Abstract
Tunicamycins (TUN) are inhibitors of the UDP-HexNAc: polyprenol-P HexNAc-1-P transferase family of enzymes, which initiate the biosynthesis of bacterial peptidoglycan and catalyze the first step in eukaryotic protein N-glycosylation. The TUN are therefore general and potent toxins to both eukaryotes and prokaryotes. Screening a library of synthetic TUN against Bacillus and yeast identified TUN that are antibacterial, but have significantly reduced eukaryotic toxicity. One of these (Tun-15:0) differs from the native TUN control only by the lack of the conjugated double bond in the tunicaminyl N-acyl group. Tun-15:0 also showed reduced inhibition for protein N-glycosylation in a Pichia-based bioassay. Natural TUN was subsequently modified by chemically reducing the N-acyl double bond (TunR1) or both the N-acyl and uridyl double bonds (TunR2). TunR1 and TunR2 retain their antibacterial activity, but with considerably reduced eukaryotic toxicity. In protein N-glycosylation bioassays, TunR1 is a less potent inhibitor than native TUN and TunR2 is entirely inactive. Importantly, the less toxic TunR1 and TunR2 both enhance the antibacterial activity of β-lactams: oxacillin by 32- to 64-fold, comparable with native TUN, and with similar enhancements for methicillin and penicillin G. Hence, the modified TUNs, TunR1 and TunR2, are potentially important as less-toxic synergistic enhancers of the β-lactams.
Similar content being viewed by others
Introduction
Antimicrobial resistance is one of the most serious health threats to both animals and humans.1, 2, 3, 4 The penicillins were developed by Howard Florey and coworkers in collaboration with the USDA in the 1940’s and now constitute over 850 different antibiotics.5, 6 Penicillins are a sub-class of the broad-spectrum β-lactam antibiotics that work by inhibiting the biosynthesis and cross-linking of the bacterial cell wall. Unfortunately, within 2 years of their introduction, Abraham and Chain7 had already reported ‘an enzyme from bacteria able to destroy penicillin’ (β-lactamase), which has all too quickly led to widespread resistance. More recently, Walker and others8, 9, 10, 11, 12 have shown that blocking the assembly of bacterial cell wall teichoic acid results in a concomitant attenuation of penicillin resistance. This signifies a possible new target for synergistic compounds that ‘re-sensitize’ resistant bacterial cells to β-lactam antibiotics.8, 12 Among these leads is the natural product tunicamycin (TUN), which inhibits the first step of bacterial wall teichoic acid biosynthesis, but which is also toxic to mammalian cells.12, 13, 14, 15 TUN is the most potent compound identified for this purpose and improves the antibacterial activity of oxacillin by 64-fold, with a similar enhancement for other β-lactams.8 A large-scale screen for effective wall teichoic acid inhibitors identified only two synthetic drugs, tarocin A and B, plus the TUNs from 2.8 million small molecules.12 TUN has potent wall teichoic acid pathway-specific inhibitory effects at ⩽0.1 mM, whereas tarocin A and B were notably less active (3–26 mM). However, the tarocins are less cytotoxicity against human HeLa cells (IC50, >100 mM) compared with natural TUN (IC50, 0.2 mM) and Lee et al.12 concluded that this toxicity precludes TUN as a viable β-lactam enhancing agent.
TUN is a potent inhibitor of the UDP-HexNAc: polyprenol-P HexNAc-1-P transferase family of enzymes.16, 17, 18 In bacteria, several polyprenol-P HexNAc-1-P transferase family members catalyze the early steps in cell wall biosynthesis, including peptidoglycan (by MraY family members) and teichoic acid (TagO transferases), and TUN is potently antibacterial by the inhibition of these activities. In eukaryotes, polyprenol-P HexNAc-1-P transferase homologs (Alg7p, GPT) catalyze the transfer of GlcNAc-1-phosphate to dolichol phosphate, an early step in protein N-glycosylation. Inhibition of this step by TUN leads to incomplete protein glycosylation and cellular death through activation of the unfolded protein response.18
In the present study we show that TUNs can be structurally modified to make them less toxic against eukaryotic cells, while retaining the potent antibacterial activity. More important, we show that these less toxicity TUNs (TunR1 and TunR2) enhance the antimicrobial activity of the β-lactams, the most important group of antibacterial agents in current usage.
