syphilis, a disease caused by bacterium Treponema pallidum. For nearly 40 years Salvarsan was the standard therapy for this disease, with new versions (Neosalvarsan and Solu-Salvarsan) being introduced later as improved drugs,23, 24, 25, 26 allowing for easier application and causing less side effects. Interestingly, the precise molecular structure of arsphenamine remained vague, with Ehrlich assuming it was dimeric in nature with the two As-atoms doubly bonded. A recent mass spectrometric investigation, however, revealed both trimeric (see 2, Figure 1) and pentameric species.27
Staphylococci, a summer sojourn, and the serendipitous scrutiny of a Scottish scientist are the three ingredients that led to one of the most impactful discoveries in the history of medicine: penicillin (3, Figure 1). After returning from his vacation in September 1928, Alexander Fleming, a bacteriologist at St Mary’s Hospital, went back to his laboratory in Paddington (Westminster, London) and found a Staphylococcus aureus colony he had left on his bench contaminated with a fungus (Penicillium notatum, now P. chrysogenum) in an intriguing pattern. This finding had occurred already to other scientists before him, but Fleming did not dispose of the contaminated petri dish without further examination. He realized that in proximity to the mold, the Staphylococci underwent lysis while colonies further away appeared to be normal and unaffected. Fleming went on to grow the fungus as a pure culture and to treat several pathogenic bacterial strains with its extract to descry similar effects as previously observed with the Staphylococci, especially for Gram-positive ones. He concluded that the fungus must have excreted a substance that killed the bacteria, and named it penicillin in March 1929.28 Fleming attempted early on to collaborate with a number of researchers in an effort to isolate and purify the unknown substance, hoping to demystify the puzzle of penicillin, but without success. In 1939, pathologist Howard Walter Florey and biochemists Ernst Boris Chain and Norman Heatley at Oxford University started to work on the isolation of the mystery substance, and within a year managed to test the isolated compound in a mouse model, revealing phenomenal results. In early 1941, the first human, policeman Albert Alexander who was suffering from a severe face infection causing a sepsis, was treated with penicillin. His condition improved significantly during the first five days of therapy, but unfortunately the limited drug supply run out and the patient died from the prevailing sepsis. However, it became clear to the clinicians that penicillin was a powerful drug with auspicious properties. Later, and after painfully securing adequate supplies of the drug, several other patients were treated successfully and with full recovery. Out of necessity, and motivated by the outbreak of the Second World War, the collaborative Anglo-American penicillin project was launched. Its aim was to produce sufficient amounts of the new antibiotic for the needs of the allied forces fighting and dying from infections in the battle fields of Europe, Africa and Asia. In 1945, penicillin became available to the public and was no longer an exclusive medication for the military, consummating in the transformation of an allegedly failed experiment to a life-saving drug for millions. The same year, the structure of penicillin was elucidated through X-ray crystallographic analysis by Dorothy Crowfoot Hodgkin,29 allowing it to be classified as the first member of the β-lactam family of naturally occurring antibiotics. Later that year, Alexander Fleming, Ernst Boris Chain and Howard Walter Florey were jointly awarded the Nobel Prize in Physiology or Medicine for the discovery of penicillin and its curative effect in various infectious diseases. The first total synthesis of a member of the penicillin family (that is, penicillin V) was reported in 1957 by John Sheehan.30, 31 It featured five linear steps and resulted in 1.4% overall yield of the product. Even though not useful for industrial production, the synthesis provided the first opportunity to prepare analogs of penicillin from 6-aminopenicillanic acid, an intermediate that emerged from the endeavor. In contrast to the mode of action of the sulfonamides, penicillin and its other β-lactam siblings act bactericidally and, through a mechanism that involves inhibition of serine-type D-alanyl-D-alanine carboxypeptidase, an enzyme crucial to the peptidoglycan layer of the bacterial cell wall.
Exerting its antibiotic activities (bactericidal) through the same mode of action as penicillin, cephalosporin C (3a, Figure 1), the first member of the cephalosporins (a sub-class of β-lactams), was originally discovered in 1945 by Giuseppe Brotsu32, 33 from Cephalosporium acremonium (now Acremonium chrysogenum). Cephalosporin C was rediscovered in 195534 and structurally elucidated by chemical degradation studies35 and X-ray crystallographic analysis in 1961.36 Its first total synthesis was achieved by Robert B. Woodward in 1965 and reported in his Nobel lecture before it was published in 1966.37 Although cephalosporin C did not become a clinical agent, its structure inspired numerous synthetic efforts that led to designed analogs from which emerged powerful antibacterial drugs, with cefalotin being the first clinically approved cephalosporin antibacterial agent.
The next advance in the antibiotics area was marked by the discovery and deployment of the sulfonamide drugs. First synthesized as a dye by Fritz Mietzsch and Josef Klarer at Bayer in December 1932, sulfamidochrysoidin (4, Prontosil, Figure 1) was recognized as an antibacterial agent by Gerhard Domagk,38 a fortunate event that led to its marketing as a drug in 1935, and a Nobel Prize in Physiology or Medicine to him in 1939. Before the Nobel Prize and the approval of the drug, Domagk and his family were rewarded with the saving of the life of his 6-year-old daughter who was suffering from a life-threatening sepsis. He decided to use the then still experimental drug, code named D 4145 (Streptozon), to treat her, a decision that must have given him the most happiness of all his prizes. In the same year that Prontosil was approved, Jacques and Thérèse Tréfouël, Federico Nitti, and Daniel Bovet at the Pasteur Institute determined that sulfamidochrysoidin acts as a prodrug, releasing in vivo the active antibacterial agent p-aminophenylsulfonamide (through reductive cleavage of the diazo linkage).39 The latter compound, however, had already been synthesized by the Austrian chemist Paul Gelmo in 1908,40 and hence this molecular entity and compounds releasing it were not patentable. This predicament led to a surge of activities in the 1930s and 1940s, primarily in Europe and the USA, that resulted in the discovery and development of several new and patentable drugs. The sulfonamides are bacteriostatic agents, acting through a mechanism that involves inhibition of folic acid biosynthesis in bacteria, leaving normal human cells unharmed since the latter have no folic acid biosynthesis machinery.41
A new chapter on antibiotics was opened in 193942, 43, 44 when French microbiologist René Dubos isolated tyrothricin45, 46, 47 from Bacillus brevis. It was later shown that tyrothricin was a mixture consisting of gramicidin D (itself a mixture of gramicidins A1/2–C1/2, all linear pentadecapeptides differing at positions one and/or eleven of the amino acid sequence) and tyrocidine (a mixture consisting of tyrocidines A–D). In 1942, Soviet biologist Georgyi Frantsevich Gause and his wife Maria Brazhnikova discovered another gramicidin from the same bacterial strain (Bacillus brevis) and named it gramicidin S (5, Figure 1).48 A symmetrical macrocyclic homodetic decapeptide [cyclo(D-Phe-Pro-Val-Orn-Leu-)2], gramicidin S is also classified as a member of the polypeptide antibiotics mentioned above but differs from the polymyxins, another family of cyclic decapeptides, in that it lacks the characteristic extended lipophilic side chain of the latter. Gramicidin S was found to be active against Gram-positive and some Gram-negative bacteria. It was used for the first time in the year of its discovery (1942) in military hospitals, and starting in 1943 in the eastern front during the Second World War, making it the Soviet counterpart of penicillin.49 And, interestingly just like penicillin, the structure of gramicidin S was solved in Great Britain, this time by Richard Synge who employed classical derivatization/degradation methods,50, 51, 52 and Hodgkin who used X-ray crystallography to decipher the structure,53 after the Red Cross brought a sample from Moscow.49 In 1964, Hodgkin was awarded the Nobel Prize in Chemistry for her determinations by X-ray techniques of the structures of important biochemical substances. The total synthesis of gramicidin S was reported in 1957 by Robert Schwyzer54, 55 following previously published methods.56 The clinical application of gramicidin S is limited to topical infections (for example, eye, ear) due to its undesired hemolytic properties originating from membrane lysis.57 Its mechanism of action as a bactericidal antibiotic relies on its ability to form ion channels in the bacterial cell wall upon dimerization, causing collapse of the electrochemical gradient.58, 59
Another important milestone in the history of antibiotics was the discovery of streptomycin (6, Figure 1) from Actinomyces griseus (now Streptomyces griseus), the first aminoglycoside antibiotic, in 1943 by Albert Schatz and Selman Waksman.60, 61 Significantly, it was shown that, in addition to being active against Gram-positive strains, the newly isolated antibiotic was also potent against multiple Gram-negative bacteria as well as against Mycobacterium tuberculosis.62 Since the latter strain was not affected by penicillin, streptomycin was meeting an unmet medical need, offering physicians and patients suffering from tuberculosis hope for a potential cure for the first time.63 The development of streptomycin as a powerful drug against this dreaded disease eventually led to the Nobel Prize in Physiology or Medicine to Waksman in 1952, but not without creating a bitter dispute between Waksman and Schatz over their individual contributions and the wealth of royalties from the streptomycin patent.64, 65 The first human cure of an infection by streptomycin was reported in 1944, and was followed by the successful treatment of a patient suffering from tuberculosis 2 months later.65 Upon completion of successful clinical trials, the drug was approved for broad clinical use in 1946. After full structural elucidation of streptomycin,66, 67, 68 the first total synthesis of the natural product was achieved in 1974 by Sumio Umezawa et al.,69 the group having first completed the synthesis of dihydrostreptomycin.70 The mode of action of streptomycin (bactericidal) involves inhibition of bacterial protein biosynthesis71 through binding to the 30S subunit of the prokaryotic ribosome.72
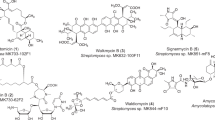
The first member of one of the most important classes of antibiotics, that of the tetracyclines, was unearthed in 1945, being isolated from Strep. aureofaciens by American plant physiologist Benjamin M. Duggar. It was originally named aureomycin due to its golden color, but is known today as chlortetracycline (7, Figure 1).73 In 1948, it was approved for clinical use as a broad spectrum antibiotic against both Gram-positive and Gram-negative bacteria.74 Soon thereafter, several other members of the tetracycline family were isolated at Pfizer (New York, NY, USA) by a group headed by Alexander C. Finlay, including oxytetracyclin (from Strep. rimosus) and tetracycline (from Strep. aureofaciens).75 The gross structure of chlortetracycline was first determined by Woodward through chemical derivatization methods in 1954.76 This was followed by X-ray crystallographic studies in 195977, 78 and 196379, which led to the elucidation of the relative stereochemical structure of chlortetracycline. Although a total synthesis of chlortetracycline (7), oddly enough, has not been reported as yet, a formal total synthesis was reported in 1973 by Hans Muxfeldt et al.80, 81 The formal total synthesis of tetracycline (7-dechloro-7) itself was achieved by Mikhail M. Shemyakin in 196782, 83, 84, 85 while Woodward had already succeeded in synthesizing the first biologically active tetracycline derivative in 1962.86, 87 Furthermore, several other tetracyclines have been synthesized over the years.88, 89, 90, 91, 92, 93, 94, 95, 96, 97, 98 Like streptomycin, the tetracyclines exert their bacteriostatic properties through a mechanism involving inhibition of bacterial protein biosynthesis by binding to the ribosome at the 30S site.74 Numerous tetracycline derivatives have been synthesized in efforts to overcome the emergence of resistance to the original drugs, including tigecycline (7a, Figure 2a), which was approved by the US Food and Drug Administration in 2005.99
Before the end of the decade of the 1940s, yet another original antibiotic made its entrance, chloramphenicol (8, Figure 1), a member of the phenylpropanoids. Discovered initially by John Ehrlich in 1947 (from Strep. venezuelae),100 and shortly thereafter and independently by David Gottlieb et al.101, 102 and Umezawa et al.,103 chloramphenicol proved to be a broad spectrum antibiotic and was used clinically for the first time, and with remarkable success during the 1947 typhus epidemic in Bolivia.75 Its chemistry was investigated in 1948104 and its structure was proposed105 and confirmed by total synthesis106 in 1949. Chloramphenicol was approved for clinical use as an antibacterial agent in the same year and became one of the best-selling drugs at the time. Its mechanism of action (bacteriostatic) involves binding to the 50S subunit of the bacterial ribosome, resulting in protein biosynthesis inhibition.107, 108 It is notable that chloramphenicol became the first naturally occurring antibiotic produced by chemical synthesis, rather than fermentation. This may be a reflection of a combination of factors, including its relative structural simplicity and the increasing capabilities of organic synthesis at the time.
Besides the rush for the discovery of antibiotics from nature triggered by penicillin’s spectacular success, the 1950s saw the re-emergence of interest in synthetic designed antibacterial agents such as Salvarsan and Prontosil, two of the very first antibacterial agents to be introduced for clinical use. The opportunity arose when a rather simple natural antibiotic, azomycin (2-nitroimidazole) was originally discovered (1953)109 and structurally elucidated (1954).110 Azomycin proved too toxic for clinical use, but nevertheless inspired scientists at Rhône-Poulenc in France as a lead compound to design and synthesize analogs of it, from which metronidazole (9, Figure 1) was selected as a drug candidate for further development due to its high potency and low toxicity.111, 112, 113 Metronidazole was approved as a clinical agent in France in 1960 and in the United States in 1963.113 It exerts its antibacterial properties through a distinctively different mechanism than the other antibiotics discussed above in that it damages directly the genetic material of the bacteria through intracellular generation of nitroso radicals.114 These reactive nitroso radicals are produced reductively through a single-electron transfer to the nitro group. This mechanism is expected to operate especially in anaerobic bacteria such as Bacteroides, Clostridium, Helicobacter or Campylobacter, but hardly in human cells, endowing the drug with the desired bactericidal properties against bacteria without side effects to the patient.114
Another inspirational antibiotic that did not actually make it to the clinic is pleuromutilin (10, Figure 1).115 Isolated in 1951 by Frederick Kavanagh from basidiomycetes Pleurotus passeckerianus (now Clitopilus passeckerianus) and Pleurotus mutilus (Fr.) Sacc (now Clitopilus scyphoides).116, 117 Found active predominantly against Gram-positive bacteria, but also against a number of Gram-negative, pleuromutilin was structurally elucidated in 1962 after extensive chemical studies118 conducted independently at Manchester (UK)119, 120 and Zürich (Switzerland).121, 122 Its structure was confirmed by X-ray crystallographic analysis and ORD studies in 1968.123 Pleuromutilin was synthesized first, in racemic form (1982),124, 125 and thence enantioselectively (2013) by David J. Procter.126 Numerous semisynthetic analogs of pleuromutilin have been synthesized and tested, one of which, retapamulin (carrying a thiotropine moiety at the glycolic acid side chain), was approved as a clinical antibacterial drug in 2007 in the United States. The mechanism of (bacteriostatic) action of the pleuromutilin family of antibiotics is based on their ability to bind the 50S subunit of the bacterial ribosome.127
The 1950s would usher in a new crop of novel antibiotics, among the most prominent of which were the erythromycins. Endowed with complex 14-membered macrolide structures, erythromycins A and B were isolated by Eli Lilly (Indianapolis, IN, USA) scientists led by James M. McGuire in 1952 from Saccharopolyspora erythraea, found in a soil sample imported to the USA from the Philippines. Erythromycins A and B were marketed in the same year under the name Ilotycin128, 129 as a mixture of compounds. This mixture was later separated and the main components were renamed as erythromycins A–D, with A being the most abundant. The molecular structure of erythromycin A (11, Figure 1) was first determined in 1957130, 131, 132, 133, 134, 135, 136, 137, 138 with the X-ray crystallographic analysis of the dihydrate reported in 1997.139 The total synthesis of erythromycin A was accomplished and published by the Woodward group (posthumously for Woodward) in 1981.140, 141, 142 It was preceded by the total synthesis of erythronolide B (the aglycon of erythromycin B) which was accomplished by the Corey group in 1978.143, 144 The mechanism of action of the erythromycins (bacteriostatic or bactericidal, depending on medicated strain and concentration) involves binding to the 50S subunit of the ribosome and inhibition of bacterial protein biosynthesis.145, 146
Soon after the discovery of the erythromycins, another family of powerful antibiotics, that of the glycopeptides, was discovered. The first member of the class was isolated in 1953 from Strep. orientalis (now Amycolatopsis orientalis) found in a soil sample originating from Borneo.147 Later named vancomycin (from its ability to vanish bacteria) (12, Figure 1), this legendary antibiotic was found to be especially active against Gram-positive bacteria, and most significantly against penicillin-resistant Staphylococci. Vancomycin was approved as a clinical agent for the treatment of bacterial infections in 1958. Its use was later decreased in favor of other antibiotics for toxicity reasons,148 although it continues to be employed in severe cases against drug-resistant bacteria. The quest for the structural elucidation of vancomycin involved chemical studies (1977)149, 150, 151 and X-ray crystallographic analysis, first of its degradation product CDP-1 (1978)152 and thence the natural product itself (1996).153 The first total synthesis of the aglycon of vancomycin was accomplished in 1998 by the Evans group154, 155 and our group.156, 157, 158 The total synthesis of vancomycin itself was achieved in 1999 by our group.159 The mechanism of bacteriostatic action of vancomycin resembles that of the β-lactams in that it interferes with the construction of the bacterial cell wall. It exerts its disruptive activity by binding to the D-Ala-D-Ala unit of a pentapeptide moiety essential for the knitting of the peptidoglycan network of the cell wall, thereby hindering its proper formation.160
In the same year that vancomycin was discovered, another new antibiotic was isolated, ushering in the family of streptogramins. The eponymous compound, streptogramin was isolated as a secondary metabolite from a then unknown Streptomyces spp. (later named Strep. graminofaciens) and found to be especially active against Gram-positive bacterial strains.161, 162 It was subsequently found that streptogramin is a mixture of two distinct components, streptogramin A (also named pristinamycin IIA, virginiamycin M1, ostreogrycin A, mikamycin A, 13, Figure 1) and streptogramin B (also named pristinamycin IA, virginiamycin S1, mikamycin B, vernamycin Bα, 14, Figure 1).163 The structures of both components were elucidated during the mid-1960s164, 165, 166 and confirmed by X-ray crystallographic analysis in 1974167 (streptogramin A) and in 1990168 (streptogramin B). Being comprised of two structurally unrelated, but concurrently produced compounds by a single species and classified as groups A and B, this paradigm represents a special case characteristic of the streptogramins. Substances of group A feature a 23-membered polyunsaturated macrolactone framework while members of group B possess a cyclic hexa- or hepta depsipeptide scaffold. Remarkably, the streptogramins demonstrated strong synergistic effects when administered in combination.163, 169 The mechanism of their antibacterial action relies on sequential binding of first streptogramin A, and then B to the 50S subunit of the bacterial ribosome, thus inhibiting protein biosynthesis. When applied as single substances, each compound acts bacteriostatically, but when used together they develop bactericidal activities.170, 171 For a long period of time, the streptogramins were mostly used in the agricultural sector as growth promoters. They were first launched in 1973 (as a 1:1 mixture of 13 and 14) as pristinamycin in Europe for the treatment of infections caused by Streptococci and Staphylococci. Later, studies at Rhône-Poulenc Rorer directed toward improving the water solubility of streptogramins A and B led to the semisynthesis of quinupristin and dalfopristin, respectively, two analogs that were successfully developed in a 3:7 combination as an antibacterial drug172 and approved by the US Food and Drug Administration in 1999. This combination medication is used especially in severe cases of infections associated with vancomycin-resistant Enterococcus faecium. Since their discovery several synthetic approaches toward the total synthesis of select members of the streptogramin family have been reported,173, 174, 175, 176, 177, 178, 179 with some successfully reaching their targets.180, 181, 182, 183, 184, 185
Trimethoprim (15, Figure 1) is another early unnatural antibiotic. Synthesized in 1962, this compound was found to exhibit strong antibacterial activities against both Gram-positive and Gram-negative strains.186 In the same year, the first successful clinical data were reported,187 and included synergistic effects when the drug was administered to patients together with sulfonamides.188 Trimethoprim was marketed in 1974 as co-trimoxazole, a combination medication consisting of one part trimethoprim to five parts sulfamethoxazole, and later also as a single molecular entity drug.189 Trimethoprim acts bacteriostatically through inhibition of dihydrofolate reductase, an enzyme involved in the bacterial folic acid biosynthesis. It has the distinction of being the first member of the dihydrofolate reductase inhibitors.190
Nalidixic acid (16, Figure 1) was also synthesized in 1962 and found to be highly potent in vitro and in vivo,191 its activity being most pronounced against Gram-negative bacteria such as Escherichia coli, Proteus mirabilis and Shigella flexneri, while exhibiting low toxicity.191 This discovery turned out to be highly significant in that it led to the emergence of quinolones, an important family of synthetic antibiotics whose structures are related to quinoline.192 Depending on the concentration, nalidixic acid can act as a bacteriostatic or bactericidal agent.193 Its biological target is topoisomerase II,194, 195 an enzyme catalyzing the ATP-dependent negative supercoiling of double-stranded closed-circular DNA,196 thus causing interruption of bacterial DNA biosynthesis.197 Nalidixic acid was launched as a clinical antibacterial agent in 1967,192 but was later replaced by fluoroquinolones like ciprofloxacin (16a, Figure 2a) which proved superior as drugs.
