Abstract
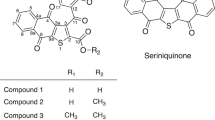
Two new rearranged linear angucycline glycosides, designated grincamycins G and H (1 and 2), together with three known congers P-1894B (vineomycin A1, 3), saquayamycin B (4) and vineomycin B2 (5), were obtained from marine-derived actinomycete Streptomyces lusitanus SCSIO LR32. The structures of 1 and 2 were elucidated by MS, 1D and 2D NMR techniques. Compounds 2–5 showed significant inhibitory effect on Jurkat T-cell proliferation with IC50 values of 3.0, 0.011, 0.037 and 0.3 μM, respectively.
Similar content being viewed by others
Introduction
Angucycline is a large group of secondary metabolites originating from the fermentation extract of different actinomycetes, mostly from the species of Streptomyces.1 Angucycline compounds possess various biological and pharmacological properties, including cytotoxicity and antitumor, antibacterial and antiviral activities, as well as enzyme-inhibition activities.1, 2 Structurally, classic angucyclines contain an angular benz[a]anthracene scaffold and non-classic congers with rearranged linear tetracyclic or tricyclic core units, and most of angucyclines exist as glycosides in nature. The observed carbohydrates in angucycline glycoside usually are deoxy sugars, such as D-olivose, L- or D-rhodinose, L-aculose, L-cinerulose A, L-cinerulose B, D-kerriose and L-amicetose.1, 2, 3 Moreover, the typical O-glycoside and C-glycoside with different types of sugars or different numbers of sugar chains have brought broad chemical diversities to natural angucycline compounds.
During our efforts to screen for novel antitumor and antimicrobial antibiotics, we reported non-classic angucycline glycosides designated as grincamycins (GCNs) B–E from deep sea-derived actinomycete Streptomyces lusitanus SCSIO LR32.4 Subsequently, the biosynthetic gene cluster of GCN cloned from S. lusitanus SCSIO LR32 was heterologously expressed in Streptomyces coelicolor M512, resulting in the isolation of P-1894B as a predominant product.5 The structural difference between P-1894B and GCN is that the terminal sugars L-cinerulose A in GCN were replaced by L-aculose units in P-1894B. In addition, the LC-MS analyses of metabolite derived from the strain SCSIO LR32 cultivated in a modified AM2 medium have detected two peaks with the molecular weight of 934.4, which were 4 units fewer than that of grincamycin and grincamycin B (molecular weight 938.4), which presumably could be P-1894B and vineomycin B2. Based on these preliminary data, strain SCSIO LR32 was further fermented on an 80 l scale. Subsequent purification of culture extract resulted in identification of five angucycline glycosides (1–5) containing L-aculose as terminal on the sugar chain (Figure 1). Hererin we report the fermentation, isolation, structure elucidation and cytotoxic activities of these compounds.
Results
Structure elucidation
Compound 1 was isolated as a yellowish powder. (–)HR-ESI-MS of 1 revealed a quasimolecular ion peak at 801.2776, in combination with 1H and 13C NMR data, establishing the molecular formula as C43H46O15. The UV spectrum of 1 showed characteristic absorption bands at 214, 243, 265 and 443 nm. The 13C NMR spectrum disclosed five carbonyls at δC 209.5, 198.2, 197.0, 189.9 and 189.8, 14 aromatic carbons in the downfield sp2 region and 24 aliphatic carbons in the upfield area (Table 1). The 1H NMR spectroscopic signals at δH 13.10 (2H, br s, 1-OH and 5-OH) suggested two hydrogen-bonded hydroxy groups. A pair of ortho-coupled aromatic protons at δH 8.02 (d, 8.0 Hz, H-8) and 7.90 (d, 8.0 Hz, H-7) and a single aromatic proton at δH 8.34 (s, H-12) were observed, which were almost consistent with those in grincamycin E4; inferred 1 had a linear tetracyclic anthraquinone aglycone. The HMBC correlations of H-7/C-6, C-9, C-10a, H-8/C-9, C-10, C-6a, H-12/C-1, C-4a, C-5a, C-11 and of H-2/C-1, C-4 