Abstract
A southern Australian soil isolate, Amycolatopsis sp. MST-108494, was subjected to a panel of fermentation and media optimization trials, supported by analytical chemical profiling, to detect and enhance production of a rare class of secondary metabolites. Chemical fractionation of two complementary fermentations yielded three new polyketides, identified by detailed spectroscopic analysis as the glycosylated macrolactones, amycolatopsins A (1), B (2) and C (3), closely related to the ammocidins and apoptolidins. Amycolatopsins 1 and 3 selectively inhibited growth of Mycobacterium bovis (BCG) and Mycobacterium tuberculosis (H37Rv) when compared with other Gram-positive or Gram-negative bacteria, with 3 exhibiting low levels of cytotoxicity toward mammalian cells. Thus, our data reveal promising structure activity relationship correlations where the antimycobacterial properties of amycolatopsins are enhanced by hydroxylation of the 6-Me (that is, 1 and 3), whereas mammalian cytotoxicity is decreased by hydrolysis of the disaccharide moiety (that is, 3).
Similar content being viewed by others
Introduction
Tuberculosis (TB) is one of the oldest recorded human infectious diseases and, despite much effort and investment, remains one of the most serious globally significant communicable diseases.1 Existing frontline treatment for active TB includes the antibiotics isoniazid, rifampicin, pyrazinamide and ethambutol, all of which were introduced into clinical practice >40 years ago. Standard therapy for drug-sensitive TB involves prolonged treatment (>6 months) with combinations of these vintage antibiotics, most of which exhibit toxic side effects and are compromised in the face of multidrug-resistant TB. With an estimated half a million new cases of multidrug-resistant TB annually, and the emergence of extensively and totally drug-resistant Mycobacterium tuberculosis strains, there is an urgent need for new TB antibiotics.2, 3
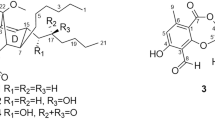
Responding to this challenge, and as an extension of our investigations into the chemistry of Australian microbial biodiversity, we screened libraries of microbial extracts and purified metabolites for growth inhibitory activity against Mycobacterium bovis (BCG), as a laboratory surrogate of M. tuberculosis. These studies detected significant growth inhibitory activity against mycobacteria from a rare class of metabolites isolated from Amycolatopsis sp. MST-108494. A detailed investigation of these metabolites identified them as amycolatopsins A (1), B (2) and C (3) (Figure 1), new analogs of the rare apoptolidin and ammocidin structure class.4, 5, 6, 7 An account of the fermentation, extraction, isolation, structure elucidation and biological properties of 1–3 is presented below.
Results and discussion
Amycolatopsis sp. MST-108494, a strain closely related to Amycolatopsis magusensis (see Supplementary Information), was isolated from soil collected in 1996 from the rural outskirts of Port Augusta, South Australia. Fermentation of MST-108494 on yeast glucose malt agar (I2A) yielded an extract with HPLC diode-array detection (HPLC-DAD) profile (Supplementary Figure S3) unique in our database of 300 000 actinomycetes, including 2500 actinomycetes type species and 40 Amycolatopsis isolates. In particular, the I2A cultivation featured low-level production of metabolites with UV–visible spectra (DAD, λmax 278, 322) unprecedented in our databases. Optimization studies in a wide range of media identified beef extract agar (LUA) as optimal for the production of these metabolites, with solvent extraction and reversed-phase preparative HPLC fractionation of a large-scale fermentation yielding 1 and 2. In subsequent analysis of antimycobacterial activity, 1 and 2 exhibited promising (>99%) growth inhibitory activity. In an effort to obtain additional analogs, we used a modified LUA media, GYA (LUA supplemented with starch), to produce and isolate the related minor metabolite 3.