The ansamycins198 are a group of macrolactam antibiotics whose first members were reported in 1959 by Piero Sensi, Maria T. Timbal and Pinhas Margalith.199 Named rifamycins, these early ansamycins were isolated from Strep. mediterranei (now Amycolatopsis rifamycinica). The structures of a number of them were elucidated through chemical degradation studies in 1964,200 while that of rifamycin B was determined in the same year by X-ray crystallographic analysis.201, 202 Naturally occurring rifamycin B failed to become a drug, but one of its slightly modified semisynthetic versions (that is, rifamycin SV) was approved as a clinical antibacterial agent especially effective against tuberculosis,203 albeit intravenously delivered. Further studies led to improved and orally active drugs,204 one of them being rifaximin (17, Figure 1).205 The latter is specifically used to treat colon-related infections without significant systemic side effects due to its poor absorption properties upon oral administration,204 and thus only effective in the intestinal lumen. The bactericidal action of the rifamycins originates from their ability to inhibit bacterial DNA-dependent RNA biosynthesis, which in turn inhibits protein biosynthesis.206, 207
After the introduction of the quinolones in the 1960s, it took, surprisingly, more than two decades for the next class of antibiotics to be ushered into the clinic. The story of linezolid (18, Figure 1) goes back to at least the 1970s when certain oxazolidinones were recognized by DuPont scientists as possessing antibacterial properties.208, 209 It was, however, a group at the Upjohn pharmaceutical company (Kalamazoo, MI, USA)210, 211 that discovered and developed linezolid as a clinically used antibacterial agent,212 approved in the United States in 2000.210 Linezolid is particularly effective in treating infections caused by Gram-positive bacterial strains such as Streptococci and Enterococci, especially in cases where other antibiotics failed. The mechanism of action of this bacteriostatic last resort antibiotic213 involves binding to the 50S subunit of the ribosome and inhibition of bacterial protein biosynthesis.214
The so-called ketolide family of antibacterial agents arose from the erythromycin class through semisynthetic studies. It was scientists at Roussel-UCLAF in France who prepared compound RU-66647 from erythromycin A (11, Figure 1) in 1997 and showed it to be active against bacteria, especially Gram-positive strains.215 Later named telithromycin (11a, Figure 1), this first member of the ketolide family was successfully developed and launched as an antibacterial drug, first in Europe (2001) and thence in the United States (2004).216 Although derived from erythromycin A, telithromycin and its siblings structurally differ, significantly, from the latter in that they lack the l-cladinose sugar moiety at C3, exhibiting instead a keto group at that position and, thereby, improving the drug stability toward acidic conditions. In addition, the ketolides include in their structures a cyclic carbamate moiety fused onto the macrolide ring at C11 and C12, with the N-atom originating from replacement of the O-atom of the erythromycin precursor of the molecule. Telithromycin specifically includes an alkyl side chain on the carbamate nitrogen ending with two N-heterocyclic rings.217 Its mechanism of action resembles its precursor compound, erythromycin A, namely inhibition of bacterial protein biosynthesis through binding to domain II of the 23S rRNA site of the ribosome.218 Telithromycin generally acts bacteriostatically, but also shows bactericidal properties at higher dosings.219
Just like telithromycin, telavancin (12a, Figure 1) was derived semisynthetically from an abundant naturally occurring antibiotic, in this case vancomycin (12, Figure 1).220 It is a newer antibacterial drug belonging to the family of lipoglycopeptides, the name being derived from the lipid chain attached on the glycopeptide domain. In addition to the lipophilic decylaminoethyl side chain attached onto the terminal carbohydrate moiety, telavancin also features a hydrophilic phosphonomethyl aminomethyl substituent on the biphenyl domain of vancomycin.221 Its bactericidal properties toward several Gram-positive strains, including methicillin-resistant Staph. aureus (MRSA),222 derive from a dual mechanism of action. On one hand, it hinders the peptidoglycan construction by binding to its D-Ala-D-Ala dipeptide terminus, thus weakening the bacterial cell wall, and on the other it interferes with the cell wall precursor lipid II, thereby causing membrane depolarization.223 Telavancin was approved for clinical use in the United States in 2009.223
Today, more or less severe infections caused by microbes are alleviated with a variety of antibiotics, albeit not always successfully, from the large arsenal we have come to discover and develop from nature and the laboratory. Among the most prevalent of these drugs are amoxicillin (3b), ciprofloxacin (16a), azithromycin (11b), cephalexin (3c), levofloxacin (16b), tobramycin (19), tigecycline (7a), fosfomycin (20), moxifloxacin (16c), trimethoprim/sulfamethoxazole (21) and amoxicillin/clavulanic acid (22), all shown in Figure 2a. As a last line of defense, humanity has vancomycin (12, Figure 2b) and its analog telavancin (12a, Figure 1), amikacin (23, Figure 2b), linezolid (18, Figure 2b), colistin (polymyxin E, 24, Figure 2b), and amphotericin B (25, an antifungal agent, Figure 2b). Isolation and synthetic organic chemists have played a major role in the discovery and development of these life-saving antibiotics. To be sure they will continue to be protagonists in our seemingly never ending fight against pathogenic microbes.
Antibiotic resistance
Antibiotic resistance is apparently an inevitable consequence of the use of antibiotics against bacteria, a war waged against them by us but also, and for a much longer time, by bacteria against each other, ironically using the same weapons in both cases. That means that bacteria had the chance to develop resistance through various mechanisms to evade the action of these antibiotics which we isolated from them in the first place.224 This explains the so-termed primary resistance of certain bacterial strains that are often unaffected by some antibiotics, whereas they are susceptible to others (for example, Gram-negative vs Gram-positive).225 More dangerous is secondary resistance, the acquired ability of bacteria to withstand the action of antibiotics that were once effective against them. Such resistance may be developed through mutations involving genetic changes that result in inactivation of the drug (for example, β-lactamases), modification of previously essential biosynthetic pathways, or alteration of the target site (cell wall construction, folic acid biosynthesis, protein biosynthesis). Expulsion of the drug from the cell (efflux) is another mechanism bacteria can use to evade the action of antibiotics. These genetic changes can later spread through lateral gene transfer via transformation, transduction or conjugation.226, 227, 228, 229 It was as early as the 1940s when the problem of resistance development and the problems of self-medication were discussed in public,230 but even today in many countries antibiotics are available over the counter without prescription and are extensively used in veterinary context worldwide. Particularly dangerous are the drug-resistant bacterial strains collectively known as ESKAPE pathogens (vancomycin-resistant E nterococci, MRSA, K lebsiella pneumoniae, A cinetobacter baumannii, P seudomonas aeruginosa, E nterobacter spp.), which can cause difficult to treat nosocomial infections.231 Although the development of resistance is inevitable, its spreading is not and that is where we can make a difference.
Bacteria surpass us massively in numbers, total mass, replication time, and time of existence on earth.232 Our inability to eradicate their pathogenic strains is, therefore, not surprising. All we can do at present is to be vigilant and alert to stay ahead of them in our efforts to avoid catastrophic epidemics and deaths by them. Antibiotic resistance is a continuous threat to society as proven before and after penicillin’s discovery, whose action on certain bacteria became obsolete even before its development as a drug during the Second World War. Certain dangerous bacterial strains developed resistance to almost all approved antibacterial drugs since (such as tetracyclines, erythromycins, methicillin, gentamicin, vancomycin, imipenem, ceftazidime, levofloxacin, linezolid, daptomycin and ceftaroline), either immediately or shortly after their approval.233 And to exacerbate the situation, the number of new antibacterial drugs approved after the golden age of antibiotics (1940s to 1960s) has been steadily declining. It was only until recently, and after a lean time period of more than 30 years, that a new antibiotic, teixobactin (26, Figure 3), was discovered in 2015 and found to be impressively active against Gram-positive bacteria. Affecting the bacterial cell wall biosynthesis234 and showing promising properties against resistance development, this new antibiotic was also accessed by total synthesis in 2016.235 Even more recently, another new antibiotic, lugdunin (27, Figure 3), was isolated from Staph. lugdunensis and structurally elucidated,236 revealing its macrocyclic peptide nature. Its total synthesis confirmed its thiazolidine-containing constitution. The prevailing doctrine of using overwhelming force of antibiotics (larger doses and prolonged times) to eradicate all pathogenic bacteria in the patent first introduced by Fleming has recently being challenged. The new theory suggests that just the opposite might by the right strategy, as lower doses may be a wiser tactic based on the competition of non-resistant bacteria and drug-resistant bacteria allowing the patient’s immune system to finish the job of killing them both.233 This may depend on the drug used, however, and the final verdict is not in yet as further research is on the way.
According to a recent review on resistance to antimicrobial drugs by O’Neil,237 on behalf of the British government and the Wellcome Trust, 700 000 people die every year from infections caused by drug-resistant pathogens, and the number can reach 10 million by 2050 if nothing is done to stop the trend. According to a survey published in 2013238 approximately one-fourth of the 40 million people with respiratory diseases were prescribed antibiotics between 2007 and 2009 in the United States, albeit only rarely indicated for their specific condition and in 78% of these cases broad spectrum versions were recommended, despite the risk for more severe side effects.239 Here lies the paradox: a chance to be healed or to heal compels the patient or the doctor to ask or prescribe an antibiotic to an individual for the possibility of a cure without concern of spreading drug resistance.
Improvements in this regard can be achieved with rapid testing first, and then deciding whether an antibiotic will help the patient or not. And it will be even more helpful if the on-the-spot testing could reveal the type of antibiotic to be used effectively. There is no question that we are in a saddle war with pathogenic bacteria, although we are cognizant that we need bacteria around us as essential symbiotic partners. A response is necessary in order to avoid major catastrophes from occurring in the future as multiple indicators predict they will. Remedies to stop the antibiotic resistance problem include: (a) stop the use of antibiotics as growth hormones in animals; (b) develop and use vaccines where appropriate for prophylaxis as opposed to treatment; (c) enforce better hygiene practices in clinics and hospitals; (d) stop the prescribing of antibiotics for viral diseases before proper diagnosis; and (e) encourage innovation to develop new antibiotics through incentives to academia and industry. Suggestions to achieve the latter initiative have been made not only for increased funding for research and development but also for ‘entry rewards’ that will guarantee to the investors substantial initial financial rewards in addition to sales.
Highlights of selected total syntheses of antibiotics
The importance of natural products and their derivatives in medicine has been amply demonstrated through their direct clinical use, but also as biological probes that unravel biological pathways and targets for pharmacological intervention. The success of penicillin intensified the search for new antibiotics and other bioactive molecules from nature as evidenced from the postwar World War II surge in the discovery of such agents of which many became drugs to treat bacterial infections and cancer,240 among other diseases. Many became challenging targets for synthetic organic chemists because of their unusual molecular architectures and the opportunities they presented for discovery and invention of new synthetic strategies and technologies for broader use. Figure 4 depicts a number of selected naturally occurring antibiotics synthesized in our laboratories during the last few decades. Herein, we highlight the total syntheses of some of these antibiotics [that is, antibiotic X-14547A (indanomycin, 28), efrotomycin (29), amphotericin B (25), vancomycin (12), everninomicin 13,384-1 (ziracin, 30), coleophomones B and C (31 and 32), thiostrepton (33), abyssomicin C and atrop-abyssomicin C (34 and 34′), marinomycins A, B and C (35, 36, 37), platensimycin (38), platencin (39), the alleged rugulin structure (40), antibiotic BE-43472B (41), hirsutellone B (42), epicoccin G, 8,8′-epi-ent-rostratin B and emethallicin E (43, 44 and 45), viridicatumtoxin B (46), and antibiotic CJ-16,264 (47)].
Antibiotic X-14547A (indanomycin)
Isolated from Strep. antibioticus NRRL 8167,241, 242 antibiotic X-14547A (28, Figure 5a) belongs to the ionophore class of antibiotics and forms, interestingly, a 2:1 (acid:base) salt with (R)-(+)-1-amino-1-(4-bromophenyl)ethane. The latter yielded suitable crystals for X-ray crystallographic analysis that led to the complete structural elucidation of the molecule.242 Its properties include antibacterial activities against Gram-positive bacteria, antihypertensive and antitumor activities and growth promotion in ruminants.241, 242, 243, 244 Its enantioselective total synthesis was achieved in our laboratories in 1981.245, 246, 247 The employed retrosynthetic analysis is shown in Figure 5a, whereas highlights of the devised total synthesis are summarized in Figure 5b. The initial synthetic maneuvers were the conversion of (−)-diethyl tartrate (48) to (S,S)-epoxide 49 (chiral pool) and the asymmetric alkylation of SAMP-hydrazone 54 to afford aldehyde 55 (asymmetric synthesis). These two intermediates were converted to the required tetrahydropyran fragment 53 (through intermediates 50–52) and tetrahydroindan 58 (through intermediates 56–57), respectively. These intermediates were coupled to afford advanced precursor 59, which was converted to carboxylic acid 60. The latter was transformed to antibiotic X-14547A (28) through a sequence involving conversion to the corresponding 2-pyridinylthioester, its reaction with pyrrolylmagnesium chloride to install the 2-ketopyrrole system, and methyl ester hydrolysis. Several other synthetic approaches248, 249, 250 and total syntheses251, 252, 253, 254, 255 of antibiotic X-14547A have been subsequently reported.
Efrotomycin
Efrotomycin (29, Figure 6a) (isolated from Nocardia lactamdurans)256 and its aglycon (aurodox or goldinomycin) are members of the elfamycin family of antibiotics. Their biological actions include activities against bacilli and dysentery, as well as growth promoting properties in ruminants.257, 258, 259 The structure of efrotomycin contains two glycoside bonds, one amide linkage, a number of olefinic units, and several other interesting structural motifs. The devised total synthesis of this antibiotic was based on the retrosynthetic disconnections indicated in Figure 6a. It proceeded from key building blocks 61, 62, 64, 68 and 73 and through advanced intermediates 63, 66, 72, 74 and 75 as summarized in Figure 6b. Key steps in this synthesis260, 261, 262 are the stereoselective glycosylation reactions of 61 and 62 to afford disaccharide 63, the cascade sequence involved in the conversion of diepoxide 69 to the tetrahydrofuran intermediate 70, the stereoselective coupling of aldehyde 72 with the phosphorane derived from 74 to form the conjugated triene precursor of advanced intermediate 75, and the final coupling of the latter with γ-lactone fragment 67, which generated efrotomycin (29) and its acetate (76, for final comparison with the acetate of the natural product), upon global deprotection and selective acetylation, respectively. This endeavor demonstrated the use of glycosyl fluorides and arylthioglycosides as glycosyl donors in complex molecule construction, pushed the frontiers of total synthesis to new levels of molecular complexity and diversity at the time, and provided opportunities for analog construction within the efrotomycin family of antibiotics.
Amphotericin B
A powerful antifungal263, 264 and antiprotozoal265 drug, initially isolated in 1954 from Strep. nodosus Trejo,266 amphotericin B (25, Figure 7a) possesses an imposing structure featuring a 38-membered macrolide ring, a conjugated heptaene system, an aminosugar, a host of hydroxyl groups and a carboxyl group. Its 19 stereogenic centers, β-glycoside bond and macrocyclic ring were the primary challenges from the synthetic point of view. Figure 7a presents the strategic bond disconnections that led to the definition of building blocks 77, ent-77, 78, ent-49 and 92 as starting materials for its total synthesis.267, 268, 269, 270 As outlined in Figure 7b, the enantiomeric building blocks 77 and ent-77 [(+)- and (−)-xylose, chiral pool approach] were selectively converted to key intermediates aldehyde 79 and ketophosphonate 80. The same building blocks were also synthesized from the prochiral allyl alcohol 78 through the then newly discovered catalytic asymmetric epoxidation reaction, furnishing epoxides 81 and ent-81 in high chemical yield and enantiomeric excess. Horner–Wadsworth–Emmons coupling of ketophosphonate 80 with aldehyde 79, followed by reduction of the resulting enone led to ketone 82. Further elaboration of the latter furnished methylthiophosphonate 83, whose reaction with aldehyde 84 (obtained from tartaric acid-derived epoxide ent-49; as shown in Figure 5b) afforded methylthioenol ether 85. This key intermediate was converted to ketophosphonate 86. Coupling of the latter with conjugated aldehyde 89 (also obtained from epoxide ent-49 through a different route through intermediates 87 and 88) furnished ketophosphonate aldehyde 90ab, which proved a worthy precursor to the coveted macrocyclic polyene 91ab under carefully defined conditions (K2CO3, 18-crown-6). Impressively, polyenone 91ab was selectively reduced and glycosylated with carbohydrate donor 92 to afford, selectively, the desired β-glycoside, whose further elaboration led to amphotericin B (25). Featuring both the chiral pool (auxiliary based) and catalysis approaches to asymmetric synthesis and highlighting the power of the ketophosphonate/aldehyde macrolactonization, this total synthesis served as facilitator to further research on amphotericin B and related compounds that led to new insights into the mechanism of action of this important antifungal agent, and novel drug candidates for clinical development.271, 272, 273, 274 For decades the presumed explanation for its antifungal action was that amphotericin B creates pores in the pathogen’s outer membrane causing leakage of monovalent ions. The newly acquired information suggested a different mode of action, including the formation of extramembranous aggregates that extract ergosterol from the lipid bilayers in a sponge-like fashion.274 Several other synthetic approaches to fragments and derivatives of amphotericin B have also been reported by other groups.275, 276, 277, 278, 279, 280, 281, 282, 283, 284, 285, 286, 287, 288, 289, 290, 291, 292, 293, 294, 295
Vancomycin
Vancomycin (12, Figure 8a), known once as the last line of defense of humanity against bacteria, was not only a vanquishing agent for them, but also a demonic challenge to synthetic chemists for a long time, the latter designation being due to its unusual macrocyclic motifs and 20 stereogenic elements. The flagship of glycopeptide antibiotics, vancomycin was isolated from Strep. orientalis (later renamed, first to Nocardia orientalis and then to Amycolatopsis orientalis).296, 297, 298 Its mechanism of action as mentioned in the previous section involves interference with the bacterial cell wall biosynthesis through binding to D-Ala-D-Ala structural motifs. Its total synthesis was reported from our laboratories in 1999.159 Figure 8a indicates the strategic bond disconnections of vancomycin that led to the devised synthetic strategy and defined key building blocks 93–99 as shown in Figure 8b. Figure 8c highlights the reactions employed for the assembly of these fragments and elaboration of the growing intermediates (100–111) into the final target. Among them are the palladium-catalyzed coupling of building block 93 and 94 to afford, upon further elaboration, azidobiaryl carboxylic acid 100. The latter was coupled with aminoester fragment 95, leading to the corresponding amide whose Boc group was removed to afford amine 101 in preparation for the next coupling. That step involved amide bond formation between carboxylic acid 99 and amino building block 101, which furnished the desired precursor for the first macrocyclization reaction, phenolic dibromotriazine 102. Intramolecular displacement of one of the two bromide residues by the phenolic hydroxyl group under the influence of CuBr·Me2S led to the macrocyclic bis-aryl ether 103, together with its atropisomer. The stage was then set for the next macrocyclization (103→104→106), and then the next (106→107→108), leading to complete vancomycin aglycon framework 109 upon further manipulation of the triazine moiety of 108 to the required phenolic group as summarized in Figure 8c. Vancomycin aglycon 110 was then generated from 109 via a seven-step sequence. Vancomycin itself was synthesized from its aglycon (110) via intermediate 111 by sequential glycosylations with glycosyl donors 98 and 97, followed by deprotections and functional group adjustments as highlighted in Figure 8c. The vancomycin endeavor provided opportunities for new methodological advances, including the triazine-driven macrocyclic arylether forming reaction to cast the two bis-arylether domains of the molecule, as well as new solid phase combinatorial chemistry for analog construction. Significantly, a number of the analogs that emerged from these studies proved to be more potent than vancomycin against drug-resistant bacterial strains.299, 300, 301
Everninomicin 13,384-1 (ziracin)
Isolated from Micromonospora carbonacea var. africana, everninomicin 13,384-1 (30, Figure 9a)302, 303, 304, 305, 306 is a member of the orthosomicin class of antibiotics.307 It exhibited promising antibacterial properties against drug-resistant bacteria, including methicillin-resistant Staphylococci and vancomycin-resistant Streptococci and Enterococci.308, 309 Its bactericidal action310 is based on its ability to inhibit protein biosynthesis by binding to the 50S subunit of the bacterial ribosome.311 Contained within its structure are a number of novel and puzzling structural motifs, including two sensitive and synthetically challenging orthoesters, a 1,1′-disaccharide bridge (an oxygen bonded to two anomeric positions), a nitrosugar, and two aromatic esters situated at the two terminal loci of the molecule. In total, everninomicin 13,384-1 possesses 13 rings and 35 stereogenic centers, making its total synthesis a formidable task. Its total synthesis, featuring an array of new synthetic methods and tactics, was reported from our laboratories in 1999.312, 313, 314, 315, 316, 317, 318 Figure 9a indicates the strategic bonds and their disconnections, defining key building blocks 112–121 (Figure 9b) that were used for the devised synthesis. Starting with commercially available and other readily prepared starting materials, these key intermediates were assembled orderly and the resulting products elaborated to advanced intermediates 126 (through compounds 122–125), 130 (through compounds 127–129) and 135 (through compounds 132–134) as summarized in Figure 9c. Advanced intermediates 126 and 130 were then joint through a glycosylation reaction to afford polycycle 131, after further elaboration (Figure 9c). Finally, the two penultimate advanced key building blocks, glycosyl fluoride 135 and hydroxyl octacycle 131, were coupled through a SnCl2-promoted α-selective glycosylation furnishing precursor 136, whose conversion to everninomicin 13,384-1 (30) was achieved in four steps as summarized in Figure 9c. This total synthesis was notable, not only for the complexity of the target molecule but also because of its featuring of new synthetic strategies and tactics as well as new synthetic methods. The latter included a series of glycosylation reactions and 1,2-phenylthio- and -phenylseleno migrations.318
Coleophomone B and C
