Abstract
Rifampicin (RIF) resistance is a risk factor for poor outcome in tuberculosis (TB). In Mycobacterium tuberculosis, both target gene mutation and efflux pumps have major roles in the resistance to anti-TB drugs. This study aimed to determine whether RIF induces efflux pump activation in RIF-monoresistant M. tuberculosis strains. Here, we took advantage of 16 RIF-monoresistant M. tuberculosis clinical isolates to evaluate the expression of 27 putative drug efflux pump genes and measured the influence of four drug efflux pump inhibitors, carbonyl cyanide m-chlorophenyl hydrazone (CCCP), verapamil (VP), thioridazine (TZ) and chlorpromazine (CPZ), on the RIF MICs of these strains. Eight of the 16 RIF-monoresistant isolates carried mutations in rpoB and overexpressed one or two of the following putative efflux pump genes: Rv2333, drrB, drrC, Rv0842, bacA and efpA. CCCP, VP, TZ and CPZ lowered the RIF MICs greater than fourfold in 6, 12, 9 and 12 isolates, respectively. The lowered RIF MICs by VP and CPZ were identical and stronger than CCCP (P-values were all 0.033). In conclusion, the efflux pumps Rv2333, DrrB, DrrC, Rv0842, BacA and EfpA may have a role in RIF resistance in addition to classical mutations in the rpoB gene, and the addition of VP and CPZ could significantly increase RIF susceptibility in RIF-monoresistant M. tuberculosis.
Similar content being viewed by others
Introduction
Tuberculosis (TB) is still a major public health problem, accounting for 8.6 million new cases and 1.3 million deaths each year.1 Moreover, the emergence of multidrug-resistant TB (MDR-TB) and extensively drug-resistant TB have hampered the effective TB treatment and control. Rifampicin (RIF) is one of the most important anti-TB antibiotics; it exerts its bactericidal activity by arresting DNA-directed RNA synthesis of Mycobacterium tuberculosis by interacting with the β subunit of RNA polymerase.2, 3 Previous research has demonstrated that >95% of RIF-resistant isolates are associated with mutations in the 81-bp region (codons 507–533) called the RIF resistance-determining region of the rpoB gene. However, several studies demonstrated that RIF induced differential expression of efflux genes in MDR-TB isolates based on large-scale transcriptional data.4, 5, 6 Some efflux pumps selectively extrude specific antibiotics, whereas MDR pumps expel a variety of structurally diverse compounds with differing antibacterial modes of action. For M. tuberculosis, the in vivo activation of both drug-specific and broadly active efflux pumps has been demonstrated for several anti-TB drugs, including isoniazid, ethambutol and fluoroquinolone.7, 8 A previous study has demonstrated that efflux pumps contribute to RIF resistance in RIF-monoresistant M. tuberculosis without mutations in rpoB.9 By eliminating the effect of polyresistance, the choice of RIF-monoresistant strains may allow us to identify RIF-specific efflux pump genes. The aim of this study was to determine the efflux pump genes contributing to RIF resistance in RIF-monoresistant clinical M. tuberculosis isolates carrying mutations in rpoB in China for the first time.
Materials and Methods
Ethical approval
This study obtained approval from the Ethics Committee of the National Institute for Communicable Disease Control and Prevention, Chinese Center for Disease Control and Prevention. The patients with TB included in this research were given a subject information sheet, and all of them gave written informed consent to participate in the study.
Bacterial strains
Sixteen RIF-monoresistant (resistant to RIF but sensitive to seven other drugs tested) M. tuberculosis clinical isolates that were isolated from 16 adult patients with pulmonary TB during the period of 2004–2006 were collected in seven provincial TB hospitals in China including Anhui, Guangxi, Henan, Hunan, Sichuan, Shanxi and Tibet. H37Rv (ATCC 27294) was included as a control.