Materials and methods
Materials, bacterial strains and culturing conditions
The chemicals, reagents and solvents used were obtained from Sigma-Aldrich, Inc., St Louis, MO, USA. Microbial strains, Saccharomyces cerevisiae strain NRRL Y2034 and Streptomyces niger NRRL B-3857 (formally Chainia nigra) were obtained from the ARS Microbial Collection housed in Peoria, IL, USA. Bacillus subtilis strain MW10 was obtained from the Bacillus Genetic Stock Center (Ohio State University, Columbus, OH, USA). Pseudomonas aeruginosa ATCC 27853 was from the American Type Culture Collection (Manassas, VA, USA). The strains were maintained on solid TYG agar (agar, 1.5%, tryptone, 2 g l−1, yeast extract, 2 g l−1, glucose, 6 g l−1 and MgCl2.6H2O, 0.3 g l−1) and grown aerobically in liquid TYG medium at 28 °C as described previously.19 Quinovosamycin was purified from S. niger NRRL B-3857 as described previously.19
Reverse-phase HPLC purification
HPLC purifications were essentially as described previously,19 using a Finnigan Surveyor instrument (ThermoFisher Scientific, West Palm Beach, FL, USA). The chromatography was achieved on a reversed phase Spheri-5 ODS column (250 mm, 5 mm particlesize) eluted with a solvent gradient from 45 to 100% aqueous acetonitrile for 15 min at a flow rate of 1 ml min−1. The detection was by diode array and samples were collected manually from multiple injection runs and subsequently analyzed by matrix-assisted laser desorption/ionization-time-of-flight MS.
Matrix-assisted laser desorption/ionization-time-of-flight MS spectra
Matrix-assisted laser desorption/ionization-time-of-flight mass spectra were obtained on a Bruker-Daltonic Microflex (Billerica, MA, USA) instrument operating in reflectron mode. The matrix use was 2,5-dihydrobenzoic acid. Ion source 1 and 2 were set to 19.0 and 14.0 kV, respectively, with lens and reflector voltages of 9.20 and 20.00 kV. The laser (337.1 nm) was typically at 60% of 150 μJ maximum output and 3000 shots are accumulated.
GC/MS carbohydrate analysis
Carbohydrate analysis of the HPLC-purified samples were acid hydrolyzed (2 M trifluoroacetic acid, 121 °C, 1 h). The residues were derivatized with hydroxylamine hydrochloride in pyridine, then peracetylated with acetic anhydride to form aldononitrile acetates. GC/MS analyses used a Shimadzu GC 2010 Plus instrument with an AOC 20i autoinjector (Shimadzu Scientific Instruments, Addison, IL, USA), interfaced with a Shimadzu QP2010 Ultra mass detector configured in electron impact mode. The chromatography used a capillary Zebron ZB-1 column (30 m; 0.25 mm) with helium (18.6 ml min−1) as the carrier gas. The oven temperature used a linear gradient from 150 to 250 °C at 4 °C min−1 with injector and detector/interface temperatures of 275 °C and 300 °C, respectively. Mass spectra were recorded in positive ion mode.
NMR spectra
NMR spectra were obtained on a Bruker Avance III instrument (Bruker BioSpin) operating at 500.11 MHz using a 5 mm z-gradient BBI probe at 27 °C. Chemical shifts are reported as p.p.m. from TMS calculated from the lock solvent. The samples under analysis were dissolved in deuterated methanol-d4 (Sigma-Aldrich, Inc.).
Microbial agar diffusion assays
Antibacterial activities were measured by an agar diffusion bioassay, essentially as described previously.19 B. subtilis strain MW10 and P. aeruginosa ATCC 27853 were used a model bacterial reporter strains and S. cerevisiae strain NRRL Y-2034 as a model eukaryote. Bacterial cultures were grown on tryptic soy broth and S. cerevisiae on yeast extract peptone dextrose medium. The cultures were adjusted to a density of 0.5 McFarland units, diluted 1:100 in 10 ml of molten (45 °C) 1% agar media (either tryptic soy broth or yeast extract peptone dextrose) and poured as soft agar overlays onto pre-set 1.5% agar plates (100 × 15 mm). After cooling, aliquots (1 μl, 0.5 mg ml−1 in dimethyl sulfoxide) of the TUN variants were applied to the microbial plates. These were incubated aerobically at 37 °C overnight and scored visually for a zone of growth inhibition.
Broth dilution assay for measuring the MIC values
MICs were determined as described previously.19 Serial two-fold dilutions of the various TUNs in growth media were introduced into the wells of a 96-well microtiter plate. Each well was inoculated with the test organism (Bacillus, yeast, or Pseudomonas; ~105 CFU ml−1) and the plates incubated for 8 h at 37 °C. Plates were scored for the lowest concentration of material that completely inhibited growth.
Protein N-glycosylation bioassays
The protein N-glycoylation bioassay uses a Pichia pastoris yeast engineered to secrete a recombinant plant chitinase (UniProt: Q9M2U5.1; NCBI accession: AAQ62423.1), as described previously.19 The Pichia cultures were grown for 1 day at 25 °C before MeOH-inducing the expression of the secrete recombinant AtchitIV N-glycoprotein for 8 h. The TUN variants under test (20 μg ml−1) were added to expression media at the same time as the methanol induction. Total secreted proteins were concentrated from cell-free media by Amicon ultra centrifugal filtered (EMD-Millipore, Billerica, MA, USA) and subsequently analyzed by SDS-polyacrylamide gel electrophoresis using oriole fluorescent stain (Bio-Rad, Hercules, CA, USA). As a positive control, the secreted recombinant AtchitIV glycoprotein from a non-treated Pichia culture was treated with PNGaseF (New England Biolabs, Ipswich, MA, USA) to remove N-glycan components before gel loading. In the bioassay, the AtchitIV5 glycoprotein is observed as a series of bands at 35–50 kDa on the SDS-polyacrylamide gel electrophoresis gels due to the mass difference of multiple N-glycosylation forms and the non-glycosylated apo-AtchitIV5 is observed as a single band of ~30 kDa.