Coleophomones B (31) and C (32) were originally isolated from the fungus Stachybothys cylindrospora RF-5900 by a Shionogi group,319 and subsequently from a Coleophoma sp. (MF6338) fungi by a Merck Sharp & Dohme group (Rahway, NJ, USA).320 Their antibacterial action is partly based on their ability to inhibit the bacterial cell wall transglycosylase activity, thus disrupting peptidoglycan biosynthesis. Furthermore, the coleophomones inhibit RNA-, DNA- and protein biosynthesis.320 Their antifungal, antibacterial and inhibitory activities against human heart chymase,319, 321 coupled with their novel molecular architectures were enticing, inspiring us to undertake their total synthesis.322, 323 The highly strained and compact nature of their 11-membered ring characterized by an (E)-olefinic bond (coleophomone B) or (Z)-olefinic bond (coleophomone C) and their unique polyoxygenated framework presented an intriguing synthetic challenge. Having attempted and failed to construct their unusual macrocycle structural motifs through traditional methods, the olefin metathesis reaction came to the rescue. As seen in Figure 10a, the retrosynthetic analysis improvised for a ring-closing metathesis reaction turned out to be pleasantly stereoselective in forming either the (E)- or (Z)-olefin within the macrocycle, depending on the exact structure of the precursor. Figure 10b highlights the total synthesis of both coleophomones B and C through an initially convergent strategy that led to advanced key intermediate 142 (from building blocks 137 and 140 and intermediates 138, 139 and 141), and which subsequently diverged to generate precursors 144 and 145 (in addition to compound 143). The structural constitution of each substrate, 144 (exocyclic methoxy enone) and 145 (endocyclic methoxy enone), proved decisively influential in forming selectively, through the indented olefin metathesis reaction (cat. 146), each desired product, 147 [(E)-olefin] and 148 [(Z)-olefin], respectively. From these precursors, the targeted natural products [coleophomones B (31) and C (32)] emerged through a short synthetic sequence as shown in Figure 10b. This achievement was a triumph for the metathesis reaction in that it demonstrated its unique power where other alternatives failed.322, 323
Thiostrepton
As the flagship of the thiopeptide class of antibiotics due to its magnificent structure,324 and because of its diverse and important biological properties325, 326 that include antibacterial, antimalarial, immunosuppressive and cytotoxic activities, and veterinary medical use, thiostrepton (33, Figure 11a) served as a challenge and inspiration to us in the early 2000s.327, 328, 329, 330, 331, 332, 333 Thiostrepton was isolated from three different species of Streptomyces (azureus ATCC 14921, hawaiiensis ATCC 12236 and laurentii ATCC 31255).334 Its mechanism of action is based on the inhibition of elongation factor G (translocase), a prokaryotic enzyme responsible for the translocation of the tRNA and mRNA down the ribosome.335, 336, 337 We reasoned that although its total synthesis might not be of urgent need due to its natural abundance it would certainly be a playground for advancing the art of organic synthesis and a means to produce otherwise unavailable designed analogs for biological investigations. Besides, there were the elements of excitement, in breaking another glass ceiling of molecular complexity and diversity for total synthesis, and serendipity for discovering new chemistry and gathering useful intelligence for future explorations and applications. The most intriguing structural motif of thiostrepton is undoubtedly its dihydropiperidine structural moiety with its surrounding substituents, including the three thiazole rings and its tetrasubstituted carbon atom, not to mention that the entire structure is loaded with reactive and unpredictable functionalities in terms of their installment and chemical behavior. Figure 11a presents the employed retrosynthetic disconnections based on which the final strategy was devised for its total synthesis. Although details of the synthesis of thiostrepton can be found in our publications on the subject,327, 328, 329, 330, 331, 332, 333 including a review,338 its highlights can be seen in Figure 11b. Among them are the biomimetic aza-Diels–Alder-based construction of the tetrasubstituted dihydropiperidine structural motif from azadiene 154 (generated from precursor 153 whose preparation from building blocks 149 and 151 proceeded through intermediates 150 and 152, see Figure 11b). Azadiene 154 reacted spontaneously upon generation via endo transition stage 155 and through intermediate 156, the latter collapsing under the reaction conditions to the desired product (158) through intermediate 157. The crucial formation of the sterically challenging amide bond in the next advanced intermediate (160) was achieved through the use of azidoacid chloride building block 159. The latter advanced intermediate was then grown to thiostrepton by crafting on it (that is, building block 160) fragments 161, 164 and 166 through a sequence of reactions involving intermediates 162, 163, 165 and 167, as summarized in Figure 11b. It is of note that the three phenylseleno groups, introduced on the growing molecule as masking devices for the two reactive dehydroalanine structural motifs, had a crucial role in ensuring the survival and eventual emergence of these moieties in the final stages of the synthesis. Another worthy feature of the synthesis was the generation, in the final step and under the desilylation conditions (HF·py), of the olefinic bond conjugated to the thiazoline ring in its correct geometrical form. We would be remiss if we did not mention that it was during this endeavor that we discovered and developed the now popular method for selective hydrolysis of methyl esters using trimethyltin hydroxide.339, 340
Abyssomicins
Abyssomicin C (34, Figure 12a) was originally isolated from the marine-derived actinomycete strain Verrucosispora AB-18-032 in 2004.341, 342 Its sibling, atrop-abyssomicin C (34′, Figure 12a) was subsequently discovered from the same species,343 but not before it was synthesized in our laboratories.344, 345, 346 Exhibiting potent antibacterial properties against drug-resistant strains due to its ability to inhibit the conversion of chorismate into p-aminobenzoic acid (thus blocking the tetrahydrofolate biosynthesis)342 and because of its novel molecular architecture, abyssomicin C presented an irresistible challenge as a synthetic target. Our devised strategy for its total synthesis was based on the retrosynthetic analysis depicted in Figure 12a and featured a Diels–Alder reaction and a ring-closing metathesis as highlighted in Figure 12b. The synthetic sequence employed building blocks 168, 171 and 175, and ligand 170 needed for the success of the stereoselective Diels–Alder reaction of 169 with 171, to afford product 173 via transition state 172. Elaboration of bicyclic product 173 then furnished tricycle 174, which was coupled under basic conditions with δ-lactone 175 to afford, upon dithioketal formation, intermediate 176. Oxidation followed by vinyl Grignard addition to the resulting aldehyde then gave metathesis precursor 177. The coveted ring-closing metathesis of precursor 177 was achieved through the use of ruthenium catalyst 178, leading, after oxidation, exclusively to macrocycle 179, from which atrop-abyssomicin C (34′, Figure 12b) was generated by exposure to phenyliodo bistrifluoroacetate. Finally, atrop-abyssomicin C was converted under acidic conditions into a separable mixture of abyssomicin C (34) and atrop-abyssomicin C (34′; 34:34′ ca. 2:1). Interestingly, in addition to being discovered in nature, atrop-abyssomicin C exhibited even higher potencies against bacteria than its sibling, abyssomicin C.343 This endeavor demonstrated and expanded further the power of the Diels–Alder and metathesis reactions in total synthesis, provided useful insights and understanding of atropisomerism, especially as it pertains to naturally occurring molecules, and resulted in a number of structure-activity relationships within the abyssomicin family. It also led to the total synthesis of a natural product in the laboratory before its discovery from nature, a rare, but noteworthy contribution of the art and science of total synthesis. Besides our own work, other groups have also attempted347, 348, 349, 350, 351 or completed the formal352 and total synthesis of the abyssomicins.353, 354, 355
Marinomycins
Marinomycin A (35, Figure 13a) is the most prominent member of the marinomycin family of marine-derived natural products isolated from the actinomycete ‘Marinispora’ CNQ-140.356 Featuring a 44-membered dimeric macrolide ring and endowed with potent antibacterial and cytotoxic properties, these scarce molecules became synthetic targets in our group not only because of their biological properties but also due to their novel and synthetically challenging architectures. Our total synthesis of marinomycin A357, 358 is summarized in Figure 13a (retrosynthetic disconnections) and 13b (highlights). The sequence employed building blocks 180, 181, 185 and 189 and proceeded through intermediates 182–184 and 186–193 as outlined in Figure 13b. The key macrocyclization reaction involved conversion of acetylenic vinyl bromide 193 to afford the corresponding vinyl borane, which underwent intramolecular Suzuki coupling furnishing, upon global desilylation, the target molecule marinomycin A (35). Interestingly, the Heck, Stille and metathesis reactions failed to produce the corresponding macrocyclic products. Another interesting outcome of this project was the preparation of the 22-membered monomeric counterpart of marinomycin A, which proved inactive as compared to the natural product against the same bacterial strains. Furthermore, marinomycin A was transformed, upon photo-irradiation, to its geometrical isomers, marinomycins B and C (as a mixture with A), which were separated as individual compounds.357, 358 In 2007, the Cossy group published their work on the synthesis of the monomeric counterpart of marinomycin A.359, 360 Two other total syntheses of marinomycin A were more recently reported, one by the Evans group361 and the other by the Hatakeyama group.362
Platensimycin
Aided by a new screening technique,363, 364 scientists at Merck Sharp & Dohme were able to isolate and characterize platensimycin (38, Figure 14a), a novel secondary metabolite from Strep. platensis that exhibited inhibitory activity against the FabH and FabF condensing enzymes of the fatty acid biosynthetic pathway crucial to bacteria.365, 366, 367 Its unique mechanism of action, coupled with its unprecedented molecular architecture, propelled platensimycin to a priority status as a synthetic target and a potential new antibiotic for investigation and further development. From the several total syntheses we developed,368, 369, 370, 371, 372 we present in Figure 14a (retrosynthetic disconnections) and 14b (highlights) an asymmetric version373 that involved an enantioselective rhodium-catalyzed cyclization of a prochiral eneyne bis-enone (that is, 196, obtained from building block 194 via 195) to afford bicyclic aldehyde 197. A samarium-induced ring closure within the latter then led to tricyclic alcohol 198, from which pentacycle 199 was formed through acid-catalyzed etherification, as shown in Figure 14b. Sequential bis-alkylation of ketone 199 then led to olefinic substrate 200, whose cross metathesis reaction with vinyl boronate 201 under the influence of ruthenium catalyst 202 furnished advanced intermediate 203, from which the coveted carboxylic acid 204 emerged in two steps. The required benzenoid domain (206) of the target molecule was secured from building block 205 through a high-yielding five-step sequence. Coupling of carboxylic acid 204 with aniline derivative 206 under the influence of HATU and Et3N led to the corresponding amide, from which platensimycin (38) was derived upon deprotection, as shown in Figure 14b. This and the other synthetic strategies and technologies we developed during this program facilitated the design, synthesis and biological evaluation of numerous analogs of platensimycin. Among them, carbaplatensimycin (207)374 and adamantaplatensimycin (208)375 are notable for their comparable potencies against drug-resistant bacteria (see Figure 14c). Besides our initial total synthesis,368 numerous other strategic approaches376, 377, 378, 379, 380 and formal and total syntheses have been reported.381, 382, 383, 384, 385, 386, 387, 388, 389, 390, 391, 392, 393, 394, 395, 396, 397, 398, 399, 400
Platencin
Isolated from Strep. platensis MA7339,401, 402 platencin (39, Figure 15a) is a relative of platensimycin (38) discussed above. Its antibacterial properties and mechanism of action are similar to those of platensimycin (38) and so is its molecular structure, except from its tricyclic domain. The latter structural site of platencin differs from that of platensimycin in that it includes a completely carbocyclic framework consisting of only three rings, instead of the tetracyclic cyclic ether-featuring framework of platensimycin. From the several strategies we developed toward platencin,403, 404, 405, 406 the one depicted in Figure 15a (retrosynthetic disconnections) and 15b (highlights) is distinguished for its enantioselectivity and novel carbocyclization to forge one of the rings of the molecule.406 Starting with building blocks 209, 211, 222 and 224, and using catalyst 214 and proceeding through intermediates 210, 213, 215–221 and 223 as shown in Figure 15b, this total synthesis of platencin delivered the natural product in its natural enantiomeric form. One of its most notable steps was the asymmetric, cobalt-catalyzed Diels–Alder reaction between diene 210 and dienophile 213 in the presence of catalyst 214 to afford cyclohexene 215, whose one-pot conversion to hydroxyenone 216 proceeded in 92% yield and 98% ee (after recrystallization). Another pleasing step was the high-yielding (95%) gold-catalyzed cyclization of acetylenic enol ether substrate 217 to afford bicyclic system 218. Several other elegant (formal) total syntheses of platencin and its analogs have been reported.394, 398, 407, 408, 409, 410, 411, 412, 413, 414, 415, 416, 417, 418, 419, 420, 421, 422, 423, 424, 425, 426, 427
Rugulin
There is a mystery surrounding the structure of rugulin, an alleged natural product isolated from Penicillium rugulosum in 1978 and claimed to possess structure 40 (Figure 16a)428 and to exhibit antibacterial properties.429 Its intriguing molecular architecture is characterized by a central core in the shape of a twisted cylinder defined by four five-membered carbocyclic rings, the latter in its naked hydrocarbon form termed ‘skyrane’.430, 431, 432 Its unusual and challenging structure, coupled with its reported biological properties, motivated us to undertake its laboratory synthesis. We achieved the total synthesis of the alleged structure of rugulin432 following the novel strategy shown in Figure 16b, which employs the so-called ‘cytoskyrin cascade’ developed in our laboratories430, 431 for the purposes of synthesizing other members of the class, to which this structure belongs, along with cytoskyrin A433 and 2,2′-epi-cytoskyrin A.432, 434 The developed synthetic strategy toward this novel structure involved two of the simplest reagents (that is, MnO2 and CF3COOH) and proceeded smoothly under thermal conditions (25–45 °C). Starting from dihydroanthracenone building block 225, this rather stunning cascade sequence gave precursor 230. It involved initial oxidation of the dihydroanthracenone 225 to the corresponding quinone (that is, 226), which served as a substrate for the next step involving enolization/dimerization to afford bis-enol 227. The latter underwent rapture of the oxygen bridge triggered by further MnO2-induced oxidation to furnish the corresponding bis-anthraquinone system 228, which entered an intramolecular Michael reaction to form the second required C–C bond of the skyrane cage, leading to intermediate 229. Upon treatment with Et3N in the same pot, this intermediate suffered a second intramolecular Michael reaction to generate advanced intermediate 230, missing only one of the four required C–C bonds to close the skyrane cage. This remaining objective was achieved by the addition of further amounts of MnO2 in the same pot forming, upon exposure to CF3COOH, the targeted rugulin structure 40. As elegant as the synthesis was, it only proved that the originally assigned structure of rugulin was wrongly assigned, leaving the true structure of the natural product in a shroud of mystery and a continuing structural puzzle. This unfortunate mystery remains to this day, due to the fact that we were not able to obtain either a sample of the natural product or its full set of spectral data.
Antibiotic BE-43472B
The bis-anthraquinone antibiotic BE-43472B (41, Figure 17a) was isolated from Streptomyces sp. A43472 found in a blue-green algae associated with the ascidian Ecteinascidia turbinate.435, 436 This structurally novel antibiotic possesses, in addition to its two anthraquinone-type structural motifs, an unusual bicyclic core featuring an internal ketal functionality and a highly sterically hindered carbon-carbon bond. Its potent activities against drug-resistant bacteria such as MRSA and tetracycline-resistant Staph. aureus and vancomycin-resistant Enterococcus faecium,437 coupled with its challenging molecular architecture prompted us to pursue its total synthesis.438, 439, 440 Our motivation was to develop novel chemistry, determine its then unknown absolute configuration, and apply the developed synthetic strategies and technologies to the synthesis and biological evaluation of designed analogs. Figure 17 displays the retrosynthetic analysis that guided the development of the devised strategy (panel a) and the highlights of the successful total synthesis of antibiotic BE-43472B (panel b) featuring two esthetically pleasing cascades. The first cascade sequence began with the Diels–Alder fusion of diene 232 and dienophile 233 (see transition state 234) and proceeded through intermediates 235–237 [involving an ipso aromatic substitution reaction (SNAr) and expulsion of a molecule of MeOH] to afford octacycle 238. The second cascade sequence involved the photo-induced rapture of epoxide 240 (prepared from 238 in six steps via intermediate 239) to form diradical 241, whose rearrangement led to isomeric radical 242, from which antibiotic BE-43472B emerged after an intramolecular H-atom shift and tautomerization of the resulting keto-enol as indicated in Figure 17b. The developed synthetic strategy allowed the total synthesis of both enantiomeric forms [(+)-41 and ent-(−)-41] of antibiotic BE-43472B, allowing its absolute configuration to be assigned as that depicted by 41 in Figure 17b. Interestingly, synthetic antibiotic BE-43472B (41) and its enantiomer (ent-41) exhibited essentially the same potencies against drug-resistant bacteria [that is, 41: MIC=0.083–0.166 μg ml−1 (methicillin-resistant Staph. aureus), 0.33–0.83 (vancomycin-resistant Enterococcus faecalis); ent-41: MIC=0.055–0.11 μg ml−1 (MRSA), 0.17–1.1 (vancomycin-resistant Enterococci)].439 In 2013, Keisuke Suzuki and his group reported a racemic total synthesis of antibiotic BE-43472B featuring an isoxazole-directed pinacol 1,2-shift.441
Hirsutellone B
A member of a large class of fungal metabolites, hirsutellone B (42, Figure 18a) rose to prominence due to its potent activity against Mycobacterium tuberculosis, the causative pathogen of tuberculosis. Isolated from fungus Hirsutella nivea BCC 2594,442 hirsutellone B includes in its structure a 6,5,6-fused tricyclic core, a hydroxy γ-lactam, and a 13-membered p-cyclophane ring encompassing an aryl ether bridge and five out of the ten stereogenic centers of the molecule. Its biological properties as a potential antituberculosis agent and novel molecular architecture elevated it to an opportunistic target molecule. Figure 18a indicates the disconnections of the identified strategic bonds onto which the devised synthetic strategy was based. A summary of the total synthesis of hirsutellone B, achieved in our laboratories in 2009,443 is depicted in Figure 18b. Thus, starting from (R)-(+)-citronellal (243), polyunsaturated epoxide 244 was prepared and subjected to the influence of Et2AlCl in CH2Cl2 at −78→25 °C, conditions that allowed its stereoselective transformation to tricyclic system 247 through a cascade sequence featuring activation of the epoxide, a chloride-induced desilylation/ring closure/epoxide opening, and an intramolecular Diels–Alder reaction. Proceeding through transition states/intermediates 245 and 246, this cascade sequence delivered its product in 50% yield and with all eight stereocenters in their desired configurations. Product 247 was then converted to dihydroxyaryl ether 248, which, on exposure to ZnI2 and AcSH, furnished in one step iodothioacetate 251 through presumed oxonium species 249 and 250. The latter intermediate was elaborated to macrocyclic sulfone 252 by sequential cyclization and oxidation. A Ramberg–Bäcklund rearrangement then led to olefin 253, from which hirsutellone B (42) was generated through intermediates 254 and 255 as shown in Figure 18b. This synthesis is marked by the elegance of its cascade reactions and its flexibility to deliver other members of the hirsutellone class444 and analogs thereof. Besides our presented synthesis, a number of other conceptual studies445, 446, 447, 448 were pursued with two culminating in formal449 and total syntheses.450
Epicoccin G, 8,8′-epi-ent-rostratin B and emethallicin E
Epidithiodiketopiperazines and bis-(methylthio)diketopiperazines are naturally occurring molecules whose biological properties include antiviral, antibacterial, antimalarial and cytotoxic activities.451, 452, 453, 454, 455, 456, 457, 458, 459, 460, 461, 462, 463 Because of their biological actions, these natural products hold potential as drug candidates or lead compounds for drug discovery, but they are often scarce for thorough biological evaluation. Their synthesis and analogs are, therefore, of crucial importance for biological investigations and drug discovery initiatives. In an effort to improve the methods for their synthesis, often suffering from the key sulfenylation step (the installment of the two required sulfur atoms onto appropriate diketopiperazine scaffolds), and in order to render them readily available for further explorations, we initiated a program directed toward their total synthesis. Attracted by the molecular architectures and biological properties of certain members of this class, and aiming to develop a new sulfenylation method, we targeted for total synthesis464 epicoccin G [43, Figure 19a, originally isolated from Epicoccum nigrum (XZC04-CS-302)],465, 466, 467 8,8′-epi-ent-rostratin B (44, Figure 19a; rostratin B was isolated from Exserohilum rostratum)468 and emethallicin E (45, Figure 19c, isolated from Emericella heterothallica ATCC 16824).469 Figure 19a shows the retrosynthetic disconnections that led to the development of the synthetic strategy toward these target molecules. Crucial to the success of these total syntheses, whose highlights are summarized in Figure 19b, were the bis-photo-oxygenation/Kornblum–DeLaMare rearrangement and the discovery of an improved method for the sulfenylation of diketopiperazines (Figure 19d, method developing box). The asymmetric synthesis of epicoccin G (43) started with the readily available building block 256 (from N-Boc-L-tyrosine), which was converted to diketopiperazine 261 through a series of synthetic steps involving intermediates 257–260 (Figure 19b). Having failed in our attempts to introduce the required sulfur atoms in the next step, we were motivated to improve existing methods previously employed in such operations.470, 471, 472, 473, 474, 475, 476, 477, 478, 479, 480, 481, 482, 483 We soon discovered that treating sulfur with NaHMDS in THF at ambient temperature for a few minutes produced a solution that contained reactive sulfur species capable of sulfenylating enolates generated from diketopiperazines (see 270→271, Figure 19d, box) under the same basic conditions and in the same pot.484 Consisting mainly of tetrasulfides, the generated products (for example, 272, formed via 271, Figure 19d, box) were easily converted to either epidithiodiketopiperazines (274) or bis-methylthio diketopiperazines (275) via dithiolate species 273 (formed by NaBH4 reduction of 272, see Figure 19d, box), as shown in Figure 19. Applying the newly developed method to diketopiperazine 261 gave epidithiodiketopiperazine intermediate 262, formed together with its 2,2′-epimer (2,2′-epi-262, see Figure 19b). The former was smoothly transformed to epicoccin G (43), whereas the latter was employed to synthesize 8,8′-epi-ent-rostratin B (44) as summarized in Figure 19b. The same key building block 256 was converted by similar chemistry to emethallicin E (45) through intermediates 276–281 as highlighted in Figure 19c. In addition to the benefit of developing the improved and valuable sulfenylation reaction, this project revealed, through biological testing, potent activities against the poliovirus of the encountered intermediates 262 and 282 (see Figure 19e, box). In addition to our own efforts several other laboratories reported on their progress toward or total syntheses of epicoccin G,485, 486 8,8′-epi-ent-rostratin B,487, 488, 489, 490 and emethallicin E.491
Retrosynthetic analyses (a) and highlights of the total syntheses of epicoccin G (43) and 8,8′-epi-ent-rostratin B (44) (b), and emethallicin E (45) (c) as well as the mechanistic rationale for the developed sulfenylation method (d), and select synthesized designed analogues and their biological activities (e).
