Abstract
A novel actinomycete, designated LIPI11-2-Ac043T, was isolated from leaf litter collected in Indonesia. According to phylogenetic analysis based on the 16S rRNA gene sequence comparisons, strain LIPI11-2-Ac043T was closely related to Actinoplanes abujensis A4029T (99.3%) and Actinoplanes brasiliensis DSM 43805T (98.8%). Spores of strain LIPI11-2-Ac043T were motile and the sporangia were spherical. The predominant menaquinone was MK-9(H6) and the principal polar lipids were phosphatidylinositol, phosphatidylethanolamine, phosphatidylglycerol and diphospatidylglycerol. The whole-cell sugars were galactose, glucose and mannose; rhibose, arabinose and xylose were also detected as minor components. The major fatty acids were anteiso-C15:0, iso-C16:0, iso-C15:0 and anteiso-C17:0. These data supported the affiliation of strain LIPI11-2-Ac043T to the genus Actinoplanes. Meanwhile, the results of DNA–DNA hybridization and physiological and biochemical tests indicated that strain LIPI11-2-Ac043T can be distinguished from its closest related species. Therefore, strain LIPI11-2-Ac043T represents a novel species of the genus Actinoplanes, for which the name Actinoplanes bogoriensis sp. nov. is proposed. The type strain is LIPI11-2-Ac043T (=InaCC A522T=NBRC 110975T).
Similar content being viewed by others
Introduction
The genus Actinoplanes is one of the actinomycete genus that is well known as producers for several bioactive compounds such as actaplanin,1 teichomycins,2 gardimycin3 and polyene anti-fungal antibiotic.4 Because of its value for drug discovery, studies on the bioprospecting and taxonomy of members of the genus are important, and the discovery of novel species will contribute toward understanding not only for the biodiversity of the genus but also their potentials for applications in drug discovery. The genus Actinoplanes was firstly described by Couch5 and is now assigned as a member of the family Micromonosporaceae.6 At the time of writing, 37 species of the genus have been found and validly described. Members of this genus produce motile spores within the spherical, digitate, cylindrical, lobate or irregular sporangia.7 The presence of meso- and/or 3-hydroxy-diaminopimelic acids, arabinose and xylose in whole-cell hydrolysates was also key characters for the members of the genus Actinoplanes.7, 8, 9 Moreover, a comprehensive phylogenetic study based on 16S rRNA gene sequences from members of the genus was reported by Tamura and Hatano.10
During the course of a study of actinobacterial diversity of tropical rain forests in Indonesia, a novel strain was discovered from a leaf-litter sample. The aim of this study was to clarify the taxonomic position of the strain by chemotaxonomic, morphological, molecular and genomic characterizations.
Materials and methods
Actinobacterial strain and isolation
Strain LIPI11-2-Ac043T was isolated from a leaf-litter sample collected in Cibodas Botanical Garden, Bogor, Indonesia, using humic acid-vitamin agar11 supplemented with cycloheximide (50 mg l−1), kabicidin (10 mg l−1) and nalidixic acid (20 mg l−1). Isolation was conducted using the rehydration and centrifugation method.12 The isolate was purified on yeast extract-starch agar and maintained in 10% (v v−1) glycerol at −80 °C. Biomass for chemotaxonomic and molecular systematic studies was obtained by cultivating the strain in shake flasks containing liquid Maltose-Bennett’s Medium (NBRC medium 231) for 5 days at 28 °C with shaking at 100 r.p.m. Actinoplanes abujensis NBRC 110769T and Actinoplanes brasiliensis NBRC 13938T were used as reference strains for comparison with the physiological, biochemical, cellular fatty acids and the DNA–DNA hybridization tests.