Microtiter plate β-lactam antibacterial enhancement assays
The enhancements of the MICs for oxacillin, methicillin and penicillin G were measured in the presence of a sub-lethal quantity of the various TUN samples under test. The wells of the plates contain B. subtilis (or P. aeruginosa) grown on tryptic soy broth medium, with a concentration gradient of the β-lactams running from the top to bottom of the plate (0.5–0.004 μg ml−1 for oxacillin, 0.25–0.002 μg ml−1 for methicillin and 0.025–0.0002 μg ml−1 for penicillin G). The TUN variants are included in triplicate: dimethyl sulfoxide-negative control in columns 1–3, native TUN (0.2 μg ml−1) in columns 4–6, TunR1 (0.4 μg ml−1) in columns 7–9 and TunR2 (0.4 μg ml−1) in columns 10–12. The growth of the Bacillus (or Pseudomonas) was scored after 12 h by inclusion of a live/dead stain (resazurin), such that non-growth is purple and growing cells are colorless. The MIC is taken as the minimum concentration of the β-lactam at which cell growth is detectable.
Cell proliferation bioassay
We used the MTT (3-(4, 5-dimethylthiazol-2-yl)-2, 5-diphenyl tetrazolium- bromide) assay to determine the cell growth.20 MDA-MB-231cells (American Type Culture Collection) were plated in a series of 2 × 104 per well in 100 μl of Dulbecco’s modified Eagle’s medium high glucose medium containing 10% fetal bovine serum and 1% Chinese hamster ovary cells (from American Type Culture Collection) were plated in a series of 1 × 104 and 2 × 104 per well in 100 μl of Dulbecco’s modified Eagle’s medium high glucose containing 10% fetal bovine serum and 1% penicillin/streptomycin. The cells were incubated with carrier (dimethyl sulfoxide) and 0.5, 1, 2 and 5 μg ml−1 of native TUN, TunR1, TunR2 or quinovosamycin respectively, for 24 h. The MTT reagent (100 μl; 5 mg ml−1; Invitrogen, Carlsbad, CA, USA) was added to each well and the plates were incubated for 2 h at 37 °C. The formazan product in cells was dissolved in 100 μl of dimethyl sulfoxide and absorbencies were read at 550 nm on a BioRad-microtiter plate reader.
TUN modification chemistry
Reduction of the conjugated double bond in the N-acyl chain of commercial TUN to give TunR1 was based on Tran et al.30 In a typical reaction, the TUN (10 mg, Sigma-Aldrich) was dissolved in methanol:toluene (1:1 v/v, 2 ml), plus glacial acetic acid (10 μl). A catalytic amount of solid palladium on carbon catalyst (10% Pd/C, 3 mg) was added followed by an addition of solid sodium borohydride (7 mg). The reaction was capped and held at 25 °C for 30 min. This was then filtered through a pad of Celite filter aid to remove the spend Pd/C catalyst. The filtrate was evaporated to dryness, and re-dissolved and refluxed in methanol (2 ml, 80 °C, 15 min). This latter step is to convert the residual sodium borohydride to methyl borate and residual acetic acid to methyl acetate, which were subsequently removed by evaporation. The material produced upon evaporation is the single reduced TunR1. Control reactions were run that either lacked the Pd/C catalyst (but included the borohydride) or lacked both Pd/C and the borohydride. The reaction was then optimized for co-reduction of the urydyl 5,6-double bond, initially using uridine as a model compound. Using the condition above at higher temperature and a longer reaction time (60 °C, 50 h) resulted in complete reduction of the uridine double bond. This optimized reaction was then applied to commercial TUN to effect the double reduction of the 2′′′,3′′′-alkene and 5,6-uridyl double bonds. Cooling and work up as described above gave the double reduced TunR2.