Antibiotics and chemicals
Middlebrook 7H9 broth and albumin-dextrose-catalase supplement were purchased from Difco (Detroit, MI, USA). Isoniazid (INH), RIF, ethambutol (EMB), streptomycin (STR), ofloxacin, kanamycin, capreomycin, amikacin, carbonyl cyanide m-chlorophenyl hydrazone (CCCP), verapamil (VP), thioridazine (TZ) and chlorpromazine (CPZ) were purchased from Sigma–Aldrich Company (St Louis, MO, USA). All solutions were prepared on the day of the experiment. Trizol was purchased from Invitrogen (Carlsbad, CA, USA). Alamar blue was purchased from AbD Serotec (Oxford, UK).
Conventional drug susceptibility testing and Mycobacterium species identification
Clinical isolates were tested for susceptibility to four first-line anti-TB drugs (INH, RIF, STR and EMB), and four second-line anti-TB drugs (kanamycin, ofloxacin, capreomycin and amikacin) using a proportion method with Lowenstein–Jensen plates as described by the World Health Organization.10 The concentrations of drugs in medium were: 0.2 μg ml−1 INH, 40 μg ml−1 RIF, 4 μg ml−1 STR, 2 μg ml−1 EMB, 30 μg ml−1 kanamycin, 2 μg ml−1 ofloxacin, 40 μg ml−1 capreomycin10 and 30 μg ml−1 amikacin.11 Lowenstein–Jensen medium containing paranitrobenzoic acid (500 μg ml−1) was used to identify M. tuberculosis complex species from non-TB mycobacteria, and medium containing thiophen-2-carboxylic acid hydrazide (5 μg ml−1) was used to exclude Mycobacterium bovis from the M. tuberculosis complex. This study included the M. tuberculosis complex but did not include M. bovis clinical isolates.
PCR amplification and DNA sequencing of the rpoB gene
Genomic DNA was extracted from freshly cultured bacteria by the cetyltrimethylammonium bromide method. A 450-bp region of the rpoB gene containing the 81-bp resistance-determining region was amplified by polymerase chain reaction (PCR). The forward primer was 5′-ACCGACGACATCGACCACTT-3′, and the reverse primer was 5′-GTACGGCGTTTCGATGAACC-3′. Using a 50-μl PCR mixture (Kangwei Biotechnology, Beijing, China), the PCR reactions were denatured at 94 °C for 5 min and subjected to 35 cycles of 94 °C for 30 s, 62 °C for 30 s and 72 °C for 30 s, followed by a final extension at 72 °C for 10 min. Partial PCR sequence was characterized by DNA sequencing using the specific primers on an ABI Prism 3730 automated DNA sequencer (ABI Prism). The resulting DNA sequences were analyzed using BLAST (http://www.ncbi.nlm.nih.gov/BLAST). The specific mutations in protein sequences of individual isolates were identified.
Spoligotyping and data analysis
Spoligotyping was performed using 43 covalently bound oligonucleotides derived from the spacer sequences of M. tuberculosis H37Rv and M. bovis BCG P3 as previously described by Kamerbeek et al.12 The results in binary format were entered in an Excel spreadsheet and compared with the spoligotyping database SpolDB4 (http://www.pasteur-guadeloupe.fr:8081/SITVITDemo/index.jsp).
IS6110 in the NTF region
All Beijing family strains identified by the spoligotyping were amplified by PCR to detect the presence or absence of IS6110 in the NTF region13 using the following primers: F-6110 (5′-CCAGATATCGGGTGTGTCGAC-3′) and R-6110 (5′-TGCCGTTGTCGAAATCTAAACCC-3′). Strains with the insert yielded an amplified product of ~1100 bp, whereas those without the insert yielded an ~300-bp PCR product.