Viridicatumtoxin B
Resembling but being more structurally complex than its siblings, viridicatumtoxin B (46, Figure 20a)492 is loosely classified as a member of the tetracycline class of antibiotics due to its linear tetracyclic structural domain. Isolated from Penicillium sp. FR11 and possessing two additional rings than the traditional tetracyclines weaved in a spirocyclic structural motif, viridicatumtoxin B exhibits impressive antibacterial activities against drug-resistant bacteria, including Staph. aureus (MRSA).492 The antibacterial properties of this impressive molecule presumably arise from its ability to inhibit UPP synthase, a key enzyme for bacterial peptidoglycan biosynthesis.493, 494 It became a synthetic target in our laboratories for its complexity and challenge, the appeal of its biological properties, and its potential as a lead compound for drug discovery. Figure 20a shows the strategic bond disconnections that guided the design of our synthetic strategy, which included two Michael–Dieckmann reactions, an SN2-type alkylation, and a Lewis acid-catalyzed spirocyclization. Requiring building blocks 283, 285, 287 and 292 as starting materials, the synthesis proceeded along the pathway depicted in Figure 20b and through intermediates 284, 286, 288–291, 293–297. Noteworthy steps in this route, which in its original form delivered racemic viridicatumtoxin B,97 included the alkylation of anthrone 286 under mild conditions, a step at which asymmetry could be introduced, in principle, in the growing molecule. Another remarkable step of this total synthesis is the final demasking of the distinguishing tetracyclic enolamide structural motif from its surrogate oxazoline functionality by hydrogenolysis, conditions that also led to the rapture of the two benzyl ethers of the ultimate precursor (297, Figure 20b). The total synthesis of viridicatumtoxin B not only rendered the racemic natural product readily available but also resulted in its structural revision.97 Furthermore, an enantioselective total synthesis of viridicatumtoxin B (and decipherment of its absolute configuration) has been achieved in these laboratories employing phase-transfer catalysis.495 Another important outcome of this total synthesis endeavor was the synthesis and biological evaluation of a series of simpler analogs of viridicatumtoxin B,98 from which compounds (±)-298 and (±)-299 were notable for their potencies as compared to those of the racemic natural product [(±)-46] against drug-resistant bacteria (see Figure 20c).
Antibiotic CJ-16,264
A member of the pyrrolizidinone family of natural products, antibiotic CJ-16,264 (47, Figure 21a) was isolated from fungus CL39457.496 It possesses a highly substituted decalin system and exhibits potent antibacterial properties against drug-resistant bacteria.496 Its total synthesis from our laboratories rendered the natural product in its naturally occurring enantiomeric form readily available and led to its structural revision (configurations at C1 and C7′).497 Figure 21a summarizes our synthetic strategy, in retrosynthetic format, toward this target molecule which included a Reformatsky-type coupling and an intramolecular Diels–Alder reaction. The forward synthesis proceeded from citronellal (ent-243, also available in the other enantiomeric form) and racemic building block (±)-305498 through intermediates 300 or 302 and 301, 303, 304 and 307 (Figure 21b). The availability of both enantiomers of these two building blocks was crucial for the final structural elucidation of antibiotic CJ-16,264. It provided several isomeric forms of the targeted molecule, allowing its structural identification as that depicted by structure 47. The most striking feature of this synthesis was the exclusive formation of diolide 303 incorporating two units of the desired decalin system from macrodiolide 301 through two consecutive, thermally induced, intramolecular Diels–Alder reactions. From the six isomers synthesized, the ones shown in Figure 21c proved the most active against methicillin-resistant Staph. aureus, Enterococcus faecalis S613 and Enterococcus faecium 105 (see Figure 21c). In addition to these CJ-16,264 isomers, the developed synthetic strategies and technologies made a number of structurally simpler analogs of this rather complex antibiotic readily available (unpublished results), whose synthesis and biological evaluation will be reported in due course.
Conclusion
Microbes have been menacing society ever since the beginning of our existence. Empowering us to create new molecules in the laboratory, the advent of organic synthesis in the nineteenth century led to the introduction of the first antibacterial drugs. But it was not until we learned more about the pathogenesis of infectious diseases and the nature of microbes that we began to strategize against them in rational ways. Indeed, it was bacteria and fungi themselves that taught us how to fight them in the most effective ways: with the same weapons by which they have been waging war against each other and continue to do so today. Chemists were able to isolate an avalanche of powerful antibiotics from natural sources in a surge that climaxed into the antibiotic rush in the 1950s after the success of penicillin. And for a while it was perceived that the war against bacteria had been won. This is not so, as we came to know shortly thereafter. And we might not ever win it completely as we attempt to control the continuously emerging bacterial resistance, a phenomenon based on natural evolution. Ironically, it is human behavior and weakness that accelerate and spread bacterial resistance, whether it is through our medical, agricultural or psychological practices. Thankfully, the ever increasing power of our abilities to isolate new antibiotics from nature and to design and synthesize others like them in the laboratory provide us with the means to catch up with them, at least temporarily, by discovering and enlisting new weapons against them. The new antibiotics included in the last section and the synthetic work associated with them, for example, may hold the secrets to one or more new weapons to add to our arsenal—if only the funds were available to advance these investigations beyond the ‘valley of death’ where, unfortunately, so many academic discoveries remain buried.
The importance of continuous research and development for new antibiotics was furthermore highlighted by a press release from the World Health Organization (WHO) in February 2017.499 It contains a list of the following 12 families of bacteria representing the greatest risk to human health.
WHO priority pathogens list for R&D of new antibiotics
Priority 1: (critical)499
-
Acinetobacter baumannii, carbapenem-resistant
-
Pseudomonas aeruginosa, carbapenem-resistant
-
Enterobacteriaceae, carbapenem-resistant, ESBL-producing
Priority 2: (high)
-
Enterococcus faecium, vancomycin-resistant
-
Staph. aureus, methicillin-resistant, vancomycin-intermediate and resistant
-
Helicobacter pylori, clarithromycin-resistant
-
Campylobacter spp., fluoroquinolone-resistant
-
Salmonellae, fluoroquinolone-resistant
-
Neisseria gonorrhoeae, cephalosporin-resistant, fluoroquinolone-resistant
Priority 3: (medium)
-
Streptococcus pneumoniae, penicillin-non-susceptible
-
Haemophilus influenzae, ampicillin-resistant
-
Shigella spp., fluoroquinolone-resistant
The severe threat emanating from these bacteria derives mainly from their resistance to known antibacterial drugs with the most dangerous strains being multidrug-resistant, and thus, capable of causing potentially untreatable, often deadly infections. The WHO urgently calls on governments worldwide to support research directed toward the discovery and development of new antibacterial agents against these dangerous strains. In parallel the organization also strongly recommends continuous improvements in preventing bacterial infections and strict enforcement of good practices in the application of currently used and to be discovered antibiotics as a means to prevent or at least curtail the emergence of resistance.499
The arms race against pathogenic microbes must continue with vigilance and prudency in order to avoid catastrophes of unpredictable and unimaginable proportions. At the same time we must restrict ourselves from practices that challenge their evolutionary instincts and propel them to develop resistance. It is this dual approach of wisdom and proactiveness that gives humanity hope to stay ahead of pathogenic microbes and, therefore, we must do what it takes to intensify our efforts in a focused and sustainable manner. We know what we have to do and how to do it. We must now gather the will and make the investment to implement our strategies to achieve the needed health security.
References
Vuillemin, J. P. Antibiose et symbiose. Assoc. Franc. pour l'Avanc. Sci. 2, 525–543 (1890).
Bentley, R., Bennett, J. W. in Advances in applied microbiology, Vol. 52 (eds Laskin, A. I., Bennett, J. W. & Gadd, G. M. 303–331 Academic Press, Cambridge, MA, USA, (2003).
Wallenfels, K. Symbiose und antibiose. Angew. Chem. 58, 1–16 (1945).
Fracastorius, H. De contagione et contagiosis morbis et curatione, Libri III, Venice, Most Serene Republic of Venice, (1546).
Henle, F. G. J. in Pathologische Untersuchungen 1–82 Verlag von August Hirschfeld, Berlin, Prussia, (1840).
Koch, R. Die Aetiologie der Tuberculose. Berl. Klin. Wochenschr. 19, 287–296 (1882).
Koch, R. Die Aetiologie der Tuberkulose. Mitth. Kais. Gesundheits 2, 1–88 (1884).
Loeffler, F. Untersuchung über die Bedeutung der Mikroorganismen für die Entstehung der Diphtherie beim Menschen, bei der Taube und beim Kalbe. Mitth. Kais. Gesundheits 2, 421–499 (1884).
Pacini, F. Osservazioni microscopiche e deduzioni patologiche sul cholera asiatico. Gazz. Med. Ital. Toscana (Serie II) 4, 397–401 (1854).
Gosio, B. Contributo all’etiologia della pellagra. Ricerche chimiche e batteriologiche sulle alterazioni del mais. G. Accad. Med. Torino 61, 464–487 (1893).
Gosio, B. Ricerche batteriologiche e chimiche sulle alterazioni del mais. Contributo all’etiologia della pellagra. Riv. d'Ig. San. Pubb. 7, 825–849 (1896).
Silverman Kitchin, J. E., Pomeranz, M. K., Pak, G., Washenik, K. & Shupack, J. L. Rediscovering mycophenolic acid: a review of its mechanism, side effects, and potential uses. J. Am. Acad. Dermatol. 37, 445–449 (1997).
Alsberg, C. L. & Black, O. F. Contributions to the study of maize deterioration. Biochemical and toxicological investigations of Penicillium puberulum and Penicillium stoloniferum. Bull. US Bur. Pl. Ind. 270, 7–48 (1913).
Birkinshaw, J. H., Raistrick, H. & Ross, D. J. Studies in the biochemistry of micro-organisms. 86. The molecular constitution of mycophenolic acid, a metabolic product of Penicillium brevi-compactum Dierckx. Part 3. Further observations on the structural formula for mycophenolic acid. Biochem. J. 50, 630–634 (1952).
Birch, A. J. & Wright, J. J. A total synthesis of mycophenolic acid. Aust. J. Chem. 22, 2635–2644 (1969).
Birch, A. J. & Wright, J. J. A total synthesis of mycophenolic acid. J. Chem. Soc. D 788–789 (1969).
Wu, J. C. Mycophenolate mofetil: molecular mechanisms of action. Perspect. Drug Discov. Des. 2, 185–204 (1994).
Ehrlich, P. & Bertheim, A. Über das salzsaure 3.3′-Diamino-4.4′-dioxy-arsenobenzol und seine nächsten Verwandten. Ber. Dtsch. Chem. Ges. 45, 756–766 (1912).
Weigert, C. Ueber Bacterien in der Pockenhaut. Centralbl. f. d. med. Wissensch. Berlin 9, 609–611 (1871).
Weigert, C. Über eine Mykose bei einem neugeborenen Kinde. Jahresb. d. schles. Gesellsch. f. vaterl. Cultur 53, 229–230 (1876).
Weigert, C. Bismarckbraun als Färbemittel. Arch. Mikrosk. Anat. 15, 258–260 (1878).
Weigert, C. Zur Technik der mikroskopischen Bakterienuntersuchungen. Arch. Pathol. Anat. Physiol. Klin. Med. 84, 275–315 (1881).
Farbwerke vorm. Meister Lucius & Brüning in Höchst Verfahren zur Darstellung von Oxyarylarsenoxyden. Deutsches Reichspatent No. 213594 (1908).
Farbwerke vorm. Meister Lucius & Brüning in Höchst Verfahren zur Darstellung von Aminoderivaten der Oxyarylarsinsäuren und deren Reduktionsprodukten. Deutsches Reichspatent No. 224953 (1909).
Farbwerke vorm. Meister Lucius & Brüning in Höchst Verfahren zur Darstellung von Aminooxyarylarsenoxyden. Deutsches Reichspatent No. 235391 (1909).
Williams, K. The introduction of ‘chemotherapy’ using arsphenamine—the first magic bullet. J. R. Soc. Med. 102, 343–348 (2009).
Lloyd, N. C., Morgan, H. W., Nicholson, B. K. & Ronimus, R. S. The composition of Ehrlich's Salvarsan: resolution of a century-old debate. Angew. Chem. Int. Ed. 44, 941–944 (2005).
Fleming, A. On the antibacterial action of cultures of a penicillium, with special reference to their use in the isolation of B. influenzæ. Br. J. Exp. Pathol. 10, 226–236 (1929).
Hodgkin, D. C. The X-ray analysis of the structure of penicillin. Adv. Sci. 6, 85–89 (1949).
Sheehan, J. C. & Henery-Logan, K. R. The total synthesis of penicillin V. J. Am. Chem. Soc. 79, 1262–1263 (1957).
Sheehan, J. C. & Henery-Logan, K. R. The total synthesis of penicillin V. J. Am. Chem. Soc. 81, 3089–3094 (1959).
Brotzu, G. Ricerche su di un nuovo antibiotico. Lavori dell'Istituto di Igiene di Cagliari 4–18 (1948).
Bo, G. Giuseppe Brotzu and the discovery of cephalosporins. Clin. Microbiol. Infect. 6, 6–8 (2000).
Newton, G. G. & Abraham, E. P. Cephalosporin C, a new antibiotic containing sulphur and D-α-aminoadipic acid. Nature 175, 548 (1955).
Abraham, E. P. & Newton, G. G. F. The structure of cephalosporin C. Biochem. J. 79, 377–393 (1961).
Hodgkin, D. C. & Maslen, E. N. The X-ray analysis of the structure of cephalosporin C.. Biochem. J. 79, 393–402 (1961).
Woodward, R. B. et al. The total synthesis of cephalosporin C. J. Am. Chem. Soc. 88, 852–853 (1966).
Domagk, G. Ein Beitrag zur Chemotherapie der bakteriellen Infektionen. Dtsch. Med. Wochenschr. 61, 250–253 (1935).
Tréfouël, J., Tréfouël, T., Nitti, F. & Bovet, D. Activité du p-aminophénylsulfamide sur l’infection streptococcique expérimentale de la souris et du lapin. C. R. Séances Soc. Biol. Ses Fil. 120, 756–758 (1935).
Gelmo, P. Über Sulfamide der p-Amidobenzolsulfonsäure. J. Prakt. Chem. 77, 369–382 (1908).
Achari, A. et al. Crystal structure of the anti-bacterial sulfonamide drug target dihydropteroate synthase. Nat. Struct. Biol. 4, 490–497 (1997).
Dubos, R. J. Studies on a bactericidal agent extracted from a soil bacillus: I. Preparation of the agent. Its activity in vitro. J. Exp. Med. 70, 1–10 (1939).
Dubos, R. J. Studies on a bactericidal agent extracted from a soil bacillus: II. Protective effect of the bactericidal agent against experimental pneumococcus infections in mice. J. Exp. Med. 70, 11–17 (1939).
Dubos, R. J. & Cattaneo, C. Studies on a bactericidal agent extracted from a soil bacillus: III. Preparation and activity of a protein-free fraction. J. Exp. Med. 70, 249–256 (1939).
Hotchkiss, R. D. & Dubos, R. J. Fractionation of the bactericidal agent from cultures of a soil bacillus. J. Biol. Chem. 132, 791–792 (1940).
Hotchkiss, R. D. & Dubos, R. J. Chemical properties of bactericidal substances isolated from cultures of a soil bacillus. J. Biol. Chem. 132, 793–794 (1940).
Hotchkiss, R. D. & Dubos, R. J. Bactericidal fractions from an aerobic sporulating bacillus. J. Biol. Chem. 136, 803–804 (1940).
Gause, G. F. & Brazhnikova, M. G. Gramicidin S and its use in the treatment of infected wounds. Nature 154, 703 (1944).
Gall, Y. M. & Konashev, M. B. The discovery of gramicidin S: the intellectual transformation of G.F. Gause from biologist to researcher of antibiotics and on its meaning for the fate of Russian genetics. Hist. Phil. Life Sci. 23, 137–150 (2001).
Synge, R. L. M. ‘Gramicidin S’: over-all chemical characteristics and amino-acid composition. Biochem. J. 39, 363–367 (1945).
Gordon, A. H., Martin, A. J. P. & Synge, R. L. M. Proceedings of the Biochemical Society. Biochem. J. 40, xliii–xliv (1946).
Consden, R., Gordon, A. H., Martin, A. J. P. & Synge, R. L. M. Gramicidin S: the sequence of the amino-acid residues. Biochem. J. 41, 596–602 (1947).
Schmidt, G. M. J., Hodgkin, D. C. & Oughton, B. M. A crystallographic study of some derivatives of gramicidin S. Biochem. J. 65, 744–750 (1957).
Schwyzer, R. & Sieber, P. Die Synthese des Gramicidin S. Angew. Chem. 68, 518 (1956).
Schwyzer, R. & Sieber, P. Die Synthese von Gramicidin S. Helv. Chim. Acta 40, 624–639 (1957).
Erlanger, B. F., Sachs, H. & Brand, E. The synthesis of peptides related to gramicidin S. J. Am. Chem. Soc. 76, 1806–1810 (1954).
Semrau, S. et al. Membrane lysis by gramicidin S visualized in red blood cells and giant vesicles. Biochim. Biophys. Acta 1798, 2033–2039 (2010).
Katsu, T., Kobayashi, H. & Fujita, Y. Mode of action of gramicidin S on Escherichia coli membrane. Biochim. Biophys. Acta 860, 608–619 (1986).
Yonezawa, H., Okamoto, K., Tomokiyo, K. & Izumiya, N. Mode of antibacterial action by gramicidin S. J. Biochem. 100, 1253–1259 (1986).
Schatz, A., Bugle, E. & Waksman, S. A. Streptomycin, a substance exhibiting antibiotic activity against Gram-positive and Gram-negative bacteria. Exp. Biol. Med. 55, 66–69 (1944).
Jones, D., Metzger, H. J., Schatz, A. & Waksman, S. A. Control of Gram-negative bacteria in experimental animals by streptomycin. Science 100, 103–105 (1944).
Schatz, A. & Waksman, S. A. Effect of streptomycin and other antibiotic substances upon Mycobacterium tuberculosis and related organisms. Exp. Biol. Med. 57, 244–248 (1944).
Hinshaw, H. C. Historical notes on earliest use of streptomycin in clinical tuberculosis. Am. Rev. Tuberc. 70, 9–14 (1954).
Waksman, S. A. & Schatz, A. Streptomycin and process of preparation. US2449866 A (1948).
Wainwright, M. Streptomycin: discovery and resultant controversy. Hist. Phil. Life Sci. 13, 97–124 (1991).
Lemieux, R. U. & Wolfrom, M. L. in Advances in Carbohydrate Chemistry, Vol. 3 (eds Pigman, W. W., Wolfrom, M. L. and Peat, S.) 337–384 (Academic Press, New York, NY, USA, 1948).
McGilveray, I. J. & Rinehart, K. L. The anomeric linkage of streptose in streptomycin and bluensomycin. J. Am. Chem. Soc. 87, 4003–4004 (1965).
Neidle, S., Rogers, D. & Hursthouse, M. B. The crystal and molecular structure of streptomycin oxime selenate. Tetrahedron Lett. 9, 4725–4728 (1968).
Umezawa, S., Takahashi, Y., Usui, T. & Tsuchiya, T. Total synthesis of streptomycin. J. Antibiot. 27, 997–999 (1974).
Umezawa, S., Tsuchiya, T., Yamasaki, T., Sano, H. & Takahashi, Y. Total synthesis of dihydrostreptomycin. J. Am. Chem. Soc. 96, 920–921 (1974).
Demirci, H. et al. A structural basis for streptomycin-induced misreading of the genetic code. Nat. Commun. 4, 1355 (2013).
Sharma, D., Cukras, A. R., Rogers, E. J., Southworth, D. R. & Green, R. Mutational analysis of S12 protein and implications for the accuracy of decoding by the ribosome. J. Mol. Biol. 374, 1065–1076 (2007).
Duggar, B. M. Aureomycin: a product of the continuing search for new antibiotics. Ann. N. Y. Acad. Sci. 51, 177–181 (1948).
Chopra, I. & Roberts, M. Tetracycline antibiotics: mode of action, applications, molecular biology, and epidemiology of bacterial resistance. Microbiol. Mol. Biol. Rev. 65, 232–260 (2001).
Landau, R., Achilladelis, B. & Scriabine, A. Pharmaceutical Innovation: Revolutionizing Human Health, Chemical Heritage Press, Philadelphia, PA, USA, (1999).
Stephens, C. R. et al. The structure of aureomycin. J. Am. Chem. Soc. 76, 3568–3575 (1954).
Hirokawa, S., Okaya, Y., Lovell, F. M. & Pepinsky, R. On the crystal structure of aureomycin hydrochloride. Acta Crystallogr. 12, 811–812 (1959).