Results
QSAR study of the antimicrobial activity (bacterial and eukaryotic) of a chemically diverse TUN library
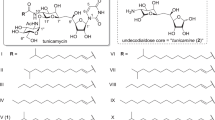
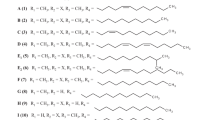
A quantitative structure–activity relationship (QSAR) study was undertaken to identify TUN structures that have potent inhibitory effects on bacteria, and have low toxicity in eukaryotes. A library of TUN analogs have been chemically synthesized using a newly developed modular approach,22 but biological testing of the library had not been undertaken. These structurally diverse synthetic TUNs (1–10) were tested for their antimicrobial activity against a model Gram-positive bacterium (B. subtilis) and a model eukaryote (S. cerevisiae, bakers’ yeast) (Figure 1a and Supplementary Table 1). The structural validity of the compounds in the library was confirmed by matrix-assisted laser desorption/ionization-time-of-flight MS (Supplementary Figure 1) and were tested at equivalent concentrations for microbial growth inhibition using a standard agar diffusion-zone of inhibition assay (Figure 1a). Native TUN (T) displayed potent toxicity (0.5 μg spotted) against Bacillus and yeast, giving a zone of clearing of ~2.5 cm. The chemically synthesized TUN 2 (Tun-15:1) gave an antimicrobial activity equivalent to the native TUN against both test organisms. These compounds both have the 2′′′,3′′′-conjugated double bond in the N-linked fatty acid chain (Figure 1d). Synthetic TUN 3 (Tun-15:0), which only differs from 2 in that it contains a fully saturated N-acyl chain, gave a comparable response on the Bacillus but, unexpectedly, was considerably less toxic than T or 2 against the yeast cells. A similar result was observed for 4 that has a 2′′-azido group replacing the 2′′-acetylamino found on the native TUN. Other analogs in the library (1 and 5–10) lacked antimicrobial activity on either organism. These include 5, which has an N-phthaloyl group in place of the N-linked fatty acid chain, 1, 6 and 9, which lack an N-acyl chain entirely, 6, 7 and 8, which have a p-methoxyphenol group in place of the N-uracil of native TUN, and 10, in which the N-uracil is replaced by N-adenine. From this study we concluded that the N-uracil group and the N-linked fatty acid chain are essential for the biological activity of TUN on prokaryotes and eukaryotes. Furthermore, the presence of the 2′′′,3′′′-unsaturated double bond in the N-linked fatty acid does not affect the antibacterial activity, but the lack of this double bond significantly reduces the eukaryotic toxicity of the TUN.
Quantitative structure–activity relationship (QSAR) studies to identify TUN structures that have potent inhibitory effects on bacteria, and have low toxicity in eukaryotes. (a) Synthetic TUN 3 (with a saturated N-acyl chain) and 4 (with a 2-azidoglucose) are less toxic on yeast, but have potent antibacterial activity on B. subtilis. Natural TUN (T) is the positive control, B is the blank control. Compounds 1 and 5–10 have no activity on either organism. The compounds (0.5 μg) were incorporated into the agar and overlaid with the Bacillus or yeast. (b) SDS-polyacrylamide gel electrophoresis (PAGE) analysis of recombinant glycoprotein secreted by Pichia yeast (glycosylated AtchitIV5, A; non-glycosylated AtchitIV5 apoprotein, (b). The TUN treatments used (20 μg ml−1 for 8 h) are synthetic TUNs, lanes 1–10; TUN natural product, lane 11; and dimethyl sulfoxide control, lane 12. Compound 3 partial inhibits protein glycosylation. (c) Relative activity of chemically reduced TUNs (TunR1 and TunR2, 0.5 μg) on Bacillus (top) or yeast (lower). MICs in the broth dilution assays are indicated by black arrows. Similar antimicrobial activity is seen on the Bacillus, whereas TunR1 and especially TunR2 are significantly less toxic on yeast. (d) Chemical structure of TUN. The fatty acid 2′′′,3′′′-acyl and the uracil 5,6-double bonds are indicated by arrows. A full colour version of this figure is available at the Journal of Antibiotics journal online.
QSAR study using a Pichia-based bioassay for protein N-glycosylation
From the QSAR antimicrobial results, it was apparent that certain synthetic TUNs in the Yu library (3 and 4) had reduced toxicity on the model eukaryote (yeast) relative to native TUN. As the known mechanism for the toxicity of TUN towards eukaryotes occurs via inhibition of the first step of protein N-glycosylation,16, 17, 18 it was decided to test the synthetic TUNs in the Yu library using a Pichia-based assay for protein glycosylation. A bioassay was developed to test this, making use of an Arabidopsis gene for a known N-glycoprotein (AtchitIV) expressed from a methanol-inducible promoter in Pichia yeast.19 This Pichia strain secretes recombinant AtchitIV N-glycoprotein into the culture medium, from which it is readily separated and visualized by SDS-polyacrylamide gel electrophoresis (Figure 1b). Synthetic TUNs (20 μg ml−1) were introduced into the Pichia cultures concomitant with the induction of AtchitIV5 expression by the addition of methanol. In the absence of TUN (Figure 1b, lane 12) the AtchitIV5 protein is apparent as a series of bands (band A) differing in gel mobility due to multiple N-glycosylation forms (Figure 1b). The addition of natural TUN results in a single band of ~30 kDa (band B) corresponding to the non-glycosylated form of AtchitIV5 (lane 11). Hence, TUN blocks the N-glycosylation of AtchitIV5. Similarly, the chemically-synthesized TUN 2 from the Yu library (Lane 2) has the same effect as the natural TUN in Lane 11. In contrast, the synthetic TUN analog 3 without the 2′′′,3′′′-unsaturated double bond in the N-linked fatty acid (Lane 3) is less effective at blocking the N-glycosylation of AtchitIV5, so that it appears on the gel as an approximate 50:50 mixture of bands A and B (Figure 1b). Noticeably, none of the other inhibitors tested showed any activity in this protein N-glycosylation assay. From this we concluded that the lack of the 2′′′,3′′′-double bond moiety on TUN significantly lowers the activity to block protein N-glycosylation and it is this mechanism that lowers the toxicity of 3 towards eukaryotes (Figure 1a).
