Abstract
Bisoxazolomycin (1), oxazolomycin A2 (2) and oxazolomycin A (3) were identified by physicochemical screening approach from a culture broth of ‘Streptomyces subflavus subsp. irumaensis’ AM-3603. Compound 2 is a hydrolyzed analog of 3 at the β-lactone ring, and 1 is a new dimeric analog of 2. Compounds 1 and 2 exhibited less potent antibacterial activity and cytotoxicity than 3, which might be due to lack of the β-lactone ring.
Similar content being viewed by others
Introduction
Our continuing search for new compounds from culture broths of microorganisms exploits innovative physicochemical (PC) screening, which is based on the physicochemical properties of compounds using LC-UV/vis diode array detection, LC/MS and color reaction.1 In the course of our PC screening for new compounds, the culture broth of ‘Streptomyces subflavus subsp. irumaensis’ AM-3603, which has been reported as a producer of irumamycin,2 was re-investigated. A new compound (1) with accurate mass ([M+H]+, m/z 1329.7126) and UV spectrum (absorption max at 228, 266, 276 and 285 nm) was produced by the strain and the physicochemical properties did not correspond to known compounds in the existing database (Dictionary of Natural Products, CRC Press, FL, USA). Compound 1 was isolated from the culture broth and determined to be a new dimeric analog of oxazolomycin A,3 namely bisoxazolomycin (1). Furthermore, a hydrolyzed analog of oxazolomycin A, namely oxazolomycin A2 (2), along with oxazolomycin A (3) were also isolated from the culture broth. In this report, we describe the fermentation, isolation, structural elucidation and some biological activity of compounds 1, 2 and 3.
Fermentation and isolation
The fermentation and isolation process is described in Supplementary Scheme S1. ‘Streptomyces subflavus subsp. irumaensis’ AM-3603, cultured on Waksman agar medium (1.0% glucose, 0.5% peptone, 0.5% meat extract, 0.3% sodium chloride and 1.2% agar, pH 7.0) was inoculated into a 500 ml-Erlenmeyer flask containing 100 ml of YD medium (1.0% glucose and 1.0% yeast extract, pH was not adjusted) and cultivated on a rotary shaker (210 r.p.m.) at 27 °C for 3 days. A 1 milliliter portion of the seed culture was inoculated into 500 ml-Erlenmeyer flasks (total 58 flasks) each containing 100 ml of 2.0% soluble starch, 0.5% glycerol, 1.0% defatted wheat germ, 0.3% meat extract, 0.3% dry yeast and 0.3% CaCO3 (pH was not adjusted) and fermentation was carried out on a rotary shaker (210 r.p.m.) at 27 °C for 4 days.
The culture broth of strain AM-3603 was centrifuged at 3000 r.p.m. for 15 min. The precipitate was extracted by ethanol and concentrated in vacuo. The extract was partitioned with ethyl acetate twice and concentrated to yield 836 mg. The ethyl acetate extract was applied to a silica gel FL100D column chromatography (32 i.d. × 40 mm; Fuji Silysia Chemical, Aichi, Japan) and eluted with a stepwise gradient of CHCl3–MeOH (1:0, 100:1, 50:1, 9:1, 1:1 and 0:1 (v/v), each 100 ml). The elute fraction of 1:1 (52.6 mg) was purified by ODS column chromatography (15 i.d. × 20 mm; Senshu Scientific, Tokyo, Japan), which was eluted with a stepwise gradient of acetonitrile (10, 20, 30, 40, 50, 60 and 100% (v/v), each 20 ml). The fractions of 50 and 30% acetonitrile were dried in vacuo to yield 1 (8.7 mg) and 2 (4.4 mg), respectively. Similarly, the 9:1 fraction (244 mg) eluted from silica gel column chromatography was applied to an ODS column (20 i.d. × 50 mm). The 30% acetonitrile fraction was dried in vacuo to yield 2 (7.4 mg). The 50% acetonitrile fraction (58.5 mg) was purified by ODS column (14 i.d × 20 mm) again, and then the 30−40% acetonitrile fraction was subsequently dried in vacuo to yield 3 (39.2 mg).