Determination of MICs
To determine the RIF MICs of 16 clinical isolates and H37Rv, the microplate alamar blue assay was performed according to the protocol of Leonard et al.14 The effect of the efflux pump inhibitors (EPIs) CCCP, VP, TZ and CPZ on the RIF MIC was also studied by incorporating the inhibitors at subinhibitory concentrations in the M. tuberculosis cultures in the assay. Twofold serial dilutions of RIF (resulting in a range of concentrations from 0.001 to 256 μg ml−1) were made directly in the wells in the absence or presence of 1.6 μg ml−1 CCCP, 64 μg ml−1 VP, 2 μg ml−1 TZ or 4 μg ml−1 CPZ. The above concentrations for four EPIs were half the MIC values against 92 M. tuberculosis isolates (data not shown). The MIC was defined as the lowest drug concentration that prevented a change in color. Strains with an MIC⩽1 μg ml−1 were considered to be susceptible to RIF.14 All tests for each strain were carried out at least in duplicate to calculate a mean MIC for each strain.
Expression of efflux pump genes
Selected RIF-monoresistant isolates and H37Rv were cultured in duplicate in enriched 7H9 medium in the presence or absence of RIF (1/4 the MIC of each strain) for 4 weeks. Total RNA was extracted by Trizol according to the instructions of the manufacturer. After treatment with DNase I (Invitrogen), the complementary DNAs were reverse-transcribed from 1.5 μg of total RNA with a TransScript First-Strand cDNA Synthesis SuperMix Kit (Transgen Bio-Technology Company, Beijing, China). Quantitative reverse transcription-PCR was performed in a 20-μl system containing 10 μl 2 × mixture supplied with SYBR Green, 10 ng complementary DNA template and 3 pmol each primer set. Primers for quantitative reverse transcription-PCR analysis are listed in Table 1. The ΔCT values were obtained by subtracting the CT value of the secA1 gene from the CT value obtained for each gene, and ΔΔCT was calculated by subtracting the ΔCT values of the non-induced strain from the corresponding RIF-induced ΔCT for each gene tested. The quantity of messenger RNA in RIF-induced strains relative to the non-induced strain was calculated by the 2−ΔΔCt method. When compared with the non-induced control, an expression equal to 1 indicated identical expression levels, and an expression ⩾4 indicated overexpressed.15, 16, 17, 18
Data analysis
SPSS 14.0 (SPSS Inc., Chicago, IL, USA) was used to perform X2 analysis, and the difference was considered to be statistically significant when P<0.05.
Results
Identification of rpoB mutations
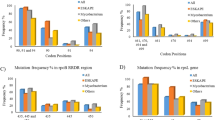
The rpoB sequence of 16 clinical isolates was examined. Only one RIF-monoresistant isolate did not have a mutation in the resistance-determining region of rpoB. Of the other RIF-monoresistant isolates, 10 carried mutations of 531 TCG-TTG (Ser–Leu), 1 carried 531 TCG-TTC (Ser–Phe), 2 carried 526 CAC-GAC (His–Asp), 1 carried 522 TCG-ATG (Ser–Met) and 1 carried 516 GAC-TTC (Asp–Phe) (Table 2).
Genotyping results
Spoligotyping results for the 16 RIF-monoresistant isolates showed that a total of 12 isolates belonged to the Beijing family, whereas 4 isolates belonged to non-Beijing families, which included the U family (2 isolates), T1 family (1 isolate) and Orphan (1 isolate) (Table 2). On the basis of the presence of IS6110 upstream of the NTF region,19, 20 10 out of 12 Beijing family strains were confirmed as modern Beijing strains and two were ancient Beijing strains (Table 2).
RIF MICs and the effect of EPIs on the RIF MICs of M. tuberculosis
As demonstrated in Table 2, levels of resistance to RIF as measured by MICs varied among RIF-monoresistant isolates. These data suggest that biological mechanisms in addition to the rpoB mutation and the genetic background of the strains are responsible for defining the respective RIF MIC values.