Morphological, physiological and biochemical tests
Cultural characteristics of strain LIPI11-2-Ac043T were investigated after incubation for 14 days at 28 °C on ISP media series.13 The morphological observation was conducted using a light microscope (model BX-51; Olympus, Tokyo, Japan) and a scanning electron microscope (model JSM-6060; JEOL, Tokyo, Japan) after cultivation on humic acid-vitamin agar for 14 days at 28 °C. Spore motility was evaluated under the light microscope after cultivation on water agar for 14 days at 28 °C. The temperature range and optimum temperature for growth were determined by incubating the cultures at 5, 10, 15, 20, 25, 28, 37, 45 and 60 °C on the plates of the NBRC medium 231. The pH range and optimum pH for initial growth were established by using a liquid NBRC medium 231 adjusted to pH 4–12 (1.0 pH interval). NaCl tolerance was observed within range 0–7% (w v−1) using a liquid NBRC medium 231. Color determination was carried out with color chips from a Mycological Color Chart.14 Utilization of carbohydrates was investigated on the ISP9 medium13 supplemented with several kinds of carbohydrates as a sole carbon source after incubation for 14 days at 28 °C. Enzyme activities were evaluated by API ZYM and API Coryne test strips (bioMérieux, Tokyo, Japan) according to the manufacturer’s instructions.
Chemotaxonomic tests
Analyses of sugar composition in whole-cell hydrolysate, amino acids and their isomers in cell-wall hydrolysate and isoprenoid quinones were determined according to the methods described by Hamada et al.15 Polar lipids were investigated by the method of Minnikin et al.16 Cellular fatty acid methyl esters were prepared and analyzed following the standard protocol of MIDI Sherlock Microbial Identification System17 using ACTINO database (version 4.0; MIDI, Newark, DE, USA) and detected using GC (model 6890N; Agilent Technologies, Santa Clara, CA, USA).
Molecular analyses and determination of phylogenetic tree
DNA was isolated using PrepMan Ultra Reagent (Applied Biosystems, Foster City, CA, USA) based on the manufacturer’s instruction. Pairs of primers, 9F (5′-GAGTTTGATCCTGGCTCAG-3′) and 1541R (5′-AAGGAGGTGATCCAGCC-3′), were used for PCR amplification of the 16S rRNA gene. The PCR product was sequenced using an automated DNA sequencer (ABI PRISM 3730 Genetic Analyzer; Applied Biosystems). Four primers, 9F, 785F (5′-GGATTAGATACCCTGGTAGTC-3′), 802R (5′-TACCAGGGTATCTAATCC-3′) and 1541R, were used to determine the almost-complete 16S rRNA gene sequence. Similarities of 16S rRNA gene sequences were calculated using the EzTaxon-e server.18 The 16S rRNA gene sequences of strain LIPI11-2-Ac043T and the members of the genus Actinoplanes were aligned using CLUSTAL_X program.19 Phylogenetic trees were reconstructed by the neighbor-joining, maximum-likelihood and maximum-parsimony algorithms.20, 21, 22 The resultant tree topology was evaluated by bootstrap analysis23 with 1000 replicates.
G+C content of DNA and DNA–DNA hybridization
The DNA G+C contents were determined by the method of Tamaoka and Komagata24 using HPLC (model LC-20A; Shimadzu, Kyoto, Japan). The microplate hybridization method developed by Ezaki et al.25 was used to determine DNA–DNA relatedness. Five replications were used for each sample, the highest and the lowest values were excluded and the mean of three values was used for calculation of DNA–DNA relatedness value.
Nucleotide sequence accession number
The GenBank/EMBL/DDBJ accession number for the 16S rRNA gene sequence of strain LIPI11-2-Ac043T is LC027115.
Results and discussion
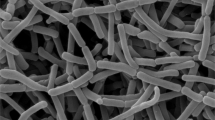
Strain LIPI11-2-Ac043T was observed to form different colors of substrate mycelia based on the kinds of media. Yellow-maple mycelia in color were observed on ISP-3 medium, orange on ISP-4 and ISP-5 media and brown copper on ISP-6 and ISP-7 media. Aerial mycelia were not observed on any tested media. The strain produced brown soluble pigment only in ISP-7 medium. The strain developed spherical sporangia, and the diameter of the sporangia ranged from 2 to 6 μm (Figure 1). Strain LIPI11-2-Ac043T could not tolerate >1% (w v−1) NaCl. The temperature range for growth was 15–28 °C (optimum 28 °C) and the pH range was pH 5.0–10.0 (optimum pH 8.0).