Structure elucidation
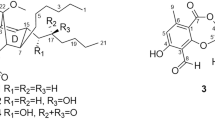
Compound 3 was identified as oxazolomycin A by 1H and 13C NMR spectra (Supplementary Information), molecular formula, UV spectrum, IR spectrum and optical rotation (Supplementary Table S1), which were consistent with those described in the literatures.4, 5 Oxazolomycin A (Figure 1) was discovered by Mori et al.3 and showed activity against P388 leukaemia cells.
Compound 2 was determined to have a molecular formula of C35H51N3O10 by HR-ESI-MS [M+H]+ m/z 674.3667 (consistent with a calculated value of 674.3647 for C35H52N3O10), which was 18 mass units (H2O) larger than 3. The UV spectrum and optical rotation of 2 were similar to those of 3, but the specific IR band of the β-lactone ring at 1825 cm−1 was not observed (Supplementary Table S1). The 1H and 13 C NMR signals of 2 were assigned by combination of COSY, HSQC and HMBC spectra (Table 1 and Supplementary Figure S1). The NMR spectra were very similar to those of 3 but a difference in chemical shifts of the oxymethylene of H2-16 (3, 4.84 and 4.51 p.p.m.; 2, 4.22 and 4.16 ppm) was observed. These results suggest that 2 is a hydrolyzed analog of 3 at the β-lactone ring (Figure 1). Furthermore, 2 could be generated from hydrolysis of 3 with 0.1 M NaOH at room temperature (Supplementary Figure S2). Although 2 has been known as one of intermediates in the total synthesis of 3,4 this is the first report of its isolation from among the metabolites of Streptomyces. Compound 2 was named as oxazolomycin A2.
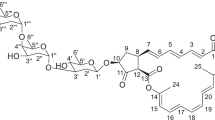
Compound 1 was determined to have a molecular formula of C70H100N6O19 by HR-ESI-MS [M+H]+ m/z 1329.7126 (consistent with a calculated value of 1329.7116 for C70H101N6O19). The UV spectra and optical rotation were similar to those of 2 and the specific IR band of β-lactone ring at 1825 cm−1 was not observed (Supplementary Table S1). The 1H and 13 C NMR signals of 1 were assigned by combination of COSY, HSQC and HMBC spectra (Table 1 and Supplementary Figure S1). The 1H and 13 C chemical shifts of oxazole-containing side chain from C-5 to C-13′ and from C-5′′ to C-13′′′ were consistent with those of 2. On the other hand, two sets of 1H and 13 C signals were observed around N-methyl-γ-lactam segment. Therefore, 1 is an asymmetric dimer of 2 consisting N-methyl-γ-lactam segments I and II (Table 1 and Supplementary Figure S1). Segment I was confirmed by the 1H-13C long-range correlations from H-2 to C-1, C-3 and C-4; from H3-14-Me to C-1 and C-3; from H2-16 to C-3, C-15 and C-17; from H3-18-Me to C-4; from H3-19-Me to C-1 and C-15 (Supplementary Figure S1). Segment II was also confirmed by the 1H-13C long-range correlations from H-2′′ to C-1′′, C-3′′ and C-4′′; from H3-14′′-Me to C-1′′ and C-3′′; from H2-16′′ to C-3′′ and C-15′′; from H3-18′′-Me to C-4′′; from H3-19′′-Me to C-1′′ and C-15′′ (Supplementary Figure S1). The chemical shifts of the oxymethylene of H2-16 (4.22 and 4.15 p.p.m.) in segment I were similar to those of 2 (4.22 and 4.16 ppm). However, the chemical shifts of the oxymethylene of H2-16′′ (4.83 and 4.74 ppm) in segment II was observed downfield. Furthermore, 1 could be hydrolyzed to 2 in 0.1M NaOH at room temperature (Supplementary Figure S2). Therefore, 1 was determined to be a new dimer of 2, which had an ester bond between C-17 and C-16′′ (Figure 1). Compound 1 was named as bisoxazolomycin.