Hirokawa, S., Okaya, Y., Lovell, F. M. & Pepinsky, R. The crystal structure of aureomycin hydrochloride. Z. Kristallogr. 112, 439–464 (1959).
Donohue, J., Dunitz, J. D., Trueblood, K. N. & Webster, M. S. The crystal structure of aureomycin (chlortetracycline) hydrochloride. Configuration, bond distances and conformation. J. Am. Chem. Soc. 85, 851–856 (1963).
Muxfeldt, H. et al. Total synthesis of anhydroaureomycin. Angew. Chem. Int. Ed. Engl. 12, 497–499 (1973).
Scott, A. I. & Bedford, C. T. Simulation of the biosynthesis of tetracyclines. A partial synthesis of tetracycline from anhydroaureomycin. J. Am. Chem. Soc. 84, 2271–2272 (1962).
Gurevich, A. I. et al. Synthesis of 12a-deoxy-5a,6-anhydrotetracycline. The first total synthesis of the naturally occuring tetracycline. Tetrahedron Lett. 8, 131–134 (1967).
Schach von Wittenau, M. Preparation of tetracyclines by photooxidation of anhydrotetracyclines. J. Org. Chem. 29, 2746–2748 (1964).
Gurevich, A. I., Karapetyan, M. G. & Kolosov, M. N. Research in the field of tetracyclines XLIII. Partial synthesis of anhydrotetracycline. Khim. Prirodn. Soedin., Akad. Nauk UzSSR 2, 141–142 (1966). Chem. Abs. 65, 13627 (1966).
Gurevich, A. I., Karapetyan, M. G. & Kolosov, M. N. Investigations in the field of tetracyclines XLIII. Partial synthesis of anhydrotetracycline. Chem. Nat. Compd. 2, 112 (1966).
Conover, L. H., Butler, K., Johnston, J. D., Korst, J. J. & Woodward, R. B. The total synthesis of 6-demethyl-6-deoxytetracycline. J. Am. Chem. Soc. 84, 3222–3224 (1962).
Korst, J. J. et al. The total synthesis of dl-6-demethyl-6-deoxytetracycline. J. Am. Chem. Soc. 90, 439–457 (1968).
Yao-Tseng, H. Experiments on the synthesis of substances related to tetracyclines. Tetrahedron 11, 52–59 (1960).
Bhati, A. Syntheses of some tetralones related to tetracyclines. Tetrahedron 18, 1519–1526 (1962).
Dürckheimer, W. Tetracyclines: chemistry, biochemistry, and structure-activity relations. Angew. Chem. Int. Ed. Engl. 14, 721–734 (1975).
Stork, G. & Hagedorn, A. A. 3-Benzyloxyisoxazole system in construction of tetracyclines. J. Am. Chem. Soc. 100, 3609–3611 (1978).
Muxfeldt, H. et al. Tetracyclines. 9. Total synthesis of dl-terramycin. J. Am. Chem. Soc. 101, 689–701 (1979).
Stork, G., La Clair, J. J., Spargo, P., Nargund, R. P. & Totah, N. Stereocontrolled synthesis of (±)-12a-deoxytetracycline. J. Am. Chem. Soc. 118, 5304–5305 (1996).
Tatsuta, K., Yoshimoto, T., Gunji, H., Okado, Y. & Takahashi, M. The first total synthesis of natural (−)-tetracycline. Chem. Lett. 29, 646–647 (2000).
Charest, M. G., Lerner, C. D., Brubaker, J. D., Siegel, D. R. & Myers, A. G. A convergent enantioselective route to structurally diverse 6-deoxytetracycline antibiotics. Science 308, 395–398 (2005).
Charest, M. G., Siegel, D. R. & Myers, A. G. Synthesis of (−)-tetracycline. J. Am. Chem. Soc. 127, 8292–8293 (2005).
Nicolaou, K. C. et al. Total synthesis and structural revision of viridicatumtoxin B. Angew. Chem. Int. Ed. 52, 8736–8741 (2013).
Nicolaou, K. C. et al. Total synthesis of viridicatumtoxin B and analogues thereof: strategy evolution, structural revision, and biological evaluation. J. Am. Chem. Soc. 136, 12137–12160 (2014).
Rose, W. E. & Rybak, M. J. Tigecycline: first of a new class of antimicrobial agents. Pharmacotherapy 26, 1099–1110 (2006).
Ehrlich, J., Bartz, Q. R., Smith, R. M., Joslyn, D. A. & Burkholder, P. R. Chloromycetin, a new antibiotic from a soil actinomycete. Science 106, 417 (1947).
Gottlieb, D., Bhattacharyya, P. K., Anderson, H. W. & Carter, H. E. Some properties of an antibiotic obtained from a species of Streptomyces. J. Bacteriol. 55, 409–417 (1948).
Carter, H. E., Gottlieb, D. & Anderson, H. W. Chloromycetin and streptothricin. Science 107, 113 (1948).
Umezawa, H., Tazaki, T., Kanari, H., Okami, Y. & Fukuyama, S. Isolation of a crystalline antibiotic substance from a strain of Streptomyces and its identity with chloromycetin. Jpn. Med. J. 1, 358–363 (1948).
Bartz, Q. R. Isolation and characterization of chloromycetin. J. Biol. Chem. 172, 445–450 (1948).
Rebstock, M. C., Crooks, H. M., Controulis, J. & Bartz, Q. R. Chloramphenicol (chloromycetin). IV. Chemical studies. J. Am. Chem. Soc. 71, 2458–2462 (1949).
Controulis, J., Rebstock, M. C. & Crooks, H. M. Chloramphenicol (chloromycetin). V. Synthesis. J. Am. Chem. Soc. 71, 2463–2468 (1949).
Drainas, D., Kalpaxis, D. L. & Coutsogeorgopoulos, C. Inhibition of ribosomal peptidyltransferase by chloramphenicol. Eur. J. Biochem. 164, 53–58 (1987).
Schlunzen, F. et al. Structural basis for the interaction of antibiotics with the peptidyl transferase centre in eubacteria. Nature 413, 814–821 (2001).
Maeda, K., Osato, T. & Umezawa, H. A new antibiotic, azomycin. J. Antibiot. 6, 182 (1953).
Nakamura, S. Structure of azomycin, a new antibiotic. Pharm. Bull. 3, 379–383 (1955).
Cosar, C. & Julou, L. Activité de l'(hydroxy-2-éthyl)-1 méthyl-2 nitro-5 imidazole (8.823R.P.) vis-à-vis des infections expérimentales Trichomonas vaginalis. Ann. Inst. Pasteur (Paris) 96, 238–241 (1959).
White, R. J. in Antibiotic Discovery and Development (eds Dougherty T. J. and Pucci M. J.) 3–31 (Springer, New York, NY, USA, 2011).
Li, J. J. & Corey, E. J. Drug Discovery: Practices, Processes, and Perspectives, John Wiley & Sons, Hoboken, NJ, USA, (2013).
Löfmark, S., Edlund, C. & Nord, C. E. Metronidazole is still the drug of choice for treatment of anaerobic infections. Clin. Infect. Dis. 50 (Suppl 1): S16–S23 (2010).
Novak, R. & Shlaes, D. M. The pleuromutilin antibiotics: a new class for human use. Curr. Opin. Investig. Drugs 11, 182–191 (2010).
Kavanagh, F., Hervey, A. & Robbins, W. J. Antibiotic substances from basidiomycetes: VIII. Pleurotus multilus (Fr.) Sacc. and Pleurotus passeckerianus Pilat. Proc. Natl. Acad. Sci. USA 37, 570–574 (1951).
Kavanagh, F., Hervey, A. & Robbins, W. J. Antibiotic substances from basidiomycetes: IX. Drosophila subtarata. (Batsch Ex Fr.) Quel. Proc. Natl. Acad. Sci. USA 38, 555–560 (1952).
Anchel, M. Chemical studies with pleuromutilin. J. Biol. Chem. 199, 133–139 (1952).
Birch, A. J., Holzapfel, C. W. & Richards, R. W. Diterpenoid nature of pleuromutilin. Chem. Ind. 14, 374–375 (1963).
Birch, A. J., Holzapfel, C. W. & Rickards, R. W. The structure and some aspects of the biosynthesis of pleuromutilin. Tetrahedron 22, 359–387 (1966).
Naegeli, P . Zur Kenntnis des Pleuromutilins. Dissertation, Eidgenössische Technische Hochschule Zürich (1961).
Arigoni, D. 75. La struttura di un terpene di nuovo genere. Gazz. Chim. Ital. 92, 884–901 (1962).
Bonavia, G . Pleuromutilin. Stereochemie und detaillierte Biosynthese. Dissertation, Eidgenössische Technische Hochschule Zürich (1968).
Gibbons, E. G. One-step synthesis of tricyclo[5.2.2.02,6]undecane derivatives: precursors to pleuromutilin. J. Org. Chem. 45, 1540–1541 (1980).
Gibbons, E. G. Total synthesis of (±)-pleuromutilin. J. Am. Chem. Soc. 104, 1767–1769 (1982).
Fazakerley, N. J., Helm, M. D. & Procter, D. J. Total synthesis of (+)-pleuromutilin. Chem. Eur. J. 19, 6718–6723 (2013).
Högenauer, G. in Mechanism of Action of Antibacterial Agents Tiamulin and Pleuromutilin (ed. Hahn F. E.) 344–360 (Springer Verlag, Berlin, Heidelberg, Federal Republic of Germany, 1979)..
McGuire, J. M. et al. Ilotycin, ein neues Antibiotikum. Schweiz. Med. Wochenschr. 82, 1064–1065 (1952).
McGuire, J. M. et al. Ilotycin, a new antibiotic. Antibiot. Chemother. (Northfield, Ill.) 2, 281–283 (1952).
Flynn, E. H., Sigal, M. V., Wiley, P. F. & Gerzon, K. Erythromycin. I. Properties and degradation studies. J. Am. Chem. Soc. 76, 3121–3131 (1954).
Flynn, E. H., Murphy, H. W. & McMahon, R. E. Erythromycin. II. Des-N-methylerythromycin and N-methyl-C14-erythromycin. J. Am. Chem. Soc. 77, 3104–3106 (1955).
Wiley, P. F. & Weaver, O. Erythromycin. III. The structure of cladinose. J. Am. Chem. Soc. 77, 3422–3423 (1955).
Wiley, P. F., Gerzon, K., Flynn, E. H., Sigal, M. V. & Quarck, U. C. Erythromycin. IV. Degradative studies. J. Am. Chem. Soc. 77, 3676–3677 (1955).
Wiley, P. F., Gerzon, K., Flynn, E. H., Sigal, M. V. & Quarck, U. C. Erythromycin. V. Isolation and structure of degradation products. J. Am. Chem. Soc. 77, 3677–3678 (1955).
Sigal, M. V. et al. Erythromycin. VI. Degradation studies. J. Am. Chem. Soc. 78, 388–395 (1956).
Wiley, P. F. & Weaver, O. Erythromycin. VII. The structure of cladinose. J. Am. Chem. Soc. 78, 808–810 (1956).
Gerzon, K. et al. Erythromycin. VIII. Structure of dihydroerythronolide. J. Am. Chem. Soc. 78, 6396–6408 (1956).
Wiley, P. F. et al. Erythromycin. X. Structure of erythromycin. J. Am. Chem. Soc. 79, 6062–6070 (1957).
Stephenson, G. A., Stowell, O. G., Toma, P. H., Pfeiffer, R. R. & Byrn, S. R. Solid-state investigations of erythromycin A dihydrate: structure, NMR spectroscopy, and hygroscopicity. J. Pharm. Sci. 86, 1239–1244 (1997).
Woodward, R. B. et al. Asymmetric total synthesis of erythromcin. 1. Synthesis of an erythronolide A secoacid derivative via asymmetric induction. J. Am. Chem. Soc. 103, 3210–3213 (1981).
Woodward, R. B. et al. Asymmetric total synthesis of erythromycin. 2. Synthesis of an erythronolide A lactone system. J. Am. Chem. Soc. 103, 3213–3215 (1981).
Woodward, R. B. et al. Asymmetric total synthesis of erythromycin. 3. Total synthesis of erythromycin. J. Am. Chem. Soc. 103, 3215–3217 (1981).
Corey, E. J. et al. Total synthesis of erythromycins. 3. Stereoselective routes to intermediates corresponding to C(1) to C(9) and C(10) to C(13) fragments of erythronolide B. J. Am. Chem. Soc. 100, 4618–4620 (1978).
Corey, E. J. et al. Total synthesis of erythromycins. 4. Total synthesis of erythronolide B. J. Am. Chem. Soc. 100, 4620–4622 (1978).
Wolfe, A. D. & Hahn, F. E. Erythromycin: mode of action. Science 143, 1445–1446 (1964).
Schönfeld, W. & Kirst, H. A. Macrolide Antibiotics, Birkhäuser, Basel, Switzerland, (2012).
Griffith, R. S. Introduction to vancomycin. Rev. Infect. Dis. 3, S200–S204 (1981).
Levine, D. P. Vancomycin: a history. Clin. Infect. Dis. 42 (Suppl 1): S5–S12 (2006).
Smith, K. A., Williams, D. H. & Smith, G. A. Structural studies on the antibiotic vancomycin; the nature of the aromatic rings. J. Chem. Soc., Perkin Trans. 1 2369–2376 (1974).
Smith, G. A., Smith, K. A. & Williams, D. H. Structural studies on the antibiotic vancomycin: evidence for the presence of modified phenylglycine and β-hydroxytyrosine units. J. Chem. Soc., Perkin Trans. 1 2108–2115 (1975).
Williams, D. H. & Kalman, J. R. Structural and mode of action studies on the antibiotic vancomycin. Evidence from 270-MHz proton magnetic resonance. J. Am. Chem. Soc. 99, 2768–2774 (1977).
Sheldrick, G. M., Jones, P. G., Kennard, O., Williams, D. H. & Smith, G. A. Structure of vancomycin and its complex with acetyl-D-alanyl-D-alanine. Nature 271, 223–225 (1978).
Schäfer, M., Schneider, T. R. & Sheldrick, G. M. Crystal structure of vancomycin. Structure 4, 1509–1515 (1996).
Evans, D. A. et al. Total syntheses of vancomycin and eremomycin aglycons. Angew. Chem. Int. Ed. 37, 2700–2704 (1998).
Evans, D. A. et al. Nonconventional stereochemical issues in the design of the synthesis of the vancomycin antibiotics: challenges imposed by axial and nonplanar chiral elements in the heptapeptide aglycons. Angew. Chem. Int. Ed. 37, 2704–2708 (1998).
Nicolaou, K. C. et al. Total synthesis of vancomycin aglycon—Part 1: synthesis of amino acids 4–7 and construction of the AB-COD ring skeleton. Angew. Chem. Int. Ed. 37, 2708–2714 (1998).
Nicolaou, K. C. et al. Total synthesis of vancomycin aglycon—Part 2: synthesis of amino acids 1–3 and construction of the AB-COD-DOE ring skeleton. Angew. Chem. Int. Ed. 37, 2714–2716 (1998).
Nicolaou, K. C. et al. Total synthesis of vancomycin aglycon—Part 3: final stages. Angew. Chem. Int. Ed. 37, 2717–2719 (1998).
Nicolaou, K. C. et al. Total synthesis of vancomycin. Angew. Chem. Int. Ed. 38, 240–244 (1999).
Courvalin, P. Vancomycin resistance in Gram-positive Cocci. Clin. Infect. Dis. 42 (Suppl 1): S25–S34 (2006).
Charney, J., Fisher, W. P., Curran, C., Machlowitz, R. A. & Tytell, A. A. Streptogramin, a new antibiotic. Antibiot. Chemother. (Northfield, Ill.) 3, 1283–1286 (1953).
Mukhtar, T. A. & Wright, G. D. Streptogramins, oxazolidinones, and other inhibitors of bacterial protein synthesis. Chem. Rev. 105, 529–542 (2005).
Vazquez, D. Studies on the mode of action of the streptogramin antibiotics. J. Gen. Microbiol. 42, 93–106 (1966).
Bodanszky, M. & Ondetti, M. A. Structures of the vernamycin B group of antibiotics. Antimicrob. Agents Chemother. 161, 360–365 (1963).
Delpierre, G. R. et al. Antibiotics of the ostreogrycin complex. Part II. Structure of ostreogrycin A. J. Chem. Soc. C 1653–1669 (1966).
Kingston, D. G. I., Todd, L. & Williams, D. H. Antibiotics of the ostreogrycin complex. Part III. The structure of ostreogrycin A. Evidence based on nuclear magnetic double resonance experiments and high-resolution mass spectrometry. J. Chem. Soc. C 1669–1676 (1966).
Durant, F., Evrard, G., Declercq, J. P. & Germain, G. Virginiamycin: factor M-dioxane: C32H43N3O9 . Cryst. Struct. Commun. 3, 503–510 (1974).
Karle, I. L. & Flippen-Anderson, J. L. Vernamycin Bα . Acta Crystallogr., Sect. C: Cryst. Struct. Commun. 46, 303–306 (1990).
Noeske, J. et al. Synergy of streptogramin antibiotics occurs independently of their effects on translation. Antimicrob. Agents Chemother. 58, 5269–5279 (2014).
Ennis, H. L. Inhibition of protein synthesis by polypeptide antibiotics I. Inhibition in intact bacteria. J. Bacteriol. 90, 1102–1108 (1965).
Cocito, C. Metabolism of macromolecules in bacteria treated with virginiamycin. Microbiology 57, 179–194 (1969).
Rubinstein, E. & Bompart, F. Activity of quinupristin/dalfopristin against Gram-positive bacteria: clinical applications and therapeutic potential. J. Antimicrob. Chemother. 39 (Suppl A): 139–143 (1997).
Meyers, A. I., Lawson, J., Amos, R. A., Walker, D. G. & Spohn, R. F. Studies on the total synthesis of streptogramin antibiotics: griseoviridin and madumycin (A-2315A). Pure Appl. Chem. 54, 2537–2544 (1982).
Meyers, A. I., Lawson, J. P., Walker, D. G. & Linderman, R. J. Synthetic studies on the streptogramin antibiotics. Enantioselective synthesis of the oxazole dienyl amine moiety. J. Org. Chem. 51, 5111–5123 (1986).
Helquist, P. et al. Synthesis of macrocyclic lactam/lactone derivatives having antimicrobial activity. Pure Appl. Chem. 66, 2063–2066 (1994).
Wood, R. D. & Ganem, B. A simple solution to the oxazole problem in virginiamycin M. Tetrahedron Lett. 24, 4391–4392 (1983).
Schlessinger, R. H., Iwanowicz, E. J. & Springer, J. P. Highly diastereoselective alkylation reactions of vinylogous urethanes derived from simple tetronic acids. Tetrahedron Lett. 29, 1489–1492 (1988).
Liu, L., Tanke, R. S. & Miller, M. J. Electrophilic sulfur transfer reactions in organic synthesis. Preparation of a diastereomer of the key macrocyclic component of griseoviridin. J. Org. Chem. 51, 5332–5337 (1986).
Adjé, N., Breuilles, P. & Uguen, D. Desymmetrisation of meso-propargylic diols. Tetrahedron Lett. 34, 4631–4634 (1993).
Ghosh, A. K. & Liu, W. A convergent, enantioselective total synthesis of streptogramin antibiotic (−)-madumycin II. J. Org. Chem. 62, 7908–7909 (1997).
Dvorak, C. A. et al. The synthesis of streptogramin antibiotics: (−)-griseoviridin and its C-8 epimer. Angew. Chem. Int. Ed. 39, 1664–1666 (2000).
Schlessinger, R. H. & Li, Y.-J. Total synthesis of (−)-virginiamycin M2 using second-generation vinylogous urethane chemistry. J. Am. Chem. Soc. 118, 3301–3302 (1996).
Entwistle, D. A., Jordan, S. I., Montgomery, J. & Pattenden, G. Total synthesis of the virginiamycin antibiotic 14,15-anhydropristinamycin IIB . J. Chem. Soc., Perkin Trans. 1 1315–1317 (1996).
Wu, J. & Panek, J. S. Total synthesis of (−)-virginiamycin M2 . Angew. Chem. Int. Ed. 49, 6165–6168 (2010).
Wu, J. & Panek, J. S. Total synthesis of (−)-virginiamycin M2: application of crotylsilanes accessed by enantioselective Rh(II) or Cu(I) promoted carbenoid Si–H insertion. J. Org. Chem. 76, 9900–9918 (2011).
Roth, B., Falco, E. A., Hitchings, G. H. & Bushby, S. R. M. 5-Benzyl-2,4-diaminopyrimidines as antibacterial agents. I. Synthesis and antibacterial activity in vitro. J. Med. Pharm. Chem. 5, 1103–1123 (1962).
Noall, E. W. P., Sewards, H. F. G. & Waterworth, P. M. Successful treatment of a case of Proteus Septicaemia. Br. Med. J. 2, 1101–1102 (1962).
Bushby, S. R. & Hitchings, G. H. Trimethoprim, a sulphonamide potentiator. Br. J. Pharmacol. Chemother. 33, 72–90 (1968).
Gleckman, R., Blagg, N. & Joubert, D. W. Trimethoprim: mechanisms of action, antimicrobial activity, bacterial resistance, pharmacokinetics, adverse reactions, and therapeutic indications. Pharmacotherapy 1, 14–19 (1981).
Brogden, R. N., Carmine, A. A., Heel, R. C., Speight, T. M. & Avery, G. S. Trimethoprim: a review of its antibacterial activity, pharmacokinetics and therapeutic use in urinary tract infections. Drugs 23, 405–430 (1982).
Lesher, G. Y., Froelich, E. J., Gruett, M. D., Bailey, J. H. & Brundage, R. P. 1,8-Naphthyridine derivatives. A new class of chemotherapeutic agents. J. Med. Pharm. Chem. 5, 1063–1065 (1962).