To test the hypothesis that efflux pumps were involved in modulating MICs, RIF-monoresistant strains were cultured in the presence of RIF together with the EPIs CCCP (1.6 μg ml−1), VP (64 μg ml−1), TZ (2 μg ml−1) or CPZ (4 μg ml−1). CCCP, VP, TZ and CPZ decreases RIF MICs greater than or equal to fourfold in 6, 12, 9 and 12 isolates, respectively (Table 2). The ability to decrease RIF MICs more than fourfold by VP and CPZ was identical and stronger than CCCP (P-values were all 0.033) (Table 2). The RIF MIC of 14 RIF-monoresistant isolates was decreased by at least eightfold by at least one out of four EPIs. We also found that the increased RIF susceptibility was independent of both the rpoB mutation and the genetic background (Table 2). However, the RIF susceptibility of none of the 16 RIF-resistant isolates was restored by the EPIs. For strain H37Rv, the RIF MIC of 0.0625 μg ml−1 decreased to 0.0156 μg ml−1 by CPZ and to 0.0313 μg ml−1 by CCCP; TZ and VP had no effect on MIC.
Effect of RIF induction on the expression of drug efflux pumps
For strain H37Rv, RIF did not increase expression for 24 of 27 genes, but resulted in a ± twofold increase in expression for three genes. The expression of drrC showed the highest induction (2.81-fold).
Of the 16 RIF-monoresistant isolates, 8 isolates overexpressed (⩾ fourfold induction) one or two of the following genes: Rv0842, bacA, Rv2333, efpA, drrB and drrC. Among the eight isolates with overexpressed drug efflux pump genes, seven carried mutations of rpoB 531 TCG-TTG (Ser–Leu), six belonged to the modern Beijing family, six had RIF MICs ⩾128 μg ml−1 and six had ⩾8-fold decrease in the RIF MICs with at least one of the four EPIs (Table 2).
Of 27 efflux pump genes tested, 15 (Rv0037c, Rv0191, Rv0783, Rv0876c, pstB, mmpL13a, Rv1672c, bacA, Rv2209, Rv2294, Rv2459, P55 (Rv1258c), drrA, drrB and drrC) were induced two- to threefold by RIF in at least one RIF-monoresistant M. tuberculosis isolate (Table 3). Thirteen of 16 isolates showed two- to threefolds induction in at least one of the 15 induced genes. Three (SHX05124, XZ06125 and SC06157) of 16 isolates (18.8%) showed no induction in any of the 27 efflux pump genes under RIF stress.
Discussion
Previous research indicated that mutations in the resistance-determining region of rpoB are responsible for up to 95% of RIF-resistant M. tuberculosis strains. In this study, resistance-determining region mutations were detected in 15 of 16 clinical isolates (93.8%). However, half of the RIF-monoresistant isolates that carried mutations in rpoB overexpressed one or two of the following putative efflux pump genes: Rv2333, drrB, drrC, Rv0842, bacA and efpA. In addition, the level of RIF resistance varied independently of the mutations in the rpoB gene and the genetic background of the clinical isolates of M. tuberculosis. One possible explanation for this is that classical mutations in rpoB and efflux pumps both confer RIF resistance in RIF-monoresistant M. tuberculosis.