Biological activity
Cytotoxic activity of 1, 2 and 3 was measured by Cell Counting Kit-8 (Dojindo Laboratories, Kumamoto, Japan) in HL60 (Human promyelocytic leukemia cells). Briefly, HL60 (3 × 105 cells per well) cell lines were seeded in 96-well plates and cultured in RPMI1640 (Nacalai Tesque, Kyoto, Japan) supplemented with 10% fetal bovine serum and 1% sodium pyruvate at 37 °C under 5% CO2. After culturing for 1 h, 1, 2 or 3 was dissolved in MeOH at appropriate concentrations was added into each well. After 48 h of incubation at 37 °C, WST-8 solution was added to each well and incubated at 37 °C for 4 h. The absorbance at 460 nm of each well was measured using a Corona Grating Microplate Reader SH-9000 (Corona Electric, Ibaraki, Japan). Compound 1, 2 and 3 displayed cell growth inhibition against HL60 cells with IC50 values of 7 μM, 20 μM and 0.6 μM, respectively.
Compounds 1, 2 and 3 were investigated for antifungal activity, being tested against Candida albicans ATCC 64548, Mucor racemosus IFO 4581, Aspergillus niger ATCC 6275 and Saccharomyces cerevisiae ATCC 9763. Their antibacterial activity was examined against Kocuria rhizophila ATCC 9341, Bacillus subtilis ATCC 6633, Escherichia coli NIHJ, Xanthomonas campestis pv. oryzae KB 88, Pseudomonas aeruginosa IFO 3080, Staphylococcus aureus ATCC 6538p and Mycobacterium smegmatis ATCC 607 using the paper disc method (φ 6 mm, Advantec, Tokyo, Japan) at the concentrations of 30, 10, 3 and 1 μg per disc. The compounds displayed no antifungal activity. Compound 3 showed antibacterial activity against K. rhizophila, B. subtilis, E. coli, X. campestis and S. aureus (Supplementary Table S2). Although 1 and 2 did exhibit antibacterial activity, it was less potent than 3. It seems likely that the β-lactone ring of 3 plays an important role in the antibacterial activity.
Conclusion
We have discovered two new natural products, bisoxazolomycin (1) and oxazolomycin A2 (2), via PC screening technique. Both compounds showed cytotoxic and mildly antibacterial activities. PC screening is a powerful and comparatively simple technique to facilitate and expedite the discovery of new compounds from culture broths of microorganisms.
References
Nakashima, T., Takahashi, Y. & Ōmura, S. Search for new compounds from Kitasato microbial library by physicochemical screening. Biochem. Pharmacol. 15, 42–55 (2017).
Ōmura, S. et al. Irumamycin, an antifungal 20-membered macrolide produced by a Streptomyces: Taxonomy, fermentation and biological properties. J. Antibiot. 37, 1572–1578 (1984).
Mori, T et al. Structure of oxazolomycin, a novel β-lactone antibiotic. Tetrahedron Lett. 26, 1073–1076 (1985).
Kawai, S., Kawabata, G., Kobayashi, A. & Kawazu, K. Inhibitory effect of oxazolomycin on Crown Gall formation. Agric. Biol. Chem. 53, 1127–1133 (1989).
Eto, K., Yoshino, M., Takahashi, K., Ishihara, J. & Hatakeyama, S. Total synthesis of Oxazolomycin A. Org. Lett. 13, 5398–5401 (2011).
Acknowledgements
We are grateful to Dr Masato Iwatsuki for helpful advice. This project was supported by JSPS and NRCT under the Japan - Thailand Research Cooperative Program, the Thailand Research Fund under Royal Golden Jubilee Ph.D. Program (Grant No. PHD/0205/2553) and the Institute for Fermentation, Osaka (IFO), Japan.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies the paper on The Journal of Antibiotics website
Supplementary information
Rights and permissions
About this article
Cite this article
Koomsiri, W., Inahashi, Y., Kimura, T. et al. Bisoxazolomycin A: a new natural product from ‘Streptomyces subflavus subsp. irumaensis’ AM-3603. J Antibiot 70, 1142–1145 (2017). https://doi.org/10.1038/ja.2017.113
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ja.2017.113