The whole-cell hydrolysate of strain LIPI11-2-Ac043T contained galactose, glucose and mannose; minor amounts of ribose, arabinose and xylose were also detected. The peptidoglycan sample of the strain contained d-alanine, d-glutamic acid, glycine and meso-diaminopimelic acid. The predominant menaquinone was MK-9(H6). The polar lipids were found to be phosphatidylinositol, phosphatidylethanolamine, phosphatidylglycerol, diphosphatidylglycerol, one unidentified phosphoglycolipid, three unidentified glycolipids and four unidentified polar lipids (Supplementary Figure S1). This result indicated that the strain has the phospholipid type II according to Lechevalier and Lechevalier.26 The major fatty acids were anteiso-C15:0 (23.45 %), iso-C16:0 (23.27 %), iso-C15:0 (21.05 %) and anteiso-C17:0 (10.15 %). The detailed results of fatty acid profiles of strain LIPI11-2-Ac043T and two closest phylogenetic relatives were shown in Table 1. The DNA G+C content of strain LIPI11-2-Ac043T was determined to be 71 mol%.
Phylogenetic analysis based on the 16S rRNA gene sequence comparisons indicated that strain LIPI11-2-Ac043T was closely related to members of the genus Actinoplanes and had the highest sequence similarities to A. abujensis A4029T (99.3%) and A. brasiliensis DSM 43805T (98.8%). The phylogenetic tree reconstructed by the neighbor-joining algorithm showed that this strain formed a monophyletic clade with A. abujensis and A. brasiliensis and this clade was supported by 89% of bootstrap value (Figure 2). Meanwhile, in the maximum-likelihood and maximum-parsimony trees, the strain did not form a reliable clade with any members of the genus Actinoplanes.
Phylogenetic tree derived from the 16S rRNA gene sequences of strain LIPI11-2-Ac043T and the recognized members of the genus Actinoplanes. Root position of the tree was determined using Streptomyces ambofaciens ATCC 23877T (M27245) as the outgroup. The tree was reconstructed using the neighbor-joining method and Knuc values. Numbers at branch nodes indicate bootstrap percentages (1000 replicates); only values of 50% or above are shown. Filled circles indicate that the corresponding nodes were also recovered with the maximum-likelihood and maximum-parsimony algorithms. Bar, 0.01Knuc.
The DNA–DNA hybridization value between strain LIPI11-2-Ac043T and A. abujensis NBRC 110796T was 19–30%, and that between strain LIPI11-2-Ac043T and A. brasiliensis NBRC 13938T was 28–39% (Supplementary Table S1). These values were well below the 70% cutoff point, which is the criterion for the assignment of bacterial strains to a different genomic species.27 Furthermore, strain LIPI11-2-Ac043T could be differentiated from A. abujensis by utilizations of fructose, enzyme activities of trypsin, acid phosphatase, nitrate reductase and pyrazinamidase and acid production from glucose and xylose. Meanwhile, strain LIPI11-2-Ac043T could be differentiated from A. brasiliensis by utilizations of fructose and inositol and enzyme activities of α-chymotrypsin, acid phosphatase, nitrate reductase and N-acetyl-β-glucosaminidase, as well as acid production from glucose, xylose, maltose and lactose (Table 2). On the basis of the results of phenotypic and genotypic characterization and DNA–DNA hybridization test, strain LIPI11-2-Ac043T represents a novel species of the genus Actinoplanes, for which the name Actinoplanes bogoriensis sp. nov. is proposed.
Description of A. bogoriensis sp. nov
A. bogoriensis (bo.go.ri.en′sis N.L. masc. adj. bogoriensis; of, or pertaining to Bogor, West Java, Indonesia, where the organism was originally isolated).