The relative antimicrobial activity of chemically modified TUNs on bacteria and eukaryotes
Having identified the TUN N-acyl double bond as a chemophore, we chemically reduced the two double bond positions in native TUN (Figure 1d). Treatment with sodium borohydride in the presence of palladium on carbon catalyst (pH 4.0, r.t., 30 min) selectively and quantitatively reduced the 2′′′,3′′′-double bond in the TUN N-acyl chain (Figure 2). This compound is referred to as acyl-reduced TunR1. Longer reaction time at elevated temperature (50 h; 60 °C) with the same reagents gave TUN reduced in both the 2′′′,3′′′-double bond and the uracil 5,6-double bond, giving a double reduced TUN referred to as TunR2 (Figure 2). We then tested the chemically-modified TUNs, TunR1 and TunR2, for antimicrobial activity against B. subtilis, and the model eukaryote S. cerevisiae using the same agar diffusion-zone of inhibition assay as used in the QSAR study, plus a quantitative broth dilution assay (Figure 1c). In the agar diffusion assay (Figure 1c., right panel) the native TUN (T) gives a zone of clearing of~2.5 cm against both organisms. TunR1 and TunR2 both have similarly potent activity against the Bacillus. However, TunR1 is about 10-fold less active against the yeast (zone of clearing~0.3 cm), similar to that seen for 3 from the Yu synthetic library (Figure 1a). Both of these compounds (TunR1 and 3), although produced in different ways, have an N-acyl group that lacks the 2′′′,3′′′-double bond. More dramatically, and unexpectedly, we observed that the modified double-reduced TunR2 was completely non-toxic to the yeast, giving no zone of clearing, but retaining full activity against the B. subtilis (Figure 1c).
Selective chemical reduction of the two double bonds in TUN. (a) Proton NMR spectra of the 5.6–8.2 p.p.m. region of modified TUNs. Characteristic signals for the TUN fatty acid H-2′′′ and H-3′′′ protons (TUN) are absent after mild reduction (24 °C, 30 min; TunR1), indicating loss of the acyl double bond. These signals and the uracil H-5 and H-6 signals disappear after prolonged reduction (60 °C, 72 h; TunR2), showing that double reduction has occurred. The alpha-H-1′ anomeric signal due to the TUN sugar motif is unaffected by the reductions. (b) Matrix-assisted laser desorption/ionization (MALDI) MS of native TUN (control), and modified TUNs (TunR1 and TunR2). The MS peaks for Tuns 14, 15, 16 and 17 correspond to the TUNs with increasing fatty acid chain length (m/z 839, 853, 867 and 881). The mass increases of 2 Da (on TunR1) corresponds to the reduction of the 2,3-acyl double bonds and of +4 Da (on TunR2) for reduction of both the N-acyl and the uracil double bonds. (c) Reverse-phase HPLC analysis of double reduced TunR2 (Tun-14, 3.2 min; Tun-15, 4.2 min; Tun-16, 5.4 min; Tun-17, 6.5 min. (d) MALDI-MS analysis of HPLC-purified TunR2 components.
In a quantitative broth dilution assay native TUN has MICs of 0.15 and 0.3 μg ml−1 on the Bacillus and yeast, respectively (Figure 1c, left). TunR1 has a comparable antibacterial activity (MIC of 0.3 μg ml−1 on the Bacillus), but has considerably reduced activity on the yeast (MIC of 2.5 μg ml−1). The double reduced TunR2 also has a similar antibacterial activity (MIC of 0.3 μg ml−1), but it is effectively non-toxic (MIC >10 μg ml−1) against the eukaryotic yeast. Hence, the broth dilution assays confirm the results found with the agar diffusion assay, and show that the modified TunR1 and TunR2 have greatly reduced toxicity against eukaryotes, but retain their antibacterial activity.
Relative inhibitory activity of TUN, TunR1 and TunR2, in the Pichia-based protein N-glycosylation bioassay
To determine a mechanism for this reduced eukaryotic toxicity we assayed native TUN, TunR1, and TunR2 for their relative ability to inhibit protein N-glycosylation (Figure 3a). Pichia-expressed AtchitIV5 is secreted as the non-glycosylated apoprotein (B; Figure 3a) in the presence of tunicamycin. With acyl-reduced TunR1 only ~50% of AtchitIV5 is non-glycosylated, which is similar to the result obtained with the synthetic 3 from the Yu library (Figure 1c, lane 3). Importantly, the double reduced TunR2 was entirely inactive in this assay (Figure 3a), suggesting that the lower toxicity on yeast cells is due to a reduced efficacy to block protein N-glycosylation.