Emmerson, A. M. & Jones, A. M. The quinolones: decades of development and use. J. Antimicrob. Chemother. 51 (Suppl 1): 13–20 (2003).
Hamatake, R. K., Mukai, R. & Hayashi, M. Role of DNA gyrase subunits in synthesis of bacteriophage φX174 viral DNA. Proc. Natl. Acad. Sci. USA 78, 1532–1536 (1981).
Sugino, A., Peebles, C. L., Kreuzer, K. N. & Cozzarelli, N. R. Mechanism of action of nalidixic acid: purification of Escherichia coli nalA gene product and its relationship to DNA gyrase and a novel nicking-closing enzyme. Proc. Natl. Acad. Sci. USA 74, 4767–4771 (1977).
Gellert, M., Mizuuchi, K., O'Dea, M. H., Itoh, T. & Tomizawa, J. I. Nalidixic acid resistance: a second genetic character involved in DNA gyrase activity. Proc. Natl. Acad. Sci. USA 74, 4772–4776 (1977).
Reece, R. J. & Maxwell, A. DNA gyrase: structure and function. Crit. Rev. Biochem. Mol. Biol. 26, 335–375 (1991).
Fournier, B., Zhao, X., Lu, T., Drlica, K. & Hooper, D. C. Selective targeting of topoisomerase IV and DNA gyrase in Staphylococcus aureus: different patterns of quinolone-induced inhibition of DNA synthesis. Antimicrob. Agents Chemother. 44, 2160–2165 (2000).
Rinehart, K. L. Jr. Antibiotics with ansa rings. Acc. Chem. Res. 5, 57–64 (1972).
Sensi, P., Margalith, P. & Timbal, M. T. Rifomycin, a new antibiotic; preliminary report. Farmaco. Sci. 14, 146–147 (1959).
Oppolzer, W., Prelog, V. & Sensi, P. Konstitution des Rifamycins B und verwandter Rifamycine. Experientia 20, 336–339 (1964).
Brufani, M., Fedeli, W., Giacomello, G. & Vaciago, A. The X-ray analysis of the structure of rifamycin B. Experientia 20, 339–342 (1964).
Leitich, J., Oppolzer, W. & Prelog, V. Über die Konfiguration des Rifamycins B und verwandter Rifamycine. Experientia 20, 343–344 (1964).
Bifani, P. et al. The evolution of drug resistance in Mycobacterium tuberculosis: from a mono–rifampin-resistant cluster into increasingly multidrug-resistant variants in an HIV-seropositive population. J. Infect. Dis. 198, 90–94 (2008).
Descombe, J. J., Dubourg, D., Picard, M. & Palazzini, E. Pharmacokinetic study of rifaximin after oral administration in healthy volunteers. Int. J. Clin. Pharmacol. Res. 14, 51–56 (1994).
Coppi, G., Mazzola, D. & Moiana, S. (Friulchem SpA). New process for the synthesis of rifaximin and a new pseudo-crystalline form of rifaximin obtained thereby. WO2012155981 A1 (2012).
Calvori, C., Frontali, L., Leoni, L. & Tecce, G. Effect of rifamycin on protein synthesis. Nature 207, 417–418 (1965).
Campbell, E. A. et al. Structural mechanism for rifampicin inhibition of bacterial RNA polymerase. Cell 104, 901–912 (2001).
Fugitt, R. B. & Luckenbaugh, R. W. 5-Halomethyl-3-phenyl-2-oxazolidinones. US4128654 A (1978).
Slee, A. M. et al. Oxazolidinones, a new class of synthetic antibacterial agents: in vitro and in vivo activities of DuP 105 and DuP 721. Antimicrob. Agents Chemother. 31, 1791–1797 (1987).
Ford, C. W., Zurenko, G. E. & Barbachyn, M. R. The discovery of linezolid, the first oxazolidinone antibacterial agent. Curr. Drug Targets Infect. Disord. 1, 181–199 (2001).
Barbachyn, M. R. & Ford, C. W. Oxazolidinone structure–activity relationships leading to linezolid. Angew. Chem. Int. Ed. 42, 2010–2023 (2003).
Barbachyn, M. R., Brickner, S. J. & Hutchinson, D. K. Substituted oxazine and thiazine oxazolidinone antimicrobials. US5688792 A (1997).
Wilson, A. P. R. et al. In vitro susceptibility of Gram-positive pathogens to linezolid and teicoplanin and effect on outcome in critically ill patients. J. Antimicrob. Chemother. 58, 470–473 (2006).
Livermore, D. M. Linezolid in vitro: mechanism and antibacterial spectrum. J. Antimicrob. Chemother. 51 (Suppl 2): ii9–ii16 (2003).
Jones, R. N. & Biedenbach, D. J. Antimicrobial activity of RU-66647, a new ketolide. Diagn. Microbiol. Infect. Dis. 27, 7–12 (1997).
Johnson, A. P. Telithromycin. Aventis Pharma. Curr. Opin. Investig. Drugs 2, 1691–1701 (2001).
Zhanel, G. G. et al. The ketolides: a critical review. Drugs 62, 1771–1804 (2002).
Berisio, R. et al. Structural insight into the antibiotic action of telithromycin against resistant mutants. J. Bacteriol. 185, 5027 (2003).
Ackermann, G. & Rodloff, A. C. Drugs of the 21st century: telithromycin (HMR 3647)—the first ketolide. J. Antimicrob. Chemother. 51, 497–511 (2003).
Judice, J. K. & Pace, J. L. Semi-synthetic glycopeptide antibacterials. Bioorg. Med. Chem. Lett. 13, 4165–4168 (2003).
Leadbetter, M. R. et al. Hydrophobic vancomycin derivatives with improved ADME properties: discovery of telavancin (TD-6424). J. Antibiot. 57, 326–336 (2004).
Saravolatz, L. D., Stein, G. E. & Johnson, L. B. Telavancin: a novel lipoglycopeptide. Clin. Infect. Dis. 49, 1908–1914 (2009).
Corey, G. R., Stryjewski, M. E., Weyenberg, W., Yasothan, U. & Kirkpatrick, P. Telavancin. Nat. Rev. Drug Discov. 8, 929–930 (2009).
Spellberg, B., Bartlett, J. G. & Gilbert, D. N. The future of antibiotics and resistance. N. Engl. J. Med. 368, 299–302 (2013).
Dever, L. A. & Dermody, T. S. Mechanisms of bacterial resistance to antibiotics. Arch. Intern. Med. 151, 886–895 (1991).
Blair, J. M. A., Webber, M. A., Baylay, A. J., Ogbolu, D. O. & Piddock, L. J. V. Molecular mechanisms of antibiotic resistance. Nat. Rev. Micro. 13, 42–51 (2015).
Li, X.-Z. & Nikaido, H. Efflux-mediated drug resistance in bacteria: an update. Drugs 69, 1555–1623 (2009).
Aminov, R. I. & Mackie, R. I. Evolution and ecology of antibiotic resistance genes. FEMS Microbiol. Lett. 271, 147–161 (2007).
Ochman, H., Lawrence, J. G. & Groisman, E. A. Lateral gene transfer and the nature of bacterial innovation. Nature 405, 299–304 (2000).
Penicillin's finder assays its future. N.Y. Times, 21 (26 June 1945).
Boucher, H. W. et al. Bad bugs, no drugs: no ESKAPE! An update from the Infectious Diseases Society of America. Clin. Infect. Dis. 48, 1–12 (2009).
Spellberg, B. et al. The epidemic of antibiotic-resistant infections: a call to action for the medical community from the Infectious Diseases Society of America. Clin. Infect. Dis. 46, 155–164 (2008).
Kupferschmidt, K. Resistance fighters. Science 352, 758–761 (2016).
Ling, L. L. et al. A new antibiotic kills pathogens without detectable resistance. Nature 517, 455–459 (2015).
Giltrap, A. M. et al. Total synthesis of teixobactin. Org. Lett. 18, 2788–2791 (2016).
Zipperer, A. et al. Human commensals producing a novel antibiotic impair pathogen colonization. Nature 535, 511–516 (2016).
O’Neil, J. Antimicrobial resistance: tackling a crisis for the future health and wealth of nations (2014). Available at: https://amr-review.org/sites/default/files/AMR%20Review%20Paper%20-%20Tackling%20a%20crisis%20for%20the%20health%20and%20wealth%20of%20nations_1.pdf.
Shapiro, D. J., Hicks, L. A., Pavia, A. T. & Hersh, A. L. Antibiotic prescribing for adults in ambulatory care in the USA, 2007–09. J. Antimicrob. Chemother. 69, 234–240 (2014).
de Lalla, F. et al. Third generation cephalosporins as a risk factor for Clostridium difficile-associated disease: a four-year survey in a general hospital. J. Antimicrob. Chemother. 23, 623–631 (1989).
Nicolaou, K. C. & Rigol, S. The evolution and impact of total synthesis on chemistry, biology and medicine. Isr. J. Chem. 57, 179–191 (2017).
Liu, C. M. et al. X-14547A, a new ionophorous antibiotic produced by Streptomyces antibioticus NRRL 8167. Discovery, fermentation, biological properties and taxonomy of the producing culture. J. Antibiot. 32, 95–99 (1979).
Westley, J. W. et al. Isolation and characterization of antibiotic X-14547A, a novel monocarboxylic acid ionophore produced by Streptomyces antibioticus NRRL 8167. J. Antibiot. 32, 100–107 (1979).
Westley, J. & Liu, C.-M. Antibiotic X-14547. US4100171 A (1978).
Westley, J. & Liu, C.-M. (Hoffmann-La Roche Inc.). Antibiotic X-14547 and its use for increasing feed efficiency in ruminants. US4167579 A (1979).
Nicolaou, K. C. & Magolda, R. L. Ionophore antibiotic X-14547A. Degradation studies and stereoselective construction of the ‘right wing‘ (C11-C25 fragment) by an intramolecular Diels–Alder reaction. J. Org. Chem. 46, 1506–1508 (1981).
Nicolaou, K. C., Papahatjis, D. P., Claremon, D. A. & Dolle, R. E. Total synthesis of ionophore antibiotic X-14547A. 1. Enantioselective synthesis of the tetrahydropyran and tetrahydroindan building blocks. J. Am. Chem. Soc. 103, 6967–6969 (1981).
Nicolaou, K. C., Claremon, D. A., Papahatjis, D. P. & Magolda, R. L. Total synthesis of ionophore antibiotic X-14547A. 2. Coupling of the tetrahydropyran and tetrahydroindan systems and construction of the butadienyl and ketopyrrole moieties. J. Am. Chem. Soc. 103, 6969–6971 (1981).
Roush, W. R. & Myers, A. G. Antibiotic X-14547A: total synthesis of the right-hand half. J. Org. Chem. 46, 1509–1511 (1981).
Roush, W. R. & Peseckis, S. M. Studies on the total synthesis of antibiotic X-14547A: the pentaene approach. Tetrahedron Lett. 23, 4879–4882 (1982).
Ho, P.-T. Studies toward polyether antibiotics: stereospecific synthesis of polysubstituted tetrahydropyrans. Can. J. Chem. 60, 90–94 (1982).
Edwards, M. P., Ley, S. V., Lister, S. G. & Palmer, B. D. Total synthesis of the structurally unique ionophore antibiotic X-14547A. J. Chem. Soc., Chem. Commun. 630–633 (1983).
Roush, W. R., Peseckis, S. M. & Walts, A. E. Synthesis of antibiotic X-14547A. J. Org. Chem. 49, 3429–3432 (1984).
Edwards, M. P., Ley, S. V., Lister, S. G., Palmer, B. D. & Williams, D. J. Total synthesis of the ionophore antibiotic X-14547A (indanomycin). J. Org. Chem. 49, 3503–3516 (1984).
Boeckman, R. K., Enholm, E. J., Demko, D. M. & Charette, A. B. An efficient enantioselective total synthesis of (−)-X-14547A (indanomycin). J. Org. Chem. 51, 4743–4745 (1986).
Burke, S. D. et al. Total synthesis of ionophore antibiotic X-14547A (indanomycin). J. Org. Chem. 59, 332–347 (1994).
Dewey, R. S., Arison, B. H., Hannah, J., Shih, D. H. & Albers-Schönberg, G. The structure of efrotomycin. J. Antibiot. 38, 1691–1698 (1985).
Frost, B. M., Valiant, M. E., Weissberger, B. & Dulaney, E. L. Antibacterial activity of efrotomycin. J. Antibiot. 29, 1083–1091 (1976).
Clabots, C. R., Shanholtzer, C. J., Peterson, L. R. & Gerding, D. N. In vitro activity of efrotomycin, ciprofloxacin, and six other antimicrobials against Clostridium difficile. Diagn. Microbiol. Infect. Dis. 6, 49–52 (1987).
Foster, A. G. et al. Effect of efrotomycin on gain and feed efficiency for pigs from weaning until market weight. J. Anim. Sci. 65, 877–880 (1987).
Dolle, R. E. & Nicolaou, K. C. Total synthesis of elfamycins: aurodox and efrotomycin. 1. Strategy and construction of key intermediates. J. Am. Chem. Soc. 107, 1691–1694 (1985).
Dolle, R. E. & Nicolaou, K. C. Total synthesis of elfamycins: aurodox and efrotomycin. 2. Coupling of key intermediates and completion of the synthesis. J. Am. Chem. Soc. 107, 1695–1698 (1985).
Dolle, R. E. & Nicolaou, K. C. Carbohydrate-based syntheses of the goldinonolactone and the tetrahydrofuran fragments of aurodox and efrotomycin. J. Chem. Soc., Chem. Commun. 1016–1018 (1985).
Ellis, D. Amphotericin B: spectrum and resistance. J. Antimicrob. Chemother. 49 (Suppl 1): 7–10 (2002).
Lewis, R. E., Kontoyiannis, D. P., Darouiche, R. O., Raad, I. I. & Prince, R. A. Antifungal activity of amphotericin B, fluconazole, and voriconazole in an in vitro model of Candida catheter-related bloodstream infection. Antimicrob. Agents Chemother. 46, 3499–3505 (2002).
den Boer, M. & Davidson, R. N. Treatment options for visceral leishmaniasis. Expert Rev. Anti Infect. Ther. 4, 187–197 (2006).
Dutcher, J. D., William, G., Pagano, J. F. & Vandeputte, J. (Olin Mathieson Chemical Corporation). Amphotericin B, its production, and its salts. US2908611 A (1959).
Nicolaou, K. C., Daines, R. A., Chakraborty, T. K. & Ogawa, Y. Total synthesis of amphotericin B. J. Am. Chem. Soc. 109, 2821–2822 (1987).
Nicolaou, K. C. et al. Total synthesis of amphoteronolide B and amphotericin B. 1. Strategy and stereocontrolled construction of key building blocks. J. Am. Chem. Soc. 110, 4672–4685 (1988).
Nicolaou, K. C., Daines, R. A., Chakraborty, T. K. & Ogawa, Y. Total synthesis of amphoteronolide B and amphotericin B. 2. Total synthesis of amphoteronolide B. J. Am. Chem. Soc. 110, 4685–4696 (1988).
Nicolaou, K. C., Daines, R. A., Ogawa, Y. & Chakraborty, T. K. Total synthesis of amphotericin B. 3. The final stages. J. Am. Chem. Soc. 110, 4696–4705 (1988).
Palacios, D. S., Dailey, I., Siebert, D. M., Wilcock, B. C. & Burke, M. D. Synthesis-enabled functional group deletions reveal key underpinnings of amphotericin B ion channel and antifungal activities. Proc. Natl. Acad. Sci. USA 108, 6733–6738 (2011).
Gray, K. C. et al. Amphotericin primarily kills yeast by simply binding ergosterol. Proc. Natl. Acad. Sci. USA 109, 2234–2239 (2012).
Wilcock, B. C., Endo, M. M., Uno, B. E. & Burke, M. D. C2′-OH of amphotericin B plays an important role in binding the primary sterol of human cells but not yeast cells. J. Am. Chem. Soc. 135, 8488–8491 (2013).
Anderson, T. M. et al. Amphotericin forms an extramembranous and fungicidal sterol sponge. Nat. Chem. Biol. 10, 400–406 (2014).
Masamune, S., Kaiho, T. & Garvey, D. S. Aldol methodology: synthesis of versatile intermediates, 3-hydroxy-2-vinylcarbonyl compounds. J. Am. Chem. Soc. 104, 5521–5523 (1982).
Brooks, D. W. & Kellogg, R. P. Synthetic studies of polyene macrolides, synthesis of a C29–37 fragment for amphotericin B and nystatin. Tetrahedron Lett. 23, 4991–4994 (1982).
Boschelli, D., Ellingboe, J. W. & Masamune, S. Aldol methodology: synthesis of syn-3-hydroxy-2-vinylcarbonyl compounds. Tetrahedron Lett. 25, 3395–3398 (1984).
Masamune, S., Ma, P., Okumoto, H., Ellingboe, J. W. & Ito, Y. Synthesis of amphotericin B. 1. Fragment A of the aglycon. J. Org. Chem. 49, 2834–2837 (1984).
Boschelli, D., Takemasa, T., Nishitani, Y. & Masamune, S. Synthesis of amphotericin B. 2. Fragment C-D of the aglycone. Tetrahedron Lett. 26, 5239–5242 (1985).
McGarvey, G. J., Williams, J. M., Hiner, R. N., Matsubara, Y. & Oh, T. L-Aspartic acid in acyclic stereoselective synthesis. Synthetic studies on amphotericin B. J. Am. Chem. Soc. 108, 4943–4952 (1986).
Solladié, G. & Hutt, J. Asymmetric synthesis of polyhydroxylated natural products II. The C-1/C-12 unit of amphotericin B. Tetrahedron Lett. 28, 797–800 (1987).
Hanessian, S., Sahoo, S. P. & Botta, M. Methodology for the polyene and related antibiotics—enantiospecific synthesis of chiral structural units of amphotericin B from a common progenitor: the C1–C13 polyol segment. Tetrahedron Lett. 28, 1143–1146 (1987).
Hanessian, S., Sahoo, S. P. & Botta, M. Methodology for the polyene and related antibiotics—enantiospecific synthesis of chiral structural units of amphotericin B from a common progenitor: the C14–C20 and C32–C38 segments. Tetrahedron Lett. 28, 1147–1150 (1987).
Hanessian, S. & Botta, M. Methodology for the polyene and related antibiotics—versatile and practical access to bifunctional all-trans polyolefinic systems. Tetrahedron Lett. 28, 1151–1154 (1987).
Kennedy, R. M., Abiko, A., Takemasa, T., Okumoto, H. & Masamune, S. A synthesis of 19-dehydroamphoteronolide B. Tetrahedron Lett. 29, 451–454 (1988).
Brückner, R. Stereocontrolled synthesis of a C14-C20 building block for amphotericin B using a novel [2,3] Wittig rearrangement. Tetrahedron Lett. 29, 5747–5750 (1988).
McGarvey, G. J. et al. Synthetic studies on the polyene macrolide antibiotics. Development of syn- and anti-1,3-diol subunits and assembly of the polyacetate region of amphotericin B. J. Org. Chem. 60, 7778–7790 (1995).
McGarvey, G. J., Mathys, J. A. & Wilson, K. J. Synthesis of amphotericin B. A convergent strategy to the polyol segment of the heptaene macrolide antibiotics. J. Org. Chem. 61, 5704–5705 (1996).
Krüger, J. & Carreira, E. M. Convergent synthesis of the amphotericin polyol subunit employing asymmetric dienolate addition reactions. Tetrahedron Lett. 39, 7013–7016 (1998).
Solladié, G., Wilb, N. & Bauder, C. A chiral β,δ-dioxo-ε-sulfinyl ester in a convergent enantioselective synthesis towards the C1–C13 polyol fragment of amphotericin B. Eur. J. Org. Chem. 1999, 3021–3026 (1999).
BouzBouz, S. & Cossy, J. Enantioselective allyltitanation. Efficient synthesis of the C1−C14 polyol subunit of amphotericin B. Org. Lett. 2, 3975–3977 (2000).
Bonini, C., Chiummiento, L., Martuscelli, A. & Viggiani, L. A convergent preparation of the C1–C13 fragment of amphotericin B from a single chiral precursor. Tetrahedron Lett. 45, 2177–2179 (2004).
Adediran, S. A. et al. Synthesis of a highly water-soluble derivative of amphotericin B with attenuated proinflammatory activity. Mol. Pharm. 6, 1582–1590 (2009).
Gray, K. C. Semisynthesis of amphotericin B and its derivatives via iterative cross-coupling (PhD thesis, Univ. Illinois at Urbana-Champaign, (2011).
Janout, V., Bienvenu, C., Schell, W., Perfect, J. R. & Regen, S. L. Molecular umbrella–amphotericin B conjugates. Bioconjug. Chem. 25, 1408–1411 (2014).
McCormick, M. H., McGuire, J. M., Pittenger, G. E., Pittenger, R. C. & Stark, W. M. Vancomycin, a new antibiotic. I. Chemical and biologic properties. Antibiot. Annu. 3, 606–611 (1955–1956).
McGuire, J. M., Wolfe, R. N. & Ziegler, D. W. Vancomycin, a new antibiotic. II. In vitro antibacterial studies. Antibiot. Annu. 3, 612–618 (1955–1956).
Griffith, R. S. & Peck, F. B. Jr. Vancomycin, a new antibiotic. III. Preliminary clinical and laboratory studies. Antibiot. Annu. 3, 619–622 (1955–1956).
Nicolaou, K. C. et al. Target-accelerated combinatorial synthesis and discovery of highly potent antibiotics effective against vancomycin-resistant bacteria. Angew. Chem. Int. Ed. 39, 3823–3828 (2000).