High-resolution electrospray ionization MS (HRESI(+)MS) analysis of 1 returned a sodium adduct ion consistent with a molecular formula (C60H98O23, Δmmu +1.1) requiring 12 double bond equivalents. Analysis of the 1D NMR (MeOH-d4) data (Table 1 and Supplementary Table S1) revealed resonances for an ester/lactone carbonyl (δC 171.2), and 12 olefinic sp2 carbons (δC 129.6–144.0), accounting for seven double bond equivalents and requiring that 1 incorporate five rings. Detailed analysis of 2D NMR (MeOH-d4) data (Figure 2) revealed diagnostic correlations for a 20-membered macrocyclic lactone (C-1 to C-19) incorporating two triene moieties (C-2 to C-7, and C-10 to C-15), a pendant polysubstituted tetrahydropyran (C-20 to C-28) and three pyranose sugar residues (C-1′ to C-6′, C1′′ to C-6′′ and C-1′′′ to C-6′′′). Double bond geometries (E Δ,2, 3 Z Δ,4, 5 Z Δ,6, 7 E Δ,8, 9 E Δ10, 11 and E Δ12, 13) were assigned by consideration of diagnostic rotating-frame Overhauser effect spectroscopy (ROESY) correlations (Figure 2). The relative configuration of the tetrahydropyran moiety was established by ROESY correlations (Figure 2) that positioned H-23 and H-25 on the opposite face to H-22 and H-20. Further consideration of a Newman projection and ROESY correlations from 20-Me to 22-OMe suggested the C-20 to C-21 relative configuration as indicated in Figure 2, whereas the axial configuration of 24-Me was determined by comparison with the co-metabolite 2, as shown in Figure 3. The relative configuration of the C-1′ to C-6′ pyranose was established by ROESY correlations from H-2′ to both H-1′ and H-4′, and from H-4′ to H3-6′, and from J-values that confirmed an equatorial orientation for H-1′ (J1′,2′ 3.8 Hz), and axial orientations for H-2′, H-3′, H-4′ and H-5′ (J2′,3′ 9.7 Hz, J3′,4′ 9.7 Hz and J4′,5′ 9.7 Hz). The relative configuration of the C-1′′ to C-6′′ pyranose was established by ROESY correlations from 1,3-diaxial disposed H-1′′ and H-5′′, and from J-values that confirmed an equatorial orientation for H-3′′ (J3′′,4′′ 2.8 Hz) and axial orientations for H-1′′, H-4′′ and H-5′′ (J1′′,2a′′ 9.5 Hz and J4′′,5′′ 9.6 Hz). The relative configuration of the C-1′′′ to C-6′′′ pyranose was established by ROESY correlations from 1,3-diaxial disposed H-5′′′ and both H-1′′′ and 3′′′-Me, and from J-values that confirmed axial orientations for H-1′′′, H-4′′′ and H-5′′′ (J1′′′,2a′′′ 9.9 Hz and J4′′′,5′′′ 9.6 Hz). As absolute configurations for the pyranose residues have not been determined, the structure for amycolatopsin A (1) is arbitrarily represented with D-hexose sugar configurations.
HRESI(+)MS analysis of 2 revealed a sodium adduct ion consistent with a molecular formula (C60H98O22, Δmmu –1.3) for a deoxy analog of 1. This hypothesis was confirmed by comparison of the 1D NMR (MeOH-d4) data for 2 with 1 (Table 1 and Supplementary Tables S1 and S2) that revealed the only significant difference as conversion of the 6-CH2OH moiety in 1 (δH 4.39 and 4.49; δC 58.1) to a 6-Me moiety in 2 (δH 2.06; δC 13.8). Detailed analysis of the 2D NMR data for 2 (Figure 3), applying the arguments outlined above for 1, permitted assignment of all double bond geometries, and relative configurations across C-20 to C-28, C-1′ to C-6′, C-1′′ to C-6′′ and C-1′′′ to C-6′′′. Significantly, a ROESY correlation from H-25 to H-1′′ in 2 established the axial orientation of 24-Me that we also attribute to 1 on the basis of excellent NMR comparisons between 1 and 2 (see above).
HRESI(+)MS analysis of 3 revealed a sodium adduct ion consistent with a molecular formula (C47H75O17 Δmmu –0.3) for an aglycone analog of 1. Comparison of the 1D NMR (MeOH-d4) data for 3 with 1 (Table 1 and Supplementary Tables S1 and S3) revealed significant differences attributed to hydrolysis of the C-24 disaccharide residue. Detailed analysis of the 2D NMR data for 3 (Figure 4), applying the arguments outlined above for 1, permitted assignment of all double bond geometries, and relative configurations across C-20 to C-28 and C-1′ to C-6′.