Four EPIs (CCCP, VP, TZ and CPZ) were used to determine the efflux pump contribution to RIF resistance. Our observation that the inhibition of VP and CPZ significantly increased RIF susceptibility in RIF-monoresistant strains suggests the importance of efflux pump proteins in defining the intracellular concentration of anti-TB drugs. This finding is concordant with findings of previous studies, which also showed that EPIs increased susceptibility to RIF.6, 21 It also implies that EPIs have the potential to improve the efficacy of anti-TB drug treatment.6 In this study, the effect of VP and CPZ on RIF susceptibility was identical and showed stronger inhibition than CCCP, suggesting that differences in the efficacy of EPIs may be associated with the drug mode of action. CPZ and VP are ion channel blockers,22, 23 whereas CCCP is an uncoupler of proton transport that can inhibit the proton concentration gradient of the active efflux system.24
In this study, a few efflux pump genes were induced by RIF in RIF-monoresistant M. tuberculosis. drrA (Rv2936), drrB (Rv2937) and drrC (Rv2938) were combined in the open reading frame of drrABC of the ABC family. drrAB expressed in Mycobacterium smegmatis can lead to resistance to EMB, STR, norfloxacin, erythromycin, tetracycline and chloramphenicol.25 Gupta et al. reported that drrC was induced 4.1- and 7.7-fold under EMB and STR stress, respectively; however, they did not find that drrC was induced under RIF stress in MDR M. tuberculosis.4 In this study drrC was overexpressed in three RIF-monoresistant isolates that belonged to the modern Beijing family and carried the rpoB 531 TCG-TTG (Ser–Leu) mutation. drrB was also overexpressed in three RIF-monoresistant isolates; however, only one of these three isolates overexpressed drrC. Pang et al.9 reported that drrA was overexpressed among high-level RIF-resistant clinical isolates. Our study showed that RIF induced a low level (two- to threefold) increase in drrA expression in six M. tuberculosis isolates with RIF MICs that ranged from 16 to 128 μg ml−1. More studies should be performed to determine whether expression differences of drrABC lead to differences in RIF MIC.
Gupta et al.4 showed that the efpA expression was 4.5-fold greater under INH stress in five MDR isolates, but there was no induction of efpA expression under RIF stress in five MDR isolates. They hypothesized that the EfpA efflux pump presents itself as an alternative/additive mechanism of INH resistance in M. tuberculosis. Wilson et al.26 also reported a 2.5-fold induction of this gene by INH treatment along with a twofold induction by ethionamide treatment in an INH-sensitive isolate of M. tuberculosis. In our study, one RIF-monoresistant strain (XZ06115) overexpressed efpA; this strain belonged to the modern Beijing family and carried the 531 TCG-TTG (Ser–Leu) mutation of rpoB. Previous studies4, 26, 27 did not provide the molecular background of strains; therefore, the molecular background should be examined to determine whether this is the reason for expression differences.
Stp and Rv0842 are probable conserved integral membrane proteins that are similar to many antibiotic and drug efflux proteins.28, 29, 30, 31, 32 No further information about these proteins has been reported. stp and Rv0842 were overexpressed with RIF stress in one RIF-monoresistant isolate each; both strains carried the 531 TCG-TTG (Ser–Leu) mutation in rpoB, but the former belonged to the modern Beijing family and the latter belonged to the U family.
BacA is thought to be involved in the active transport of drugs across the membrane. Gupta et al.4 showed that bacA was induced in two of five MDR isolates in the present study, only one high-level RIF-monoresistant M. tuberculosis isolate overexpressed bacA.
In the present study, two RIF-monoresistant M. tuberculosis isolates (SHX05124 and SHX05178) belonged to the U family, carried an identical mutation of rpoB, and showed identical RIF MICs. However, in isolate SHX05124, none of the 27 examined efflux pump genes had a greater than twofold induction because of RIF stress. In contrast, in strain SHX05178, Rv0842 was overexpressed 10-fold under RIF stress. In strain GX06026, which has no rpoB mutations, and a RIF MIC value of 64 μg ml−1, none of the 27 efflux pump genes was overexpressed (more than a fourfold change), but the expression of three genes (drrA, drrB and drrC) was increased threefold, this result was not consistent with the result of a previous report.9 Therefore, more research should be undertaken to determine the cause of the difference.
Conclusion
In conclusion, in addition to classical mutations in rpoB, the efflux pumps Rv2333, DrrB, DrrC, Rv0842, BacA and EfpA may have a role in RIF resistance. M. tuberculosis with knockout genes (such as those belonging to certain genotypes or carrying certain mutations in rpoB) should be used to repeat experiments to determine their contributions to RIF resistance. In addition, the EPIs VP and CPZ significantly increased RIF susceptibility in RIF-monoresistant M. tuberculosis.