Nicolaou, K. C. et al. Solid- and solution-phase synthesis of vancomycin and vancomycin analogues with activity against vancomycin-resistant bacteria. Chem. Eur. J. 7, 3798–3823 (2001).
Nicolaou, K. C. et al. Synthesis and biological evaluation of vancomycin dimers with potent activity against vancomycin-resistant bacteria: target-accelerated combinatorial synthesis. Chem. Eur. J. 7, 3824–3843 (2001).
Ganguly, A. K. et al. The structure of new oligosaccharide antibiotics, 13-384 components 1 and 5. Heterocycles 28, 83–88 (1989).
Ganguly, A. K. in Topics in Antibiotic Chemistry, Vol. 2, Part B (ed. Sammes P. G.) 61–96 (Ellis Horwood, Chichester, UK, 1978).
Ganguly, A. K., McCormick, J. L., Saksena, A. K., Das, P. R. & Chan, T.-M. Chemical modifications and structure activity studies of ziracin and related everninomicin antibiotics. Bioorg. Med. Chem. Lett. 9, 1209–1214 (1999).
Ganguly, A. K., Girijavallabhan, V. M. & Sarre, O. (Schering Corporation). Novel derivatives of the oligosaccharide antibiotic complex 13-384, their preparation and pharmaceutical compositions containing them. WO8702366 (1987).
Patel, M. et al. Lipophilic oligosaccharide antibiotic salt compositions. EP0538011 (A1) (1993).
Wright, D. E. The orthosomycins, a new family of antibiotics. Tetrahedron 35, 1207–1237 (1979).
Maertens, J. A. Sch-27899 Schering-Plough. Curr. Opin. Anti-Infect. Invest. Drugs 1, 49–56 (1999).
Maertens, J. A. Sch-27899 (Schering-Plough Corp). IDrugs 2, 446–453 (1999).
Urban, C. et al. Comparative in-vitro activity of SCH 27899, a novel everninomicin, and vancomycin. J. Antimicrob. Chemother. 37, 361–364 (1996).
McNicholas, P. M. et al. Evernimicin binds exclusively to the 50S ribosomal subunit and inhibits translation in cell-free systems derived from both Gram-positive and Gram-negative bacteria. Antimicrob. Agents Chemother. 44, 1121–1126 (2000).
Nicolaou, K. C. et al. Total synthesis of everninomicin 13,384-1—Part 1: synthesis of the A1B(A)C fragment. Angew. Chem. Int. Ed. 38, 3334–3339 (1999).
Nicolaou, K. C., Rodríguez, R. M., Fylaktakidou, K. C., Suzuki, H. & Mitchell, H. J. Total synthesis of everninomicin 13,384-1—Part 2: synthesis of the FGHA2 fragment. Angew. Chem. Int. Ed. 38, 3340–3345 (1999).
Nicolaou, K. C., Mitchell, H. J., Rodríguez, R. M., Fylaktakidou, K. C. & Suzuki, H. Total synthesis of everninomicin 13,384-1—Part 3: synthesis of the DE fragment and completion of the total synthesis. Angew. Chem. Int. Ed. 38, 3345–3350 (1999).
Nicolaou, K. C. et al. Total synthesis of everninomicin 13,384-1—Part 1: Retrosynthetic analysis and synthesis of the A1B(A)C fragment. Chem. Eur. J. 6, 3095–3115 (2000).
Nicolaou, K. C., Mitchell, H. J., Fylaktakidou, K. C., Rodríguez, R. M. & Suzuki, H. Total synthesis of everninomicin 13,384-1—Part 2: synthesis of the FGHA2 fragment. Chem. Eur. J. 6, 3116–3148 (2000).
Nicolaou, K. C. et al. Total synthesis of everninomicin 13,384-1—Part 3: synthesis of the DE fragment and completion of the total synthesis. Chem. Eur. J. 6, 3149–3165 (2000).
Nicolaou, K. C. et al. Total synthesis of everninomicin 13,384-1—Part 4: explorations of methodology; stereocontrolled synthesis of 1,1′-disaccharides, 1,2-seleno migrations in carbohydrates, and solution- and solid-phase synthesis of 2-deoxy glycosides and orthoesters. Chem. Eur. J. 6, 3166–3185 (2000).
Kamigakinai, T., Nakashima, M. & Tani, H. (Shionogi and Co., Ltd.). New benzoxacyclotridecyne compound and medicinal composition containing the same. JP10101666 (1998).
Wilson, K. E. et al. Isolation and structure elucidation of coleophomones A and B, novel inhibitors of bacterial cell wall transglycosylase. Tetrahedron Lett. 41, 8705–8709 (2000).
Urata, H., Kinoshita, A., Misono, K. S., Bumpus, F. M. & Husain, A. Identification of a highly specific chymase as the major angiotensin II-forming enzyme in the human heart. J. Biol. Chem. 265, 22348–22357 (1990).
Nicolaou, K. C., Vassilikogiannakis, G. & Montagnon, T. The total synthesis of coleophomones B and C. Angew. Chem. Int. Ed. 41, 3276–3281 (2002).
Nicolaou, K. C., Montagnon, T., Vassilikogiannakis, G. & Mathison, C. J. N. The total synthesis of coleophomones B,C and D. J. Am. Chem. Soc. 127, 8872–8888 (2005).
Anderson, B., Hodgkin, D. C. & Viswamitra, M. A. The structure of thiostrepton. Nature 225, 233–235 (1970).
Donovick, R., Pagano, J. F., Stout, H. A. & Weinstein, M. J. Thiostrepton, a new antibiotic. I. In vitro studies. Antibiot. Annu. 3, 554–559 (1955–1956).
Jambor, W. P., Steinberg, B. A. & Suydam, L. O. Thiostrepton, a new antibiotic. III. In vivo studies. Antibiot. Annu. 3, 562–565 (1955–1956).
Nicolaou, K. C., Safina, B. S., Funke, C., Zak, M. & Zécri, F. J. Stereocontrolled synthesis of the quinaldic acid macrocyclic system of thiostrepton. Angew. Chem. Int. Ed. 41, 1937–1940 (2002).
Nicolaou, K. C., Nevalainen, M., Safina, B. S., Zak, M. & Bulat, S. A biomimetically inspired synthesis of the dehydropiperidine domain of thiostrepton. Angew. Chem. Int. Ed. 41, 1941–1945 (2002).
Nicolaou, K. C. et al. Synthetic studies on thiostrepton: construction of thiostrepton analogues with the thiazoline-containing macrocycle. Angew. Chem. Int. Ed. 42, 3418–3424 (2003).
Nicolaou, K. C., Safina, B. S., Zak, M., Estrada, A. A. & Lee, S. H. Total synthesis of thiostrepton, Part 1: construction of the dehydropiperidine/thiazoline-containing macrocycle. Angew. Chem. Int. Ed. 43, 5087–5092 (2004).
Nicolaou, K. C., Zak, M., Safina, B. S., Lee, S. H. & Estrada, A. A. Total synthesis of thiostrepton, Part 2: construction of the quinaldic acid macrocycle and final stages of the synthesis. Angew. Chem. Int. Ed. 43, 5092–5097 (2004).
Nicolaou, K. C. et al. Total synthesis of thiostrepton. Retrosynthetic analysis and construction of key building blocks. J. Am. Chem. Soc. 127, 11159–11175 (2005).
Nicolaou, K. C. et al. Total synthesis of thiostrepton. Assembly of key building blocks and completion of the synthesis. J. Am. Chem. Soc. 127, 11176–11183 (2005).
Dutcher, J. D. & Vandeputte, J. Thiostrepton, a new antibiotic. II. Isolation and chemical characterization. Antibiot. Annu. 3, 560–561 (1955–1956).
Naaktgeboren, N., Roobol, K., Gubbens, J. & Voorma, H. O. The mode of action of thiostrepton in the initiation of protein synthesis. Eur. J. Biochem. 70, 39–47 (1976).
Rodnina, M. V. et al. Thiostrepton inhibits the turnover but not the GTPase of elongation factor G on the ribosome. Proc. Natl. Acad. Sci. USA 96, 9586–9590 (1999).
Walter, J. D., Hunter, M., Cobb, M., Traeger, G. & Spiegel, P. C. Thiostrepton inhibits stable 70S ribosome binding and ribosome-dependent GTPase activation of elongation factor G and elongation factor 4. Nucleic Acids Res. 40, 360–370 (2011).
Nicolaou, K. C. How thiostrepton was made in the laboratory. Angew. Chem. Int. Ed. 51, 12414–12436 (2012).
Nicolaou, K. C., Estrada, A. A., Zak, M., Lee, S. H. & Safina, B. S. A mild and selective method for the hydrolysis of esters with trimethyltin hydroxide. Angew. Chem. Int. Ed. 44, 1378–1382 (2005).
Nicolaou, K. C., Estrada, A. A., Zak, M., Lee, S. H. & Safina, B. S. A mild and selective method for the hydrolysis of esters with trimethyltin hydroxide. ChemInform 36, doi:10.1002/chin.200524057 (2005).
Bister, B. et al. Abyssomicin C–a polycyclic antibiotic from a marine Verrucosispora strain as an inhibitor of the p-aminobenzoic acid/tetrahydrofolate biosynthesis pathway. Angew. Chem. Int. Ed. 43, 2574–2576 (2004).
Riedlinger, J. et al. Abyssomicins, inhibitors of the para-aminobenzoic acid pathway produced by the marine Verrucosispora strain AB-18-032. J. Antibiot. 57, 271–279 (2004).
Keller, S. et al. Abyssomicins G and H and atrop-abyssomicin C from the marine Verrucosispora strain AB-18-032. J. Antibiot. 60, 391–394 (2007).
Nicolaou, K. C. & Harrison, S. T. Total synthesis of abyssomicin C and atrop-abyssomicin C. Angew. Chem. Int. Ed. 45, 3256–3260 (2006).
Nicolaou, K. C. & Harrison, S. T. Total synthesis of abyssomicin C, atrop-abyssomicin C, and abyssomicin D: implications for natural origins of atrop-abyssomicin C. J. Am. Chem. Soc. 129, 429–440 (2007).
Nicolaou, K. C., Harrison, S. T. & Chen, J. S. Discoveries from the abyss: the abyssomicins and their total synthesis. Synthesis 2009, 33–42 (2009).
Snider, B. B. & Zou, Y. Synthesis of the carbocyclic skeleton of abyssomicins C and D. Org. Lett. 7, 4939–4941 (2005).
Rath, J.-P., Eipert, M., Kinast, S. & Maier, M. E. Synthesis of the tetronate-containing core structure of the antibiotic abyssomicin C. Synlett 2005, 314–318 (2005).
Rath, J.-P., Kinast, S. & Maier, M. E. Synthesis of the fully functionalized core structure of the antibiotic abyssomicin C. Org. Lett. 7, 3089–3092 (2005).
Zografos, A. L., Yiotakis, A. & Georgiadis, D. Rapid access to the tricyclic spirotetronic core of abyssomicins. Org. Lett. 7, 4515–4518 (2005).
Kinast, S . Strategien zur Synthese von Abyssomicin C Derivaten. Dissertation, Eberhard Karls Universität Tübingen (2008).
Couladouros, E. A., Bouzas, E. A. & Magos, A. D. Formal synthesis of abyssomicin C. Tetrahedron 62, 5272–5279 (2006).
Zapf, C. W., Harrison, B. A., Drahl, C. & Sorensen, E. J. A Diels–Alder macrocyclization enables an efficient asymmetric synthesis of the antibacterial natural product abyssomicin C. Angew. Chem. Int. Ed. 44, 6533–6537 (2005).
Bihelović, F. & Saičić, R. N. Total synthesis of (−)-atrop-abyssomicin C. Angew. Chem. Int. Ed. 51, 5687–5691 (2012).
Bihelović, F., Karadžić, I., Matović, R. & Saičić, R. N. Total synthesis and biological evaluation of (−)-atrop-abyssomicin C. Org. Biomol. Chem. 11, 5413–5424 (2013).
Kwon, H. C., Kauffman, C. A., Jensen, P. R. & Fenical, W. Marinomycins A−D, antitumor-antibiotics of a new structure class from a marine actinomycete of the recently discovered genus “Marinispora”. J. Am. Chem. Soc. 128, 1622–1632 (2006).
Nicolaou, K. C., Nold, A. L., Milburn, R. R. & Schindler, C. S. Total synthesis of marinomycins A–C. Angew. Chem. Int. Ed. 45, 6527–6532 (2006).
Nicolaou, K. C. et al. Total synthesis of marinomycins A−C and of their monomeric counterparts monomarinomycin A and iso-monomarinomycin A. J. Am. Chem. Soc. 129, 1760–1768 (2007).
Amans, D., Bellosta, V. & Cossy, J. An efficient and stereoselective synthesis of the monomeric counterpart of marinomycin A. Org. Lett. 9, 1453–1456 (2007).
Amans, D., Bareille, L., Bellosta, V. & Cossy, J. Synthesis of the monomeric counterpart of marinomycin A. J. Org. Chem. 74, 7665–7674 (2009).
Evans, P. A., Huang, M.-H., Lawler, M. J. & Maroto, S. Total synthesis of marinomycin A using salicylate as a molecular switch to mediate dimerization. Nat. Chem. 4, 680–684 (2012).
Nishimaru, T. et al. Total synthesis of marinomycin A based on a direct dimerization strategy. Angew. Chem. Int. Ed. 53, 8459–8462 (2014).
Singh, S. B., Phillips, J. W. & Wang, J. Highly sensitive target-based whole-cell antibacterial discovery strategy by antisense RNA silencing. Curr. Opin. Drug Discov. Dev. 10, 160–166 (2007).
Young, K. et al. Discovery of FabH/FabF inhibitors from natural products. Antimicrob. Agents Chemother. 50, 519–526 (2006).
Wang, J. et al. Platensimycin is a selective FabF inhibitor with potent antibiotic properties. Nature 441, 358–361 (2006).
Singh, S. B. et al. Isolation, structure, and absolute stereochemistry of platensimycin, a broad spectrum antibiotic discovered using an antisense differential sensitivity strategy. J. Am. Chem. Soc. 128, 11916–11920 (2006).
Häbich, D. & von Nussbaum, F. Platensimycin, a new antibiotic and ‘superbug challenger’ from nature. ChemMedChem 1, 951–954 (2006).
Nicolaou, K. C., Li, A. & Edmonds, D. J. Total synthesis of platensimycin. Angew. Chem. Int. Ed. 45, 7086–7090 (2006).
Nicolaou, K. C., Tang, Y. & Wang, J. Formal synthesis of (±)-platensimycin. Chem. Commun. 1922–1923 (2007).
Nicolaou, K. C., Pappo, D., Tsang, K. Y., Gibe, R. & Chen, D. Y. K. A chiral pool based synthesis of platensimycin. Angew. Chem. Int. Ed. 47, 944–946 (2008).
Nicolaou, K. C., Li, A., Ellery, S. P. & Edmonds, D. J. Rhodium-catalyzed asymmetric enyne cycloisomerization of terminal alkynes and formal total synthesis of (−)-platensimycin. Angew. Chem. Int. Ed. 48, 6293–6295 (2009).
Nicolaou, K. C., Li, A., Edmonds, D. J., Tria, G. S. & Ellery, S. P. Total synthesis of platensimycin and related natural products. J. Am. Chem. Soc. 131, 16905–16918 (2009).
Nicolaou, K. C., Edmonds, D. J., Li, A. & Tria, G. S. Asymmetric total syntheses of platensimycin. Angew. Chem. Int. Ed. 46, 3942–3945 (2007).
Nicolaou, K. C. et al. Total synthesis and antibacterial properties of carbaplatensimycin. J. Am. Chem. Soc. 129, 14850–14851 (2007).
Nicolaou, K. C., Lister, T., Denton, R. M., Montero, A. & Edmonds, D. J. Adamantaplatensimycin: a bioactive analogue of platensimycin. Angew. Chem. Int. Ed. 46, 4712–4714 (2007).
Heretsch, P. & Giannis, A. An efficient entry to amino-substituted resorcylic acid derivatives for the synthesis of platensimycin and analogues. Synthesis 2007, 2614–2616 (2007).
Kaliappan, K. P. & Ravikumar, V. An expedient enantioselective strategy for the oxatetracyclic core of platensimycin. Org. Lett. 9, 2417–2419 (2007).
Xing, S., Pan, W., Liu, C., Ren, J. & Wang, Z. Efficient construction of oxa- and aza-[n.2.1] skeletons: Lewis acid catalyzed intramolecular [3+2] cycloaddition of cyclopropane 1,1-diesters with carbonyls and imines. Angew. Chem. Int. Ed. 49, 3215–3218 (2010).
Wang, J. & Sintim, H. O. Dialkylamino-2,4-dihydroxybenzoic acids as easily synthesized analogues of platensimycin and platencin with comparable antibacterial properties. Chem. Eur. J. 17, 3352–3357 (2011).
Beaulieu, M.-A., Guérard, K. C., Maertens, G., Sabot, C. & Canesi, S. Oxidative Prins-pinacol tandem process mediated by a hypervalent iodine reagent: scope, limitations, and applications. J. Org. Chem. 76, 9460–9471 (2011).
Tiefenbacher, K. & Mulzer, J. Protecting-group-free formal synthesis of platensimycin. Angew. Chem. Int. Ed. 46, 8074–8075 (2007).
Zou, Y., Chen, C.-H., Taylor, C. D., Foxman, B. M. & Snider, B. B. Formal synthesis of (±)-platensimycin. Org. Lett. 9, 1825–1828 (2007).
Ghosh, A. K. & Xi, K. Enantioselective synthesis of (−)-platensimycin oxatetracyclic core by using an intramolecular Diels−Alder reaction. Org. Lett. 9, 4013–4016 (2007).
Lalic, G. & Corey, E. J. An effective enantioselective route to the platensimycin core. Org. Lett. 9, 4921–4923 (2007).
Li, P., Payette, J. N. & Yamamoto, H. Enantioselective route to platensimycin: an intramolecular Robinson annulation approach. J. Am. Chem. Soc. 129, 9534–9535 (2007).
Matsuo, J.-i., Takeuchi, K. & Ishibashi, H. Stereocontrolled formal synthesis of (±)-platensimycin. Org. Lett. 10, 4049–4052 (2008).
Kim, C. H., Jang, K. P., Choi, S. Y., Chung, Y. K. & Lee, E. A carbonyl ylide cycloaddition approach to platensimycin. Angew. Chem. Int. Ed. 47, 4009–4011 (2008).
McGrath, N. A., Bartlett, E. S., Sittihan, S. & Njardarson, J. T. A concise ring-expansion route to the compact core of platensimycin. Angew. Chem. Int. Ed. 48, 8543–8546 (2009).
Yun, S. Y., Zheng, J.-C. & Lee, D. Stereoelectronic effect for the selectivity in C−H insertion of alkylidene carbenes and its application to the synthesis of platensimycin. J. Am. Chem. Soc. 131, 8413–8415 (2009).
Ghosh, A. K. & Xi, K. Total synthesis of (−)-platensimycin, a novel antibacterial agent. J. Org. Chem. 74, 1163–1170 (2009).
Magnus, P., Rivera, H. & Lynch, V. Concise formal total synthesis of platensimycin mediated by a stereoselective autoxidation and hydroxyl group directed conjugative reduction. Org. Lett. 12, 5677–5679 (2010).
Eey, S. T. C. & Lear, M. J. A bismuth(III)-catalyzed Friedel−Crafts cyclization and stereocontrolled organocatalytic approach to (−)-platensimycin. Org. Lett. 12, 5510–5513 (2010).
Oblak, E. Z. & Wright, D. L. Highly substituted oxabicyclic derivatives from furan: synthesis of (±)-platensimycin. Org. Lett. 13, 2263–2265 (2011).
Hirai, S. & Nakada, M. Enantioselective divergent approaches to both (−)-platensimycin and (−)-platencin. Tetrahedron 67, 518–530 (2011).
Ueda, Y., Iwahashi, K., Iguchi, K. & Ito, H. Enantioselective synthesis of the tetracyclic core of platensimycin. Synthesis 2011, 1532–1536 (2011).
Horii, S., Torihata, M., Nagasawa, T. & Kuwahara, S. Stereoselective approach to the racemic oxatetracyclic core of platensimycin. J. Org. Chem. 78, 2798–2801 (2013).
Zhu, L., Han, Y., Du, G. & Lee, C.-S. A bifunctional Lewis acid induced cascade cyclization to the tricyclic core of ent-kaurenoids and its application to the formal synthesis of (±)-platensimycin. Org. Lett. 15, 524–527 (2013).
Zhu, L. et al. Formal syntheses of (±)-platensimycin and (±)-platencin via a dual-mode Lewis acid induced cascade cyclization approach. J. Org. Chem. 78, 7912–7929 (2013).
Eey, S. T. C. & Lear, M. J. Total synthesis of (−)-platensimycin by advancing oxocarbenium- and iminium-mediated catalytic methods. Chem. Eur. J. 20, 11556–11573 (2014).
Jiao, Z.-W. et al. Formal synthesis of (−)-platensimycin. Org. Chem. Front. 2, 913–916 (2015).
Wang, J. et al. Discovery of platencin, a dual FabF and FabH inhibitor with in vivo antibiotic properties. Proc. Natl. Acad. Sci. USA 104, 7612–7616 (2007).
Jayasuriya, H. et al. Isolation and structure of platencin: a FabH and FabF dual inhibitor with potent broad-spectrum antibiotic activity. Angew. Chem. Int. Ed. 46, 4684–4688 (2007).
Nicolaou, K. C., Tria, G. S. & Edmonds, D. J. Total synthesis of platencin. Angew. Chem. Int. Ed. 47, 1780–1783 (2008).
Nicolaou, K. C., Toh, Q.-Y. & Chen, D. Y. K. An expedient asymmetric synthesis of platencin. J. Am. Chem. Soc. 130, 11292–11293 (2008).