In our hands, 1–3 did not inhibit growth (IC50 >30 μM) of the Gram-positive bacteria Bacillus subtilis (ATCC 6633) and Staphylococcus aureus (ATCC 25923), the Gram-negative bacteria Escherichia coli (ATCC 25922) and Pseudomonas aeruginosa (ATCC 27853), or the fungus Candida albicans (ATCC 90028) (Table 2 and Supplementary Figure S11). In comparison, 1 and 3 inhibited growth of M. bovis (BCG, IC50 0.4 and 2.7 μM) and M. tuberculosis (H37Rv, IC50 4.4 and 5.7 μM) in liquid culture (Table 2). It is also noteworthy that 1 and 2 exhibited significant cytotoxicity toward human colon (SW620, IC50 0.08 and 0.14 μM) and lung (NCIH-460, IC50 1.2 and 0.28 μM) carcinoma cells, whereas the aglycone 3 was 5- to 100-fold less cytotoxic (IC50 10 and 5.9 μM) (Table 2 and Supplementary Figure S12).
The amycolatopsins A–C (1–3) are new examples of a rare class of glycosylated polyketide macrolides, structurally and biosynthetically related to the apoptolidins reported from Amycolatopsis14, 15 and Nocardiopsis4, 8, 9, 10 spp., and ammocidins reported from Saccharothrix sp.5, 6, 7 The amycolatopsins share a common macrolide scaffold with the apoptolidins and ammocidins, and combine other unique structure features from both. For example, amycolatopsins share 4′-O-methylation with apoptolidins,4, 8, 9, 10, 14, 15 and 25-methoxypropyl and 20-methyl moieties with ammocidins.5, 6, 7 With respect to the latter, our suggested assignment of configurations about the C-20 to C-25 moiety in 1–3 provides a basis for making similar assignments in all known ammocidins (where these configurations are currently unassigned).5, 6, 7 Although neither absolute nor relative configurations have been assigned to chiral centers within the macrolactone ring system of any known ammocidins, in 1998 NOESY correlations were used to assign absolute configurations about comparable centers in the closely related apoptolidin.11 Subsequent researchers extrapolated these assignments, and applied them to all other known examples of the apoptolidin structure class.4, 8, 9, 10, 12, 14, 15 To the best of our knowledge there are no published validations of these assignments that, in our view, remain tentative at best. In the case of the amycolatopsins, careful analysis of ROESY data revealed evidence of macrolactone conformational flexibility, cautioning against the use of such approaches to configurational assignment. For example, in the case of amycolatopsin B and C we observed ROESY correlations between 2-Me and H-5, and between 3-H and H-5, suggestive of rotation flexibility about the 3–4 bond. Erring on the side of caution, configurations about C-8, C-9, C-16, C-17 and C-19 in 1–3 remain unassigned. Our preliminary structure activity relationship analysis of amycolatopsins revealed noteworthy correlations. First, the aglycone 3 exhibits comparatively lower mammalian cytotoxicity than the corresponding disaccharride glycosides 1 and 2, consistent with literature reports where the aglycone apoptolidin F exhibits reduced cytotoxicity compared with all other known disaccharride glycoside apoptolidins.13 Second, and arguably of greater importance, the antimycobacterial activity of amycolatopsins correlate strongly with hydroxylation of the 6-Me (that is, 1 and 3), a site of hydroxylation not previously observed among apoptolidins or ammocidins. Our observations on the amycolatopsins are encouraging in that they suggest a strategy (selective hydroxylation and glycoside hydrolysis) for enhancing antimycobacterial activity across this class of polyketide macrolide.