confirmed the existence of the linear tetracyclic anthraquinone unit (Figure 2). The Me-13 was attached at C-3 based on the HMBC correlations of H3-13/C-2, C-3, C-4. Additionally, four doublet methyl signals at δH 1.34, δC 18.4 (CH3-6′), δH 1.28, δC 17.4 (CH3-6′′), δH 0.64, δC 18.4 (CH3-6′′′) and δH 1.21, δC 16.4 (CH3-6′′′′), as well as four anomeric methine signals at δH 5.07 (d, 9.5), δC 73.3 (CH-1′), δH 5.26 (d, 3.0), δC 93.2 (CH-1′′), δH 5.17 (br s), δC 91.9 (CH-1′′′) and δH 5.13 (d, 3.4), δC 96.9 (CH-1′′′′), indicated the presence of four deoxysugars in 1. Detailed comparison showed that the 1H and 13C NMR spectroscopic data of the sugar units were similar to those of grincamycin D,4 except that the signals of two methines in cinerulose A disappeared, while a set of signals ascribed to a double bond at δH 6.93 (10.3, 3.4 Hz), δC 146.0 (CH-2′′′′), δH 5.97 (10.0 Hz) and δC 128.1 (CH-3′′′′) was observed. Moreover, the 13C NMR resonance of the carbonyl in cinerulose A was shifted upfield from δC 210.9 in grincamycin D to δC 198.2 in 1, indicating that the cinerulose A unit in grincamycin D was replaced by an aculose in 1. The HMBC correlations of H-1′′′/C-4′′′′ confirmed the existence of an aculose-(1→4)-rhodinosyl unit. The relative configurations of the anomeric carbons were defined as β for olivose and α for cinerulose B, rhodinose and aculose on the basis of the large coupling constant (J=9.5 Hz) of H-1′ and the small coupling constants of H-1′′, H-1′′′ and H-1′′′′, respectively. Being similar to the conversion of grincamycin into grincamycin E under UV irradiation,4 saquayamycin B (4) could be transformed to 1 in the same manner (Supplemantary Figure S15). Therefore, the absolute configuration of C-3 was established to be R, and the sugars were elucidated as β-D-olivose, α-L-cinerulose B, α-L-rhodinose and α-L-aculose, which were identical with those of saquayamycin B (4).6 Compound 1 was named grincamycin G.
Compound 2 was isolated as a dark-red powder. Its molecular formula was determined to be C37H38O12 by analyses of (−)HR-ESI-MS, as well as 1H and 13C NMR spectra, indicating 19 degrees of unsaturation. Detailed analyses of the 1H and 13C NMR spectroscopic data showed that the structure of 2 was similar to those of quanolirones and galtamycinone,7, 8, 9 which contained 1,5,10-trihydroxy-naphthacenequinone scaffold. The HMBC spectrum of 2 confirmed the position of a pair of ortho-coupled protons at δH 7.79 (d, 7.9 Hz, H-7) and 7.85 (d, 7.9 Hz, H-8) and three singlet protons at δH 6.97 (H-2), 7.55 (H-4) and 8.45 (H-12) (Figure 2). A methyl group (δH 2.43, δC 21.8, Me-13) was attached at C-3 based on the HMBC correlations from the methyl protons to C-2 (δC 116.1), C-3 (δC 141.6) and C-4 (δC 114.2). The 1H and 13C NMR spectroscopic data also revealed three anomeric carbon signals at δH 4.80 (d, 11.9) and δC 70.5 (CH-1′), δH 4.94 (br s) and δC 97.3 (CH-1′′), δH 5.35 (d, 3.1) and δC 94.3 (CH-1′′′), three methyl groups at δH 1.30 and δC 18.3 (Me-6′), δH 1.10 and δC 16.7 (Me-6′′), δH 1.24 and δC 14.9 (Me-6′′′), as well as (Z)-α, β-unsaturated carbonyl at δH 7.08 (10.2, 3.1 Hz) and δC 144.7 (=CH-2′′′), δH 6.08 (10.2 Hz) and δC 126.1 (=CH-3′′′) and δC 196.6 (C-4′′′), indicating the presence of a trisaccharide comprising three deoxysugars. Detailed analyses of the HMBC associations (Figure 2) revealed an α-aculose-(1→4)-α-rhodinosyl-(1→4)-β-olivosyl linked at C-9 of the naphthacenequinone to form a C-glycoside (Figure 2).10 The absolute configurations of the three deoxysugars were presumed to be identical with those in vineomycin B2 (5), which were α-L-aculose, α-L-rhodinose and β-D-olivose based on the 13C NMR data for the trisaccharide in 2 being almost consistent with those in vineomycin B2 (5) (Supplementary Table S1), as well as that 2 and 5 shared a biosynthesis pathway. Compound 2 was designated grincamycin H.