Nicolaou, K. C., Toh, Q.-Y. & Chen, D. Y. K. An expedient asymmetric synthesis of platencin. J. Am. Chem. Soc. 130, 14016 (2008).
Nicolaou, K. C., Tria, G. S., Edmonds, D. J. & Kar, M. Total syntheses of (±)-platencin and (−)-platencin. J. Am. Chem. Soc. 131, 15909–15917 (2009).
Austin, K. A. B., Banwell, M. G. & Willis, A. C. A formal total synthesis of platencin. Org. Lett. 10, 4465–4468 (2008).
Hayashida, J. & Rawal, V. H. Total synthesis of (±)-platencin. Angew. Chem. Int. Ed. 47, 4373–4376 (2008).
Tiefenbacher, K. & Mulzer, J. Short formal synthesis of (−)-platencin. Angew. Chem. Int. Ed. 47, 6199–6200 (2008).
Yun, S. Y., Zheng, J.-C. & Lee, D. Concise synthesis of the tricyclic core of platencin. Angew. Chem. Int. Ed. 47, 6201–6203 (2008).
Waalboer, D. C. J., Schaapman, M. C., van Delft, F. L. & Rutjes, F. P. J. T. High-pressure entry into platencin. Angew. Chem. Int. Ed. 47, 6576–6578 (2008).
Tiefenbacher, K. & Mulzer, J. A nine-step total synthesis of (−)-platencin. J. Org. Chem. 74, 2937–2941 (2009).
Varseev, G. N. & Maier, M. E. Formal total synthesis of platencin. Angew. Chem. Int. Ed. 48, 3685–3688 (2009).
Barykina, O. V., Rossi, K. L., Rybak, M. J. & Snider, B. B. Synthesis and antibacterial properties of (−)-nor-platencin. Org. Lett. 11, 5334–5337 (2009).
Ghosh, A. K. & Xi, K. A symmetry-based concise formal synthesis of platencin, a novel lead against “superbugs”. Angew. Chem. Int. Ed. 48, 5372–5375 (2009).
Singh, V., Sahu, B. C., Bansal, V. & Mobin, S. M. Intramolecular cycloaddition in 6,6-spiroepoxycyclohexa-2,4-dienone: simple aromatics to (±)-platencin. Org. Biomol. Chem. 8, 4472–4481 (2010).
Li, P. & Yamamoto, H. Formal synthesis of platencin. Chem. Commun. 46, 6294–6295 (2010).
Tiefenbacher, K., Gollner, A. & Mulzer, J. Syntheses and antibacterial properties of iso-platencin, Cl-iso-platencin and Cl-platencin: identification of a new lead structure. Chem. Eur. J. 16, 9616–9622 (2010).
Waalboer, D. C. J., Leenders, S. H. A. M., Schülin-Casonato, T., van Delft, F. L. & Rutjes, F. P. J. T. Total synthesis and antibiotic activity of dehydrohomoplatencin. Chem. Eur. J. 16, 11233–11236 (2010).
Hirai, S. & Nakada, M. An enantioselective approach to (−)-platencin via catalytic asymmetric intramolecular cyclopropanation. Tetrahedron Lett. 51, 5076–5079 (2010).
Leung, G. Y. C. et al. Total synthesis and biological evaluation of the Fab-inhibitory antibiotic platencin and analogues thereof. Eur. J. Org. Chem. 2011, 183–196 (2011).
Yoshimitsu, T., Nojima, S., Hashimoto, M. & Tanaka, T. Total synthesis of (±)-platencin. Org. Lett. 13, 3698–3701 (2011).
Palanichamy, K., Subrahmanyam, A. V. & Kaliappan, K. P. A radical cyclization approach to the formal total syntheses of platencin. Org. Biomol. Chem. 9, 7877–7886 (2011).
Yadav, J. S., Goreti, R., Pabbaraja, S. & Sridhar, B. Short route to platencin. Org. Lett. 15, 3782–3785 (2013).
Chang, E. L., Schwartz, B. D., Draffan, A. G., Banwell, M. G. & Willis, A. C. A chemoenzymatic and fully stereocontrolled total synthesis of the antibacterial natural product (−)-platencin. Chem. Asian J. 10, 427–439 (2015).
Wang, J. et al. A concise formal synthesis of platencin. Org. Chem. Front. 2, 674–676 (2015).
Muhammad, R. N., Draffan, A. G., Banwell, M. G. & Willis, A. C. A second-generation chemoenzymatic total synthesis of platencin. Synlett 27, 61–66 (2016).
Sedmera, P., Podojil, M., Vokoun, J., Betina, V. & Nemec, P. 2,2′-Dimethoxy-4a,4a′-dehydrorugulosin (rugulin), a minor metabolite from Penicillium rugulosum. Folia Microbiol. 23, 64–67 (1978).
Betina, V. & Nemec, P. Spôsob prípravy antibiotika rugulínu a metabolitov skyrínu a rugulozínu z mikroorganizmu Penicillium rugulosum. Czechoslovakian patent No. 187049 (1978).
Nicolaou, K. C., Papageorgiou, C. D., Piper, J. L. & Chadha, R. K. The cytoskyrin cascade: a facile entry into cytoskyrin A, deoxyrubroskyrin, rugulin, skyrin, and flavoskyrin model systems. Angew. Chem. Int. Ed. 44, 5846–5851 (2005).
Nicolaou, K. C., Lim, Y. H., Papageorgiou, C. D. & Piper, J. L. Total synthesis of (+)-rugulosin and (+)-2,2′-epi-cytoskyrin A through cascade reactions. Angew. Chem. Int. Ed. 44, 7917–7921 (2005).
Nicolaou, K. C., Lim, Y. H., Piper, J. L. & Papageorgiou, C. D. Total syntheses of 2,2′-epi-cytoskyrin A, rugulosin, and the alleged structure of rugulin. J. Am. Chem. Soc. 129, 4001–4013 (2007).
Brady, S. F., Singh, M. P., Janso, J. E. & Clardy, J. Cytoskyrins A and B, new BIA active bisanthraquinones isolated from an endophytic fungus. Org. Lett. 2, 4047–4049 (2000).
Agusta, A., Ohashi, K. & Shibuya, H. Bisanthraquinone metabolites produced by the endophytic fungus Diaporthe sp. Chem. Pharm. Bull. 54, 579–582 (2006).
Kushida, H. et al (Banyu Pharmaceuticals Co., Ltd.). Antitumor substance BE-43472 JP08143569 (1996).
Socha, A. M., Garcia, D., Sheffer, R. & Rowley, D. C. Antibiotic bisanthraquinones produced by a streptomycete isolated from a cyanobacterium associated with Ecteinascidia turbinata. J. Nat. Prod. 69, 1070–1073 (2006).
Socha, A. M., LaPlante, K. L. & Rowley, D. C. New bisanthraquinone antibiotics and semi-synthetic derivatives with potent activity against clinical Staphylococcus aureus and Enterococcus faecium isolates. Biorg. Med. Chem. 14, 8446–8454 (2006).
Nicolaou, K. C., Lim, Y. H. & Becker, J. Total synthesis and absolute configuration of the bisanthraquinone antibiotic BE-43472B. Angew. Chem. Int. Ed. 48, 3444–3448 (2009).
Nicolaou, K. C. et al. Total synthesis and biological evaluation of (+)- and (−)-bisanthraquinone antibiotic BE-43472B and related compounds. J. Am. Chem. Soc. 131, 14812–14826 (2009).
Hayden, A. E. et al. Origins of regioselectivity of Diels−Alder reactions for the synthesis of bisanthraquinone antibiotic BE-43472B. J. Org. Chem. 75, 922–928 (2010).
Yamashita, Y., Hirano, Y., Takada, A., Takikawa, H. & Suzuki, K. Total synthesis of the antibiotic BE-43472B. Angew. Chem. Int. Ed. 52, 6658–6661 (2013).
Isaka, M. et al. Hirsutellones A–E, antimycobacterial alkaloids from the insect pathogenic fungus Hirsutella nivea BCC 2594. Tetrahedron 61, 5577–5583 (2005).
Nicolaou, K. C., Sarlah, D., Wu, T. R. & Zhan, W. Total synthesis of hirsutellone B. Angew. Chem. Int. Ed. 48, 6870–6874 (2009).
Nicolaou, K. C., Sun, Y.-P., Sarlah, D., Zhan, W. & Wu, T. R. Bioinspired synthesis of hirsutellones A, B, and C. Org. Lett. 13, 5708–5710 (2011).
Tilley, S. D., Reber, K. P. & Sorensen, E. J. A rapid, asymmetric synthesis of the decahydrofluorene core of the hirsutellones. Org. Lett. 11, 701–703 (2009).
Huang, M., Huang, C. & Liu, B. Studies toward the total synthesis of the hirsutellones. Tetrahedron Lett. 50, 2797–2800 (2009).
Halvorsen, G. T. & Roush, W. R. Stereoselective synthesis of the decahydrofluorene core of the hirsutellones. Tetrahedron Lett. 52, 2072–2075 (2011).
Song, L., Huang, C., Huang, M. & Liu, B. Toward the synthesis of hirsutellone B via an intramolecular Diels–Alder/ketene-trapping strategy. Tetrahedron 71, 3603–3608 (2015).
Reber, K. P., Tilley, S. D., Carson, C. A. & Sorensen, E. J. Toward a synthesis of hirsutellone B by the concept of double cyclization. J. Org. Chem. 78, 9584–9607 (2013).
Uchiro, H., Kato, R., Arai, Y., Hasegawa, M. & Kobayakawa, Y. Total synthesis of hirsutellone B via Ullmann-type direct 13-membered macrocyclization. Org. Lett. 13, 6268–6271 (2011).
Waring, P., Eichner, R. D. & Müllbacher, A. The chemistry and biology of the immunomodulating agent gliotoxin and related epipolythiodioxopiperazines. Med. Res. Rev. 8, 499–524 (1988).
Kamei, H. et al. Piperafizines A and B, potentiators of cytotoxicity of vincristine. J. Antibiot. 43, 1018–1020 (1990).
Waring, P. & Beaver, J. Gliotoxin and related epipolythiodioxopiperazines. Gen. Pharmacol. 27, 1311–1316 (1996).
Bull, S. D., Davies, S. G., Parkin, R. M. & Sanchez-Sancho, F. The biosynthetic origin of diketopiperazines derived from D-proline. J. Chem. Soc., Perkin Trans. 1 2313–2320 (1998).
Greiner, D., Bonaldi, T., Eskeland, R., Roemer, E. & Imhof, A. Identification of a specific inhibitor of the histone methyltransferase SU(VAR)3-9. Nat. Chem. Biol. 1, 143–145 (2005).
Gardiner, D. M., Waring, P. & Howlett, B. J. The epipolythiodioxopiperazine (ETP) class of fungal toxins: distribution, mode of action, functions and biosynthesis. Microbiology 151, 1021–1032 (2005).
Řezanka, T., Sobotka, M., Spížek, J. & Sigler, K. Pharmacologically active sulfur-containing compounds. Antiinfect. Agents Med. Chem. 5, 187–224 (2006).
Isham, C. R. et al. Chaetocin: a promising new antimyeloma agent with in vitro and in vivo activity mediated via imposition of oxidative stress. Blood 109, 2579–2588 (2007).
Ding, G. et al. Pestalazines and pestalamides, bioactive metabolites from the plant pathogenic fungus Pestalotiopsis theae. J. Nat. Prod. 71, 1861–1865 (2008).
Huang, R., Zhou, X., Xu, T., Yang, X. & Liu, Y. Diketopiperazines from marine organisms. Chem. Biodivers. 7, 2809–2829 (2010).
Cornacchia, C. et al. 2,5-Diketopiperazines as neuroprotective agents. Mini-Rev. Med. Chem. 12, 2–12 (2012).
Jiang, C.-S., Müller, W. E. G., Schröder, H. C. & Guo, Y.-W. Disulfide- and multisulfide-containing metabolites from marine organisms. Chem. Rev. 112, 2179–2207 (2012).
Borthwick, A. D. 2,5-Diketopiperazines: synthesis, reactions, medicinal chemistry, and bioactive natural products. Chem. Rev. 112, 3641–3716 (2012).
Nicolaou, K. C. et al. Synthesis and biological evaluation of epidithio-, epitetrathio-, and bis-(methylthio)diketopiperazines: synthetic methodology, enantioselective total synthesis of epicoccin G, 8,8′-epi-ent-rostratin B, gliotoxin, gliotoxin G, emethallicin E, and haematocin and discovery of new antiviral and antimalarial agents. J. Am. Chem. Soc. 134, 17320–17332 (2012).
Guo, H. et al. Diketopiperazines from the Cordyceps-colonizing fungus Epicoccum nigrum. J. Nat. Prod. 72, 2115–2119 (2009).
Wang, J.-M. et al. Thiodiketopiperazines produced by the endophytic fungus Epicoccum nigrum. J. Nat. Prod. 73, 1240–1249 (2010).
Nicolaou, K. C., Totokotsopoulos, S., Giguère, D., Sun, Y.-P. & Sarlah, D. Total synthesis of epicoccin G. J. Am. Chem. Soc. 133, 8150–8153 (2011).
Tan, R. X., Jensen, P. R., Williams, P. G. & Fenical, W. Isolation and structure assignments of rostratins A−D, cytotoxic disulfides produced by the marine-derived fungus Exserohilum rostratum. J. Nat. Prod. 67, 1374–1382 (2004).
Kawahara, N., Nozawa, K., Yamazaki, M., Nakajima, S. & Kawai, K.-i. Novel epidithiodioxopiperazines, emethallicins E and F, from Emericella heterothallica. Heterocycles 30, 507–515 (1990).
Trown, P. W. Antiviral activity of N,N'-dimethyl-epidithiapiperazinedione, a synthetic compound related to the gliotoxins, LL-S88α and β, chetomin and the sporidesmins. Biochem. Biophys. Res. Commun. 33, 402–407 (1968).
Shimazaki, N., Shima, I., Hemmi, K., Tsurumi, Y. & Hashimoto, M. Diketopiperazine derivatives, a new series of platelet-activating factor inhibitors. Chem. Pharm. Bull. 35, 3527–3530 (1987).
Poisel, H. & Schmidt, U. Synthesis of 2,5-piperazinediones having sulfur-containing bridges between C-3 and C-6. Angew. Chem. Int. Ed. Engl. 10, 130–131 (1971).
Poisel, H. & Schmidt, U. Syntheseversuche in der Reihe der 3.6-Epidithio-2.5-dioxo-piperazin-Antibiotika Gliotoxin, Sporidesmin, Aranotin und Chaetocin, II. Chem. Ber. 104, 1714–1721 (1971).
Öhler, E., Poisel, H., Tataruch, F. & Schmidt, U. Syntheseversuche in der Reihe der 3.6-Epidithio-2.5-dioxo-piperazin-Antibiotika Gliotoxin, Sporidesmin, Aranotin und Chaetocin, IV. Synthese des Epidithio-L-prolyl-L-prolinanhydrids. Chem. Ber 105, 635–641 (1972).
Kishi, Y., Fukuyama, T. & Nakatsuka, S. New method for the synthesis of epidithiodiketopiperazines. J. Am. Chem. Soc. 95, 6490–6492 (1973).
Yoshimura, J., Nakamura, H. & Matsunari, K. A new synthesis of 3,6-dialkyl-1,4-dimethyl-3,6-epithio- and -3,6-epidithio-2,5-piperazinediones. Bull. Chem. Soc. Jpn. 48, 605–609 (1975).
Overman, L. E. & Sato, T. Construction of epidithiodioxopiperazines by directed oxidation of hydroxyproline-derived dioxopiperazines. Org. Lett. 9, 5267–5270 (2007).
Scherer, O. & Schmidt, M. Abbau von Schwefel mit Natrium-bis-(trimethylsilyl)-amid. Naturwissenschaften 50, 302 (1963).
Schmidt, M. & Potschka, V. Über die Reaktion von Natriumphenylazetylid mit elementarem Schwefel. Naturwissenschaften 50, 302 (1963).
Scherer, O. & Schmidt, M. Zur Frage der Existenz eines silylsubstituierten Amino-rhodans. Z. Naturforsch. B 18, 415–416 (1963).
Siivari, J., Maaninen, A., Haapaniemi, E., Laitinen Risto, S. & Chivers, T. Formation and identification of bis[bis(trimethylsilyl)amino]triand tetrachalcogenides. Z. Naturforsch. B 50, 1575–1582 (1995).
Iwasa, E. et al. Total synthesis of (+)-chaetocin and its analogues: their histone methyltransferase G9a inhibitory activity. J. Am. Chem. Soc. 132, 4078–4079 (2010).
Kim, J. & Movassaghi, M. General approach to epipolythiodiketopiperazine alkaloids: total synthesis of (+)-chaetocins A and C and (+)-12,12′-dideoxychetracin A. J. Am. Chem. Soc. 132, 14376–14378 (2010).
Nicolaou, K. C., Giguère, D., Totokotsopoulos, S. & Sun, Y.-P. A practical sulfenylation of 2,5-diketopiperazines. Angew. Chem. Int. Ed. 51, 728–732 (2012).
Gross, U., Nieger, M. & Bräse, S. A unified strategy targeting the thiodiketopiperazine mycotoxins exserohilone, gliotoxin, the epicoccins, the epicorazines, rostratin A and aranotin. Chem. Eur. J 16, 11624–11631 (2010).
Lee, J . Part I. Synthetic investigations of heterocyclic natural and unnatural compounds Part II. New approach to latent fingerprint detection on paper. PhD thesis, Univ. Pennsylvania (2014).
Liu, Z. & Rainier, J. D. Ring-opening/ring-closing metathesis (RORCM) reactions of 7-azanorbornene derivatives. An entry into perhydroindolines. Org. Lett. 8, 459–462 (2006).
Liu, Z . High regioselective ring-opening/cross metathesis of norbornene derivatives and ring-opening/ring-closing metathesis and their applications towards total synthesis of rostratins and synthesis of acid sensing ion channel inhibitors. PhD thesis, Univ. Utah (2007).
Ruff, B. M. et al. A combined vinylogous Mannich/Diels–Alder approach for the stereoselective synthesis of highly functionalized hexahydroindoles. Eur. J. Org. Chem 2011, 6558–6566 (2011).
Zhong, S., Sauter, P. F., Nieger, M. & Bräse, S. Stereoselective synthesis of highly functionalized hydroindoles as building blocks for rostratins B–D and synthesis of the pentacyclic core of rostratin C. Chem. Eur. J. 21, 11219–11225 (2015).
Wang, H . Enantioselective total synthesis of diketopiperazinecontaining natural products: (–)-lansai B, (+)-nocardioazines A and B, and (–)-acetylapoaranotin. PhD thesis, California Institute of Technology (2015).
Zheng, C.-J., Yu, H.-E., Kim, E.-H. & Kim, W.-G. Viridicatumtoxin B, a new anti-MRSA agent from Penicillium sp. FR11. J. Antibiot. 61, 633–637 (2008).
Inokoshi, J. et al. Spirohexalines, new inhibitors of bacterial undecaprenyl pyrophosphate synthase, produced by Penicillium brasilianum FKI-3368. J. Antibiot. 66, 37–41 (2013).
Koyama, N., Inokoshi, J. & Tomoda, H. Anti-infectious agents against MRSA. Molecules 18, 204–224 (2013).
Nicolaou, K. C., Liu, G., Beabout, K., McCurry, M. D. & Shamoo, Y. Asymmetric alkylation of anthrones, enantioselective total synthesis of (−)- and (+)-viridicatumtoxins B and analogues thereof: absolute configuration and potent antibacterial agents. J. Am. Chem. Soc. 139, 3736–3746 (2017).
Sugie, Y. et al. New pyrrolizidinone antibiotics CJ-16,264 and CJ-16,367. J. Antibiot. 54, 917–925 (2001).
Nicolaou, K. C. et al. Total synthesis and structural revision of antibiotic CJ-16,264. Angew. Chem. Int. Ed. 54, 9203–9208 (2015).
Lambert, T. H. & Danishefsky, S. J. Total synthesis of UCS1025A. J. Am. Chem. Soc. 128, 426–427 (2006).
World Health Organization WHO publishes list of bacteria for which new antibiotics are urgently needed (2017) Available at: http://www.who.int/mediacentre/news/releases/2017/bacteria-antibiotics-needed/en/.
Acknowledgements
KCN thanks his students and post-doctoral fellows who contributed decisively to the achievements described in this article and to express his unlimited gratitude to his wife Georgette, daughter Colette, sons Alex, Christopher and Paul and grandchildren Nicolas, Gigi, Eleni, Ava Alexandra and Kyri for their unconditional love and support. We are grateful to the National Institutes of Health (USA), the Cancer Prevention & Research Institute of Texas (CPRIT), and The Welch Foundation (grant C-1819) for their generous funding of our research.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Rights and permissions
About this article
Cite this article
Nicolaou, K., Rigol, S. A brief history of antibiotics and select advances in their synthesis. J Antibiot 71, 153–184 (2018). https://doi.org/10.1038/ja.2017.62
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ja.2017.62
This article is cited by
-
Characterization, antibacterial, and cytotoxic activities of silver nanoparticles using the whole biofilm layer as a macromolecule in biosynthesis
Scientific Reports (2024)
-
Evolutionary History of Periodontitis and the Oral Microbiota—Lessons for the Future
Current Oral Health Reports (2024)
-
Cemeteries and graveyards as potential reservoirs of antibiotic resistance genes and bacteria: a review
Environmental Chemistry Letters (2024)
-
Avian reservoirs or vectors? Unravelling the relationship between waterbirds and E. coli contamination
Ornithology Research (2024)
-
A decennial study of the trend of antibiotic studies in China
Environmental Science and Pollution Research (2023)