Native TUN and less toxic NP1 and NP2 enhance the activity of β-lactams. (a) SDS-polyacrylamide gel electrophoresis (PAGE) analysis of N-glycoprotein secreted by Pichia after treatment with TUN (T), acyl reduced TunR1 or double reduced TunR2 (20 μg ml−1 for 8 h). The heterologously expressed glycoproteins (A: glycosylated AtchitIV5 and B: non-glycosylated AtchitIV5 apoprotein) were analyzed as described.19 Lane C: untreated control. The rightmost lane is molecular weight markers. (b) Relative toxicity on mammalian cells. 1. Human MDA-MB-231; 2. Chinese hamster ovary (CHO) cells. (c) Relative enhancement of β-lactams (oxacillin, methicillin and penicillin G) against B. subtilis.
Relative cytotoxicity of TUN, TunR1 and TunR2, against cultured Chinese hamster ovary and human cancer cell lines
The comparative cytotoxicity of native and modified TUNs was also assessed against mammalian cell lines. Chinese hamster ovary cells and MDA-MB-231 triple negative human breast cancer cells were chosen for study, because both have well-defined sensitivity to native TUNs.17, 23, 24 These cell types were treated with either natural TUN or chemically reduced TUNs (TunR1 or TunR2) at various concentrations. Cellular proliferation was monitored by OD (OD at 550 nm) Figure 3b and Supplementary Results). The inclusion of native TUN resulted in a 50% decrease of proliferation of the human cancer cells at 0.5 μg ml−1. TunR1 is 5–10% less toxic, but at higher concentration (5 μg ml−1) is equally toxic as the native TUN. TunR2 is substantially less toxic in this assay, and is essentially non-toxic to a concentration of 1–5 μg ml−1. Similar data were obtained on Chinese hamster ovary cells, with the observed relative toxicity TUN>TunR1>TunR2 (Supplementary Results).
Relative enhancement of the antibacterial activity of β-lactams (oxacillin, methicillin and penicillin G) against Gram-positive and Gram-negative bacteria by TUN, TunR1 and TunR2
The aim was to test whether TUNs could be produced with reduced eukaryotic toxicity, while retaining their ability to enhance the antimicrobial activity of the β-lactams. We therefore tested native TUN, TunR1 and TunR2 in β-lactam antibacterial enhancement assays, using concentration levels less than the observed toxicity against yeast cells (Figure 3c and see Figure 1c). The MICs for oxacillin (0.25 μg ml−1), methicillin (0.125 μg ml−1) and penicillin G (0.025 μg ml−1) were determined against B. subtilis using a microtiter plate dilution assay (Figure 3c). The inclusion of TUN (0.2 μg ml−1) in the assay lowered these MICs by 32-fold (0.08 μg ml−1), 4-fold (0.031 μg ml−1) and 2-fold (0.0125 μg ml−1), respectively. Similar enhancements of the β-lactams were obtained with TunR1 and TunR2 (0.4 μg ml−1) using concentrations at below the MIC for these compounds (Figure 3c). Hence, against Gram-positive bacteria the less toxic TunR1 and TunR2 are as effective as native TUN for enhancement of the β-lactams. We also note that the β-lactam enhancement was not observed against Gram-negative bacteria (P. aeruginosa), which lack either teichoic acid or the TagO target protein (Supplementary Fig. 4).
Discussion
In the present study we report that reduction of the double bonds in TUN results in lower eukaryotic toxicity while maintain the antibacterial activity and the enhancement of β-lactams. The structure of a TUN-MraY complex has recently been described in which TUN binds at a cytoplasmic cavity of MraY, a bacterial polyprenol-P HexNAc-1-P transferase transmembrane protein.25 Significantly, the TUN uracil ring is shown to bind to a cavity composed of Gly-176, Asn-221 and Phe-228, and mutating the latter two residues entirely blocks the MraY activity. Two other essential residues, Asp-175 and Glu-300, are close to the 2-carbonyl of the uracil ring and Lys-74 is coordinated to the 2-carbonyl (Figure 4 and Table 1). This TUN uracil-binding pocket (UBP) shares a partial overlapping binding mode also found for the uracil group of muraymycin D, a related nucleotide inhibitor of MraY.26, 27 Comparable π–π stacking of Phe-228 occurs with both TUN and muraymycin and also with a truncated MraY substrate, UDP-MurNAc-L-Ala, providing a mechanism for the binding of the tunicaminyl uracil motif.25, 27 This Phe residue is located in the active site of the Clostridium MraY, on the cytoplasmic face between helices 7 and 8, and is highly conserved with both the human and yeast UBP sequences (NWYPSRVFVGDTFCY and NRWPATVFVGDTYCY, respectively)25, 28 (Table 1).
The binding interaction of TUN with bacterial MraY protein based on Hakulinen et al.25. Note the π–π stacking between active site Phe-228 and the tunicaminyl uracil ring. This is potentially disrupted by the non-planar reduced 5,6-dihydrouracil ring of the reduced TUN, resulting in less efficient binding. The colors of the amino acid residues are as defined in Table 1.