Methods
General experimental procedures
Chiroptical measurements ([α]D) were obtained on a JASCO P-1010 polarimeter (Tokyo, Japan) in a 100 × 2 mm cell at 22 °C. UV–visible spectra were obtained on a Varian Cary 50 UV–visible spectrophotometer (Santa Clara, CA, USA)with 1 cm pathway quartz cells. NMR spectra were obtained on a Bruker Avance DRX600 spectrometer (Billerica, MA, USA) in the solvents indicated and referenced to residual signals (δH 3.31 and δC 49.1 ppm) in deuterated solvents. ESIMS were acquired using Agilent 1100 Series separations module equipped with an Agilent 1100 Series LC/MS mass detector (Santa Clara, CA, USA) in both positive and negative ion modes under the following LC conditions using Zorbax SB-C8 5 μm column, 150 × 4.6 mm, eluting with 1.0 ml min−1 90% H2O/MeCN to 100% MeCN, and an isocratic 0.05% HCO2H modifier over 15 min, with detection at 210 and 254 nm. HRESIMS measurements were obtained on a Bruker micrOTOF mass spectrometer by direct infusion in MeCN at 3 μl min−1 using sodium formate clusters as an internal calibrant. Analytical HPLC-DAD was performed on a gradient Shimadzu HPLC system comprising a LC-10AT VP gradient chromatograph, SPD-M10A VP DAD and SCL-10A VP system controller. Preparative HPLC was performed on a gradient Shimadzu HPLC system (Sydney, Australia) comprising two LC-8A preparative liquid pumps with a static mixer, SPD-M10AVP diode array detector and SCL-10AVP system controller with a standard Rheodyne injection port. All solvents were HPLC grade.
Isolation and identification of Amycolatopsis sp. MST-108494
The strain was isolated from a soil sample (Soil no. 3019) collected in timbered country near Woolshed Flat, 15 km north of Port Augusta, South Australia, in 1996. The soil sample was red brown in color, with low levels of organic matter and salinity (106 ppm) and of neutral pH (6.6). A subsample of soil (20 g) was treated with sterile H2O (100 ml), and 10−3 and 10−4 dilutions plated on ISP2 agar and incubated for 7 days at 28 °C. Single colonies were transferred to ISP2 agar isolation plates that were incubated at 28 °C for 2–3 weeks. The pure strain of MST-108494 was accessioned and preserved at −80 °C in 20% aqueous glycerol. MeOH extracts prepared from residual agar (1 ml g−1 agar) were used for chemical and biological profiling. Cells from an isolated colony on a 3-day ISP2 agar plate were sampled and suspended in PrepMan Ultra Sample Preparation Reagent (Sydney, Australia) (100 μl). The sample was boiled at 100 °C for 10 min and the recovered 16S rRNA sequenced at the Australian Genome Research Facility (AGRF, Sydney, Australia), and registered in the NCBI (National Center for Biotechnology Information) GenBank database with accession number MF102107. The gene sequence is provided in the Supplementary Information.
Media optimization and co-metabolite profiling
Amycolatopsis sp. MST-108494 was cultured on a range of agar, liquid and grain media. Agar media (15 ml) were prepared from yeast glucose malt agar (I2A), I2A+starch (GYA), I2A+mannitol (IMA), modified Bennetts agar (MSA), soy and beef extract agar (LUA), yeast, molasses and peptone agar (MOA) and casein glycerol agar (CGA). Liquid media (50 ml) were prepared from these same media with the omission of 2.0% bacteriological agar (that is, I2L, GYL, IML, MSL, LUL and CGL) in single baffled Erlenmeyer flasks (250 ml). Grains media (50 g) (that is, barley (PB), wheat (BL), jasmine rice (JR) and basmati rice (BS)) in H2O (30 ml) were prepared in Erlenmeyer flasks (250 ml). Sterilized agar, liquid and grain media were inoculated with a spore suspension of MST-108494 prepared from a 7-day, 28 °C, I2A agar plate. Subsamples (~1 g) recovered from each cultivation on days 7 and 10 were extracted with MeOH (2 ml), centrifuged (15 700 g for 3 min) and the supernatant analyzed by HPLC-DAD (Alltima C18 ‘rocket’ column, 100 Å, 53 × 7 mm, 3 μm, eluted with a 3 ml min−1 gradient from 90% H2O/MeCN to 100% MeCN, over 7 min, with an isocratic 0.01% TFA modifier and a 2 min hold). The resulting co-metabolite retention time and UV–visible (DAD) spectra were accessioned into the Microbial Screening Technologies (Sydney, Australia) database (COMET), and dereplicated against 5000 microbial metabolites and the co-metabolite profiles of >2500 strains of known species, patented strains and metabolically rich actinomycetes species isolated from solid samples. Identification of likely replicate cultures was undertaken against a database of 300 000 actinomycete cultures isolated at MST.16