References
WHO. Global tuberculosis report 2013 (2013).
McClure, W. R. & Cech, C. L. On the mechanism of rifampicin inhibition of RNA synthesis. J. Biol. Chem. 253, 8949–8956 (1978).
Somoskovi, A., Parsons, L. M. & Salfinger, M. The molecular basis of resistance to isoniazid, rifampin, and pyrazinamide in Mycobacterium tuberculosis. Respir. Res. 2, 164–168 (2001).
Gupta, A. K. et al. Microarray analysis of efflux pump genes in multidrug-resistant Mycobacterium tuberculosis during stress induced by common anti-tuberculous drugs. Microb. Drug Resist. 16, 21–28 (2010).
Louw, G. E. et al. A balancing act: efflux/influx in mycobacterial drug resistance. Antimicrob. Agents Chemother. 53, 3181–3189 (2009).
Louw, G. E. et al. Rifampicin reduces susceptibility to ofloxacin in rifampicin-resistant Mycobacterium tuberculosis through efflux. Am. J. Respir. Crit. Care Med. 184, 269–276 (2011).
Srivastava, S. et al. Efflux-pump-derived multiple drug resistance to ethambutol monotherapy in Mycobacterium tuberculosis and the pharmacokinetics and pharmacodynamics of ethambutol. J. Infect. Dis. 201, 1225–1231 (2010).
Pasca, M. R. et al. Rv2686c-Rv2687c-Rv2688c, an ABC fluoroquinolone efflux pump in Mycobacterium tuberculosis. Antimicrob. Agents Chemother. 48, 3175–3178 (2004).
Pang, Y. et al. Study of the rifampin monoresistance mechanism in Mycobacterium tuberculosis. Antimicrob. Agents Chemother. 57, 893–900 (2013).
WHO Policy Guidance on Drug-susceptibility Testing (DST) of Second-line Antituberculosis Drugs (WHO/HTM/TB/2008.392), (World Health Organization: Geneva, (2008).
World Health Organization. Updated interim critical concentrations for first-line and second-line DST (as for May 2012) (2012).
Kamerbeek, J. et al. Simultaneous detection and strain differentiation of Mycobacterium tuberculosis for diagnosis and epidemiology. J. Clin. Microbiol. 35, 907–914 (1997).
Wada, T., Iwamoto, T. & Maeda, S. Genetic diversity of the Mycobacterium tuberculosis Beijing family in East Asia revealed through refined population structure analysis. FEMS Microbiol. Lett. 291, 35–43 (2009).
Leonard, B. et al. Inter- and intra-assay reproducibility of microplate Alamar blue assay results for isoniazid, rifampicin, ethambutol, streptomycin, ciprofloxacin, and capreomycin drug susceptibility testing of Mycobacterium tuberculosis. J. Clin. Microbiol. 46, 3526–3529 (2008).
Rodrigues, L. et al. Contribution of efflux activity to isoniazid resistance in the Mycobacterium tuberculosis complex. Infect. Genet. Evol. 12, 695–700 (2011).
Livak, K. J. & Schmittgen, T. D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25, 402–408 (2001).
Bustin, S. A. et al. MIQE precis: practical implementation of minimum standard guidelines for fluorescence-based quantitative real-time PCR experiments. BMC. Mol. Biol. 11, 74 (2010).
DeMarco, C. E. et al. Efflux-related resistance to norfloxacin, dyes, and biocides in bloodstream isolates of Staphylococcus aureus. Antimicrob. Agents Chemother. 51, 3235–3239 (2007).
Mokrousov, I. et al. Origin and primary dispersal of the Mycobacterium tuberculosis Beijing genotype: clues from human phylogeography. Genome Res. 15, 1357–1364 (2005).
Kurepina, N. E. et al. Characterization of the phylogenetic distribution and chromosomal insertion sites of five IS6110 elements in Mycobacterium tuberculosis: non-random integration in the dnaA-dnaN region. Tuber. Lung Dis. 79, 31–42 (1998).