The known compounds, P-1894B (vineomycin A1, 3),11, 12 saquayamycin B (4)6, 13 and vineomycin B2 (5)10, 14 were identified on the basis of comparisons of MS, 1H and 13C NMR spectroscopic data with the compounds previously reported.
Cytotoxic activities
In the in vitro cytotoxic assay, the known compounds P-1894B (vineomycin A1, 3) and saquayamycin B (4) exhibited significant inhibition to Jurkat T-cell proliferation with IC50 values of 11 and 37 nM, respectively, which were more or nearly equally efficient than that of the positive control, doxorubicin. Vineomycin B2 (5) displayed powerful inhibitory effect to Jurkat T cells with an IC50 value of 0.3 μM. The new compound 2 showed cytotoxicity against Jurkat T cells with an IC50 value of 3.0 μM, while new compound 1 showed no cytotoxic activity at the concentration of 20 μM (Table 2).
Discussion
Following the evidence of heterologous expression of GCN gene cluster, five angucycline glycosides containing L-aculose at the end of sugar chains, which was different from the terminal sugar unit L-cinerulose A in our previously reported GCNs, were identified from marine-derived actinomycete S. lusitanus SCSIO LR32.4 The core skeleton of the new compounds 1 and 2 is the rearranged galtamycin-type non-classic aglycone of angucycline.1 However, compounds 1 and 2 showed lower cytotoxicity on Jurkat T cells than the known compounds 3 and 4, which possess a classic aquayamycin-type aglycone.1 Compound 5 has a tricyclic non-classic aglycone,1 which exhibited moderate cytotoxicity among the five compounds. These results indicated that the chemical and biological diversity of angucyclines derived not only from the sugar unit but also from the aglycone moiety.
Materials and methods
General experimental procedures
Column chromatography (CC) was performed by using silica gel (100–200 mesh; Jiangpeng Silica Gel Development, Inc., Yantai, China) or Sephadex LH-20 (40–70 μm; GE Healthcare, Uppsala, Sweden). TLC was conducted with precoated glass plates (0.1–0.2 mm; silica gel GF254, 10–40 nm, Jiangpeng). HPLC analyses were performed with a 1260 infinity system (Agilent, Santa Clara, CA, USA) using a Phenomenex Prodigy ODS (2) column (150 × 4.6 mm, 5 μm; Torrance, CA, USA). Semipreparative HPLC was performed with a Primaide 1110 solvent delivery module equipped with a 1430 photodiode array detector (Hitachi, Japan), using a YMC-Pack ODS-A column (250 × 10 mm, 5 μm; Kyoto, Japan). UV spectra were recorded on a U-2910 spectrometer (Shimadzu, Tokyo, Japan); IR spectra were obtained on an IRAffinity-1 spectrophotometer (Shimadzu). High-resolution mass spectral data were obtained on a MaXis Q-TOF mass spectrometer (Bruker, Rleigh, NC, USA). Optical rotations were recorded with an MCP-500 polarimeter (Anton Paar, Graz, Austria). NMR spectra were recorded on an Avance 500 spectrometer (Bruker) at 500 MHz for 1H nuclei and 125 MHz for 13C nuclei. Chemical shifts (δ) were given with reference to tetramethyl silane (TMS) or solvent residue signal. Coupling constants (J) were given in Hz.