Preparative cultivation and purification
A spore suspension of Amycolatopsis sp. MST-108494 (100 μl) applied to LUA plates (200 × 15 ml) was incubated at 28 °C for 14 days. The combined agar (2.5 kg) was extracted with acetone (2 × 2.8 l) for 2 h on a rotatory platform (150 r.p.m.) and then concentrated in vacuo to an aqueous residue (2.2 l, 24.7 g). The aqueous residue was applied to two C18 columns (2 × 10 g), washed with deionized H2O and the bound metabolites eluted with MeCN (2 × 40 ml) and concentrated in vacuo to an organic extract (2.28 g). A portion of the extract (1.0 g) was dissolved in MeCN (4 ml) and fractionated by preparative HPLC (Platinum EPS C18, gradient 30 to 60% MeCN/H2O over 20 min, with a step to 100% MeCN over 10 min) to yield amycolatopsin A (1) (tR 14.9 min, 7.3%) and amycolatopsin B (2) (tR 19.3 min, 3.5%). A second fermentation was undertaken using GYA agar and processed as described above to yield an extract (0.57 g). The extract was dissolved in MeCN (4 ml) and fractionated by preparative HPLC (Platinum EPS C18, isocratic conditions 45% MeCN/H2O over 20 min, with a step to 100% MeCN over 10 min) to yield amycolatopsin C (3) (tR 11.0 min, 1.6%) and (1) (tR 15.1 min, 6.1%) (note that % yields were determined on a mass-to-mass basis against the organic extract).
Amycolatopsin A (1): white powder; [α]21D –41 (c 0.06, MeOH); UV (MeOH) λmax (log ɛ): 248 (3.49), 276 (3.38), 320 (2.96) nm, Supplementary Figure S5; 1H NMR (600 MHz, MeOH-d4), Table 1 and Supplementary Figure S4; HRESI(+)MS m/z 1209.6380 [M+Na]+ (calcd for C60H98O23Na, 1209.6391).
Amycolatopsin B (2): white powder; [α]21D –40 (c 0.05, MeOH); UV (MeOH) λmax (log ɛ): 268 (3.59), 325 (3.15) nm, Supplementary Figure S7; 1H NMR (600 MHz, MeOH-d4), Table 1 and Supplementary Figure S6; HRESI(+)MS m/z 1193.6455 [M+Na]+ (calcd for C60H98O22Na, 1193.6442).
Amycolatopsin C (3): white powder; [α]21D –15 (c 0.06, MeOH); UV (MeOH) λmax (log ɛ): 271 (3.60), 313 (3.49) nm, Supplementary Figure S9; 1H NMR (600 MHz, MeOH-d4), Table 1 and Supplementary Figure S8; HRESI(+)MS m/z 935.4978 [M+Na]+ (calcd for C47H76O17Na, 935.4975).
Antibacterial and antifungal assays
The bacteria and the fungus to be tested were streaked onto a tryptic soy agar plate and incubated at 37 °C for 24 h and 26.5 °C for 48 h, respectively. One colony was then transferred to fresh tryptic soy broth (15 ml) and the cell density was adjusted to 104–105 CFUs per ml to OD 0.1. Amycolatopsins A–C (1–3) were dissolved in DMSO and diluted with H2O to give 300 μM stock solutions (10% DMSO). The stock solutions were then serially diluted with 10% DMSO. An aliquot (20 μl) of each dilution was transferred to a 96-well microtiter plate and freshly prepared microbial broth (180 μl) was added to each well to give final concentration of 30 to 0.01 μM in 1% DMSO. The plates were incubated at 37 °C for 24 h and the optical density of each well was measured spectrophotometrically at 600 nm (POLARstar Omega plate reader, BMG LABTECH, Offenburg, Germany). Each test compound was screened against the Gram-negative bacteria E. coli (ATCC 11775) and P. aeruginosa (ATCC 10145), the Gram-positive bacteria S. aureus (ATCC 9144 and ATCC 25923), B. subtilis (ATCC 6633 and ATCC 6051) and C. albicans (ATCC 90028). The IC50 value was calculated as the concentration of the compound or antibiotic required for 50% inhibition of the bacterial and/or fungal growth.