There are, however, noticeable differences between the bacterial and eukaryotic UBP sequences, most notably the Met residue following F228 (M229) and a subsequent G233-S234 couple four residues further away (Table 1 and Figure 4). Human, yeast, plant, slime mold and insect Alg7p proteins, for example, do not contain the conserved bacterial FMGDXGSX motif, and this is replaced by a conserved Val in place of Met229 and a Phe/Tyr-X-Tyr in place of the G233-S-234-X (Table 1). Noticeably, other family members of the bacterial UDP-HexNAc: polyprenol-P transferase enzymes including WecA, WbcO and TagO also contain the conserved FMGDXGSX sequence more typical of the bacterial UBP (Table 1). It is conceivable that it is this variation within the TUN UBP motifs, especially between that of the eukaryotic Alg7p and bacterial TagO proteins, which underlie the different biological responses to the double-reduced TUNs that we have observed. The loss of planarity of the TUN uracil ring upon reduction to 5,6-dihydrouracil would lead to a reduction in the π-bond overlap with Phe-228 and hence to reduced binding (Figure 4). This is presumably more evident for the eukaryotic-type UBP, leading to a selective reduction in toxicity of the reduced TUNs towards eukaryotic cells.
A mechanism for the lowering of eukaryotic toxicity due to reduction of the conjugated double bond in the TUN N-acyl groups (as in TunR1 and TunR2) will require further work. However, we note that bacterial members of the UDP-HexNAc: polyprenol-P transferase enzyme family (such as MraY and TagO) typically utilize undecaprenyl phosphate (bactoprenol phosphate) as the membrane-bound isoprenol acceptor substrate, whereas eukarotic Alg7p transferases use a variety of dolichol phosphates. Undecaprenyl differs structurally from dolichols in that it is shorter chain length, and also because the terminal α-isoprenol group of undecaprenol contains an unsaturated double bond that is absent for the dolichols.29 The N-acyl chain of the TUNs is a structural analog of the isoprenol phosphates in the transition state of these enzymes.25 Moreover, there is a requirement for the saturated α-prenol of dolichol for effective protein N-glycosylation to occur in eukaryotes, which is not needed for the analogous biosynthesis of bacterial cell walls.20, 21, 29 We propose that this selective requirement for saturated dolichol-P substrates by the eukaryotic UDP-HexNAc: polyprenol-P transferase enzyme family may also distinguish the binding differences between the native 2′′′,3′′′-unsaturated TUN and the modified 2′′′,3′′′-saturated TUNs (TunR1 and TunR2), and hence to the difference in their relative toxicities on eukaryotes.
Dedication
We dedicated this paper to the memory of our friend, colleague and co-author Dr Kenneth Bischoff, who sadly passed away on 18 March of this year.
References
Perros, M. Infectious disease. A sustainable model for antibiotics. Science 347, 1062–1064 (2015).
Baker, S. Infectious disease. A return to the pre-antimicrobial era? Science 347, 1064–1066 (2015).
Andersson, D. I. & Hughes, D. Antibiotic resistance and its cost: is it possible to reverse resistance? Nat. Rev. Microbiol. 8, 260–271 (2010).
Wright, G. D. Something old, something new: revisiting natural products in antibiotic drug discovery. Can. J. Microbiol. 60, 147–154 (2014).
Brown, K. Penicillin Man. Alexander Fleming and the Antibiotic Revolution (Sutton Publishing, Stroud, Gloucestershire, 2004).
Drawz, S. M. & Bonomo, R. A. Three decades of β-lactamase inhibitors. Clin. Microbiol. Rev. 23, 160–201 (2010).
Abraham, E. P. & Chain, E. An enzyme from bacteria able to destroy penicillin. Nature 146, 837–837 (1940).
Campbell, J. et al. Synthetic lethal compound combinations reveal a fundamental connection between wall teichoic acid and peptidoglycan biosyntheses in Staphylococcus aureus. ACS Chem. Biol. 6, 106–116 (2011).
Brown, S. et al. Methicillin resistance in Staphylococcus aureus requires glycosylated wall teichoic acids. Proc. Natl Acad. Sci. USA 109, 18909–18914 (2012).
Farha, M. A. et al. Inhibition of WTA synthesis blocks the cooperative action of PBPs and sensitizes MRSA to β-lactams. ACS Chem. Biol. 8, 226–233 (2013).
Reed, P. et al. Staphylococcus aureus survives with a minimal peptidoglycan synthesis machine but sacrifices virulence and antibiotic resistance. PLoS Pathog. 11, e1004891 (2015).
Lee, S. H. et al. TarO-specific inhibitors of wall teichoic acid biosynthesis restore β-lactam efficacy against methicillin-resistant staphylococci. Sci. Transl. Med. 8, 329ra32 (2016).
Takatsuki, A., Arima, K. & Tamura, G. Tunicamycin, a new antibiotic. I. Isolation and characterization of tunicamycin. J. Antibiot. (Tokyo) 24, 215–223 (1971).