Gupta, A. K. et al. Estimation of efflux mediated multi-drug resistance and its correlation with expression levels of two major efflux pumps in mycobacteria. J. Commun. Dis. 38, 246–254 (2006).
Sun, H. et al. Chronic inhibition of cardiac Kir2.1 and HERG potassium channels by celastrol with dual effects on both ion conductivity and protein trafficking. J. Biol. Chem. 281, 5877–5884 (2006).
Tolon, R. et al. Potassium depolarization-induced cAMP stimulates somatostatin mRNA levels in cultured diencephalic neurons. Brain Res. 868, 338–346 (2000).
Banerjee, S. K. et al. Involvement of an efflux system in mediating high level of fluoroquinolone resistance in Mycobacterium smegmatis. Biochem. Biophys. Res Commun. 226, 362–368 (1996).
Choudhuri, B. S. et al. Overexpression and functional characterization of an ABC (ATP-binding cassette) transporter encoded by the genes drrA and drrB of Mycobacterium tuberculosis. Biochem. J. 367, 279–285 (2002).
Wilson, M. et al. Exploring drug-induced alterations in gene expression in Mycobacterium tuberculosis by microarray hybridization. Proc. Natl Acad. Sci. USA 96, 12833–12838 (1999).
Waddell, S. J. et al. The use of microarray analysis to determine the gene expression profiles of Mycobacterium tuberculosis in response to anti-bacterial compounds. Tuberculosis (Edinb) 84, 263–274 (2004).
Cole, S. T. et al. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature 393, 537–544 (1998).
De Rossi, E. et al. The multidrug transporters belonging to major facilitator superfamily in Mycobacterium tuberculosis. Mol. Med. 8, 714–724 (2002).
TubercuList [Internet]. Accessed 2010 Nov 15. http://genolist.pasteur.fr/TubercuList.
De Rossi, E., Ainsa, J. A. & Riccardi, G. Role of mycobacterial efflux transporters in drug resistance: an unresolved question. FEMS Microbiol. Rev. 30, 36–52 (2006).
Li, X. Z., Zhang, L. & Nikaido, H. Efflux pump-mediated intrinsic drug resistance in Mycobacterium smegmatis. Antimicrob. Agents Chemother. 48, 2415–2423 (2004).
Acknowledgements
This work was financially supported by projects 2013ZX10003002-001 of the National Key Programme of Mega Infectious Diseases and the key project 2014SKLID104 of the State Key Laboratory for Infectious Disease Prevention and Control. The funders had no role in the study design, data collection and analysis, decision to publish or manuscript preparation.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Rights and permissions
About this article
Cite this article
Li, G., Zhang, J., Guo, Q. et al. Study of efflux pump gene expression in rifampicin-monoresistant Mycobacterium tuberculosis clinical isolates. J Antibiot 68, 431–435 (2015). https://doi.org/10.1038/ja.2015.9
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ja.2015.9
This article is cited by
-
Accuracy of a reverse dot blot hybridization assay for simultaneous detection of the resistance of four anti-tuberculosis drugs in Mycobacterium tuberculosis isolated from China
Infectious Diseases of Poverty (2020)
-
Scrutinizing the drug resistance mechanism of multi- and extensively-drug resistant Mycobacterium tuberculosis: mutations versus efflux pumps
Antimicrobial Resistance & Infection Control (2019)
-
Updated and standardized genome-scale reconstruction of Mycobacterium tuberculosis H37Rv, iEK1011, simulates flux states indicative of physiological conditions
BMC Systems Biology (2018)
-
Machine learning and structural analysis of Mycobacterium tuberculosis pan-genome identifies genetic signatures of antibiotic resistance
Nature Communications (2018)
-
Mycobacterium tuberculosis Major Facilitator Superfamily Transporters
The Journal of Membrane Biology (2017)