Antimycobacterial assays
M. bovis, Bacille Calmette Guerin (BCG) and M. tuberculosis (H37Rv) were grown until early-mid log phase in 7H9 liquid medium (Difco, Brisbane, Australia) containing 0.2% glycerol, 0.05% Tween-80, 5% bovine serum albumin, 2% dextrose and 0.85% sodium chloride and then diluted in complete 7H9 medium to an OD (600 nm) of 0.02. Amycolatopsins A–C (1–3) were dissolved in DMSO and diluted with H2O to give a 600 μM stock solution in 10% DMSO. The stock solution was then serially diluted with 10% DMSO and an aliquot of 10 μl of each dilution was transferred to wells of a 96-well microtiter plate together with 190 μl of bacterial suspension, resulting in final concentrations of 30 to 0.01 μM in 1% DMSO. Bacterial growth at 37 °C in the presence of amycolatopsins (1–3) was monitored by OD measurements over 7 days. Isoniazid (INH, 20 μg ml−1 in H2O) and DMSO served as positive and negative controls.
Cytotoxicity assays
The MTT (3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide) assay was modified from that previously described17, 18 using adherent cell lines SW620 and NCIH-460. SW620 and NCIH-460 were cultured in RPMI medium 1640 as adherent monolayer in flasks supplemented with 10% fetal bovine serum, 2 mM L-glutamine, 100 unit ml−1 penicillin and 100 μg ml−1 streptomycin in a humidified 37 °C incubator supplied with 5% CO2. Cells were harvested with trypsin and dispensed into 96-well microtiter assay plates at 2000 cells per well for SW620, and 5000 cells per well for NCIH-460, then incubated for 18 h at 37 °C with 5% CO2. Amycolatopsins A–C (1–3) were dissolved in 5% DMSO in phosphate-buffered saline (v/v) and aliquots (20 μl) tested over a series of final concentrations ranging from 10 nM to 30 μM. Control wells were treated with 5% aqueous DMSO. After 68 h of incubation at 37 °C with 5% CO2 an aliquot (20 μl) of MTT in phosphate-buffered saline was added to each well (final concentration of 0.4 mg ml−1), and the microtiter plates incubated for a further 4 h at 37 °C with 5% CO2. Medium was aspirated and precipitated formazan crystals dissolved in DMSO (100 μl per well). The absorbance of each well was measured at 580 nm (PowerWave XS Microplate Reader, Bio-Tek Instruments, Winooski, VT, USA). IC50 values were calculated as the concentration of analyte required for 50% inhibition of cell growth (compared with negative controls). Vinblastine (20 μg ml−1 in 5% DMSO) and DMSO served as positive and negative controls, respectively. All experiments were performed in duplicate.
Accession codes
References
Garcia, A., Bocanegra-Garcia, V., Palma-Nicolas, J. P. & Rivera, G. Recent advances in antitubercular natural products. Eur. J. Med. Chem. 49, 1–23 (2012).
World Health Organisation WHO treatment guidelines for drug- resistant tuberculosis, 2016 (ISBN 978 92 4 154963 9).
Stop TB Partnership The Global Plan to Stop TB, 2006-2015. actions for life: towards a world free of tuberculosis. Int. J. Tuberc. Lung Dis. 10, 240–241 (2006).
Kim, J. W., Adachi, H., Shin-Ya, K., Hayakawa, Y. & Seto, H. Apoptolidin, a new apoptosis inducer in transformed cells from Nocardiopsis sp. J. Antibiot. 50, 628–630 (1997).