Tamura, G. Tunicamycins, Japan Scientific Press, Tokyo, (1982).
Price, N. P. & Tsvetanova, B. Biosynthesis of the tunicamycins: a review. J. Antibiot. (Tokyo) 60, 485–491 (2007).
Heifetz, A., Keenan, R. W. & Elbein, A. D. Mechanism of action of tunicamycin on the UDP-GlcNAc:dolichyl-phosphate GlcNAc-1-phosphate transferase. Biochemistry 18, 2186–2192 (1979).
Mclachlan, K. R. & Krag, S. S. Substrate specificity of N-acetylglucosamine 1-phosphate transferase activity in chinese hamster ovary cells. Glycobiology 2, 313–319 (1992).
Lehrman, M. A. Biosynthesis of N-acetylglucosamine-P-P-dolichol, the committed step of asparagine-linked oligosaccharide assembly. Glycobiology 1, 553–562 (1991).
Price, N. P. J. et al. Quinovosamycins: new tunicamycin-type antibiotics in which the α, β-1″,11′-linked N-acetylglucosamine residue is replaced by N-acetylquinovosamine. J. Antibiot. 69, 637–646 (2016).
Cantagrel, V. et al. SRD5A3 is required for converting polyprenol to dolichol and is mutated in a congenital glycosylation disorder. Cell 142, 203–217 (2010).
Jozwiak, A. et al. Polyprenol reductase2 deficiency is lethal in Arabidopsis due to male sterility. Plant Cell 27, 3336–3353 (2015).
Li, J. & Yu, B. A modular approach to the total synthesis of tunicamycins. Angew. Chem. Int. Ed. Engl. 54, 6618–6621 (2015).
Bannerjee, A. et al. N-Acetylglucosaminyl 1-phosphate transferase: an excellent target for developing new generation breast cancer therapeutic. Adv. Exp. Med. Biol. 842, 355–374 (2015).
Banerjee, A. et al. Unfolded protein response is required in nu/nu mice microvasculature for treating breast tumor with tunicamycin. J. Biol. Chem. 286, 29127–29138 (2011).
Hakulinen, J. K. et al. MraY–antibiotic complex reveals details of tunicamycin mode of action. Nat. Chem. Biol. 13, 265–267 (2017).
Chung, B. C. et al. Crystal structure of MraY, an essential membrane enzyme for bacterial cell wall synthesis. Science 341, 1012–1016 (2013).
Chung, B. C. et al. Structural insights into inhibition of lipid I production in bacterial cell wall synthesis. Nature 533, 557–560 (2016).
Furlong, S. E. & Valvano, M. Characterization of the highly conserved VFMGD motif in a bacterial polyisoprenyl-phosphate N-acetylaminosugar-1-phosphate transferase. Protein Sci. 21, 1366–1375 (2012).
Hartley, M. D. & Imperiali, B. At the membrane frontier: a prospectus on the remarkable evolutionary conservation of polyprenols and polyprenyl-phosphates. Arch. Biochem. Biophys. 517, 83–97 (2012).
Tran, A. T., Huynh, V. A., Friz, E. M., Whitney, S. K. & Cordes, D. B. A general method for the rapid reduction of alkenes and alkynes using sodium borohydride, acetic acid, and palladium. Tetrahedron Lett. 50, 1817–1819 (2009).
Acknowledgements
BY acknowledges the support of the National Natural Science Foundation of China (21372253 and 21432012). We thank WW Metcalf (University of Illinois, Champaign, IL, USA) and JO Rich (ARS-USDA, Peoria, IL, USA) for the initially review of the manuscript. A provisional patent application (patent number 62/450,760) has been filed. Mention of any trade names or commercial products is solely for the purpose of providing specific information and does not imply recommendation or endorsement by the US Department of Agriculture. USDA is an equal opportunity provider and employer.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies the paper on The Journal of Antibiotics website
Supplementary information
Rights and permissions
About this article
Cite this article
Price, N., Hartman, T., Li, J. et al. Modified tunicamycins with reduced eukaryotic toxicity that enhance the antibacterial activity of β-lactams. J Antibiot 70, 1070–1077 (2017). https://doi.org/10.1038/ja.2017.101
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ja.2017.101
This article is cited by
-
The tunicamycin derivative TunR2 exhibits potent antibiotic properties with low toxicity in an in vivo Mycobacterium marinum-zebrafish TB infection model
The Journal of Antibiotics (2024)
-
Synergistic enhancement of beta-lactam antibiotics by modified tunicamycin analogs TunR1 and TunR2
The Journal of Antibiotics (2019)
-
Liposidomycin, the first reported nucleoside antibiotic inhibitor of peptidoglycan biosynthesis translocase I: The discovery of liposidomycin and related compounds with a perspective on their application to new antibiotics
The Journal of Antibiotics (2019)
-
Selective catalytic hydrogenation of the N-acyl and uridyl double bonds in the tunicamycin family of protein N-glycosylation inhibitors
The Journal of Antibiotics (2017)