Fermentation
The isolation and characterization of strain SCSIO LR32 have been described previously.4 A portion of SCSIO LR32 spore suspension containing about 2.16 × 107 spores was inoculated into 100 ml modified AM2 medium (0.5% soluble starch, 0.5% yeast extract, 0.2% peptone, 2.0% glucose, 0.05% KH2PO4, 0.05% MgSO4·7H2O, 0.4% NaCl, 0.2% CaCO3, 3.0% sea salt, pH 7.2 before sterilization) in a 250 ml Erlenmeyer flask and incubated at 30 °C for 48 h on a rotary shaker at 200 r.p.m. Then 600 ml of the seed culture from six flasks was transferred into 8 l modified AM2 medium in a 15 l fermentor, with the agitation speed of 200 r.p.m. and the aeration rate of 5 l min−1. After 24 h cultivation, the entire 8 l seed culture was further transferred into a 150 l fermentor containing 80 l production medium (0.5% corn flour, 2% soluble starch, 0.5% yeast extract, 1% malt extract, 1% glucose, 1% maltose, 0.2% CaCO3, 3.0% sea salt, pH 7.2 before sterilization) and cultivated at 30 °C. The dissolved oxygen of broth was maintained above 50% by adjusting the agitation speed and aeration rate. Broth sample was withdrawn every 24 h and centrifuged; the resulting supernatant was applied for the residual sugar measurement, and the mycelium pellets were extracted with methanol for further HPLC analysis. The fermentation was terminated after 7 days of cultivation. The entire 80 l broth was harvested and centrifuged to yield a mycelial cake and a liquid broth.
Extraction and isolation
Each 500 g mycelial cake was extracted with 1 l acetone three times. The acetone layers were combined and vacuum dried to yield a residue. The residue was subjected to a SiO2 CC using gradient elution with CHCl3 and MeOH mixtures (100:0, 99:1, 97:3, 95:5, 90:10, 80:20 and 50:50, v/v) to give seven fractions (Fr.A1∼Fr.A7). Fr.A1 and Fr.A2 were combined after HPLC analysis and purified by SiO2 CC eluting with petroleum ether and EtOAc mixtures (100:0, 90:10, 80:20, 70:30, 60:40, 40:60, 20:80, 0:100, v/v) to give eight fractions (Fr.B1∼Fr.B8). Fr.B3 was purified by semipreparative HPLC with an ODS column eluted with CH3CN/H2O (30:70 to 100:0 over 28 min and then held for 7 min 2.5 ml min−1) to afford 2 (8 mg) at tR 32 min. Fr.B4 was subjected to Sephadex LH-20 CC eluted with CHCl3/MeOH (1:1) to obtain 1 (6 mg) and 4 (12 mg). Fr.B5 and Fr.B6 were combined and further purified by semipreparative HPLC with an ODS column using the same elution procedure to afford 3 (220 mg) at tR 26 min and 5 (7 mg) at tR 30 min.
Grincamycin G ( 1): Yellowish powder; [α]20D=+53 (c 0.09, acetone); UV (MeOH) λmax (log ɛ) 214 (4.32), 243 (4.26), 265 (4.30), 443 (3.86); 1H NMR (500 MHz, acetone-d6) and 13C NMR (125 MHz, acetone-d6) data are listed in Table 1; (–)HR-ESI-MS m/z 801.2776 ([M–H]−, calcd for C43H45O15, 801.2764).
Grincamycin G ( 2): Dark-red powder; [α]20D=+65 (c 0.10, acetone); UV (MeOH) λmax (log ɛ) 230 (4.39), 258 (4.15), 294 (3.68), 492 (3.79); 1H NMR (500 MHz, DMSO-d6/CDCl3) and 13C NMR (125 MHz, DMSO-d6/CDCl3) data are listed in Table 1; (–)HR-ESI-MS m/z 673.2311 ([M–H]–, calcd for C37H37O12, 673.2291).
Cytotoxic activity assay
Proliferation-inhibition assay against Jurkat T cells was conducted according to the previously reported method.15 Jurkat T cells (2 × 104 cells per well) were maintained in RPMI1640 medium with 10% fetal bovine serum at 37 °C, in 5% CO2. For cytotoxic activity assay, Jurkat T cells were seeded in a 96-well plate and treated with compounds as indicated and then incubated at 37 °C for 72 h. The viability of cells were measured by adding alamarblue dye (1:10) to each well, and the plate was incubated at 37 °C for 4 h before fluorescence intensity was obtained by Flexstation3 (Molecular Devices, Sunnyvale, CA, USA). All data were processed with Prism6 (La Jolla, CA, USA).