Murakami, R. et al. Ammocidins B, C and D, new cytotoxic 20-membered macrolides from Saccharothrix sp. AJ9571. J. Antibiot. 62, 123–127 (2009).
Murakami, R. et al. Ammocidin, a new apoptosis inducer in Ras-dependent cells from Saccharothrix sp. I. Production, isolation and biological activity. J. Antibiot. 54, 710–713 (2001).
Murakami, R. et al. Ammocidin, a new apoptosis inducer in Ras-dependent cells from Saccharothrix sp. II. Physico-chemical properties and structure elucidation. J. Antibiot. 54, 714–717 (2001).
Wender, P. A., Sukopp, M. & Longcore, K. Apoptolidins B and C: isolation, structure determination, and biological activity. Org. Lett. 7, 3025–3028 (2005).
Wender, P. A. & Longcore, K. E. Apoptolidins E and F, new glycosylated macrolactones isolated from Nocardiopsis sp. Org. Lett. 11, 5474–5477 (2009).
Wender, P. A. & Longcore, K. E. Isolation, structure determination, and anti-cancer activity of apoptolidin D. Org. Lett. 9, 691–694 (2007).
Hayakawa, Y., Kim, J. W., Adachi, H., Shin-ya, K., Fujita, K. & Seto, H. Structure of apoptolidin, a specific apoptosis inducer in transformed Cells. J. Am. Chem. Soc. 120, 3524–3525 (1998).
Wender, P. A., Gulledge, A. V., Jankowski, O. D. & Seto, H. Isoapoptolidin: structure and activity of the ring-expanded isomer of apoptolidin. Org. Lett. 4, 3819–3822 (2002).
Chau, S. T., Sulikowski, G. A. & Wu, B. Studies on the synthesis of the apoptolidins. Strategies and Tactics in Organic Synthesis 8, 375–394 (2012).
Sheng, Y. et al. Succinylated apoptolidins from Amycolatopsis sp. ICBB 8242. Org. Lett. 17, 2526–2529 (2015).
Serrill, J. D. et al. Apoptolidins A and C activate AMPK in metabolically sensitive cell types and are mechanistically distinct from oligomycin A. Biochem. Pharmacol. 93, 251–265 (2015).
Lacey, E. & Tennant, S. Secondary metabolites - the focus of biodiscovery and perhaps the key to unlocking new depths in taxonomy. Microbiol. Aust. 24, 34–35 (2003).
Carmichael, J., DeGraff, W. G., Gazdar, A. F., Minna, J. D. & Mitchell, J. B. Evaluation of a tetrazolium-based semiautomated colorimetric assay: assessment of chemosensitivity testing. Cancer Res. 47, 936–942 (1987).
Henrich, C. J. et al. A high-throughput cell-based assay for inhibitors of ABCG2 activity. J. Biomol. Screen. 11, 176–183 (2006).
Acknowledgements
We thank AE Lacey (MST) for assistance with the metabolite purification. We acknowledge support from the University of Queensland Institute for Molecular Bioscience and University of Queensland Diamantina Institute. This research was funded in part by The University of Queensland (UQECR1607045).
Author contributions
EL, AC and DV were responsible for the cultivation and isolation of amycolatopsins. ZGK acquired, tabulated and interpreted all spectroscopic data, carried out all bioassays and assembled the Supplementary Information. AAS reviewed all NMR data. AB had oversight of all mycobacterial studies. RJC initiated the research program, and had oversight of all chemical studies and drafting of the manuscript. All authors have given approval to the final version of the manuscript.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies the paper on The Journal of Antibiotics website
Supplementary information
Rights and permissions
About this article
Cite this article
Khalil, Z., Salim, A., Vuong, D. et al. Amycolatopsins A–C: antimycobacterial glycosylated polyketide macrolides from the Australian soil Amycolatopsis sp. MST-108494. J Antibiot 70, 1097–1103 (2017). https://doi.org/10.1038/ja.2017.119
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ja.2017.119
This article is cited by
-
Suertides A–C: selective antibacterial cyclic hexapeptides from Amycolatopsis sp. MST-135876v3
The Journal of Antibiotics (2022)