References
Rohr, J. & Thiericke, R. Angucycline group antibiotics. Nat. Prod. Rep. 9, 103–137 (1992).
Kharel, M. K. et al. Angucyclines: biosynthesis, mode-of-action, new natural products, and synthesis. Nat. Prod. Rep. 29, 264–325 (2012).
Krohn, K. & Rohr, J. Angucyclines: total syntheses, new structures, and biosynthetic studies of an emerging new class of antibiotics. Top. Curr. Chem. 188, 127–195 (1997).
Huang, H. et al. Cytotoxic angucycline class glycosides from the deep sea actinomycete Streptomyces lusitanus SCSIO LR32. J. Nat. Prod. 75, 202–208 (2012).
Zhang, Y. et al. Identification of the grincamycin gene cluster unveils divergent roles for GcnQ in different hosts, tailoring the L-rhodinose moiety. Org. Lett. 15, 3254–3257 (2013).
Henkel, T. & Zeeck, A. Derivatives of saquayamycins A and B. Regio- and diastereoselective addition of alcohols to the L-aculose moiety. J. Antibiot. 43, 830–837 (1990).
Qian-Cutrone, F. et al. Quanolirones I and II, two new human cytomegalovirus protease inhibitors produced by Streptomyces sp. WC76535. J. Nat. Prod. 61, 1379–1382 (1998).
Alvi, K. A. et al. Identification of inhibitors of inducible nitric oxide synthase from microbial extracts. J. Antibiot. 53, 496–501 (2000).
Ströch, K. et al. Retymicin, galtamycin B, saquayamycin Z and ribofuranosyllumichrome, novel secondary metabolites from Micromonospora sp. Tü 6368. II. Structure elucidation. J. Antibiot. 58, 103–110 (2005).
Kusumi, S. et al. Total synthesis of vineomycin B2. J. Am. Chem. Soc. 135, 15909–15912 (2013).
Ohta, K. et al. The absolute configuration of P-1894B, a potent prolyl hydroxylase inhibitor. Chem. Pharm. Bull. 32, 4350–4359 (1984).
Okazaki, H. et al. A potent prolyl hydroxylase inhibitor, P-1894B, produced by a strain of Streptomyces. J. Antibiot. 34, 1355–1356 (1981).
Uchida, T. et al. Saquayamycins, new aquayamycin-group antibiotics. J. Antibiot. 38, 1171–1181 (1985).
Imamura, N. et al. The structure of vineomycin B2. J. Antibiot. 34, 1517–1518 (1981).
Zhong, W.-M. et al. Three minor new compounds from the aerial parts of Leonurus japonicas. Chin. Chem. Lett. 26, 1000–1003 (2015).
Acknowledgements
This study was supported in part by the National Science Foundation of China (41476133) and the projects of Guangdong Provincial Oceanic and Fishery Administration (A201401C05). We express our thanks to the analytical facility center (Ms Sun, Ms Zhang, Mr Li and Ms Xiao) of the South China Sea Institute of Oceanology for measuring MS and NMR data.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies the paper on The Journal of Antibiotics website
Supplementary information
Rights and permissions
About this article
Cite this article
Zhu, X., Duan, Y., Cui, Z. et al. Cytotoxic rearranged angucycline glycosides from deep sea-derived Streptomyces lusitanus SCSIO LR32. J Antibiot 70, 819–822 (2017). https://doi.org/10.1038/ja.2017.17
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ja.2017.17
This article is cited by
-
Identification and characterization of isocitrate dehydrogenase 1 (IDH1) as a functional target of marine natural product grincamycin B
Acta Pharmacologica Sinica (2021)
-
Structure and biosynthesis of mayamycin B, a new polyketide with antibacterial activity from Streptomyces sp. 120454
The Journal of Antibiotics (2018)
-
Cytotoxic antibiotic angucyclines and actinomycins from the Streptomyces sp. XZHG99T
The Journal of Antibiotics (2018)