Cells are aerobic and Gram stain positive. Optimum growth is observed on ISP-5 medium, moderate growth on ISP-4 medium and poor growth on ISP-3, ISP-6 and ISP-7 media. Substrate mycelia are yellow-maple on ISP-3 medium, orange on ISP-4 and ISP-5 media and brown copper on ISP-6 and ISP-7 media. Brown soluble pigment is produced in the ISP-7 medium. Aerial mycelia are not produced. Irregular sporangia are formed and sporangiospores are motile. Growth occurs at 15–28 °C (optimum 28 °C) and pH 5.0–10.0 (optimum pH 8.0). No growth is observed in the presence of >1 % (w v−1) NaCl. The peptidoglycan diamino acid is meso-diaminopimelic acid and the whole-cell hydrolysate contains galactose, glucose and mannose, together with minor amounts of ribose, arabinose and xylose. The predominant menaquinone is MK-9(H6). The major fatty acids are anteiso-C15:0, iso-C16:0, iso-C15:0 and anteiso-C17:0. The polar lipids consist of phosphatidylinositol, phosphatidylethanolamine, phosphatidylglycerol, diphosphatidylglycerol, one unidentified phosphoglycolid, one unidentified phospholipid, three unidentified glycolipids and four unidentified polar lipids. rhamnose, d-raffinose, d-sucrose, inositol, d-xylose and d-mannitol are used as a sole carbon source. Positive result in tests for alkaline phosphatase, α-chymotrypsin, acid phosphatase, β-galactosidase, α-glucosidase, β-glucosidase, N-acetyl-β-glucosaminidase, esterase (C4), esterase lipase (C8) and leucine arylamidase are weakly positive. Gelatin is hydrolyzed and catalase activity is positive. The DNA G+C content of the type strain is 71 mol%.
The type strain LIPI11-2-Ac043T (InaCC A522T=NBRC 110975T) was isolated from a leaf-litter sample collected from Cibodas Botanical Garden, Bogor, Indonesia.
References
Debono, M. et al. Actaplanin, new glycopeptide antibiotics produced by Actinoplanes missouriensis. The isolation and preliminary chemical characterization of actaplanin. J. Antibiot. 37, 85–95 (1984).
Bardone, M. R., Paternoster, M . & Coronelli, C. Teichomycins, new antibiotics from Actinoplanes teichomyceticus nov. sp. II. Extraction and chemical characterization. J. Antibiot. 31, 170–177 (1978).
Coronelli, C., Tamoni, G. & Lancini, G. C. Gardimycin, a new antibiotic from Actinoplanes. II. Isolation and preliminary characterization. J. Antibiot. 29, 507–510 (1976).
Wagman, G. H. et al. New polyene antifungal antibiotic produced by a species of Actinoplanes. Antimicrob. Agents Chemother. 7, 457–461 (1975).
Couch, J. N. Actinoplanes, a new genus of the Actinomycetales. J. Elisha. Mitchell. Sci. Soc. 66, 87–92 (1950).
Zhi, X. Y., Li, W. J. & Stackebrandt, E. An update of the structure and 16S rRNA gene sequence-based definition of higher ranks of the class Actinobacteria, with the proposal of two new suborders and four new families and emended descriptions of the existing higher taxa. Int. J. Syst. Evol. Microbiol. 59, 589–608 (2009).
Stackebrandt, E. & Kroppenstedt, R. M. Union of the genera Actinoplanes couch, Ampullariella couch, and Amorphosporangium couch in a redefined genus Actinoplanes. Syst. Appl. Microbiol. 9, 110–114 (1987).
Goodfellow, M. et al. Numerical and chemical classification of Actinoplanes and some related actinomycetes. J. Gen. Microbiol. 136, 19–36 (1990).
Vobis, G. in The Actinoplanetes (eds Williams, S. T. et al, 2418–2428 (Bergey’s Manual of Systematic Bacteriology: Baltimore, MD, USA, (1989) .
Tamura, T. & Hatano, K. Phylogenetic analysis of the genus Actinoplanes and transfer of Actinoplanes minutisporangius Ruan et al. 1986 and ‘Actinoplanes aurantiacus’ to Cryptosporangium minutisporangium comb. nov. and Cryptosporangium aurantiacum sp. nov. Int. J. Syst. Evol. Microbiol. 51, 2119–2125 (2001).
Hayakawa, M. & Nonomura, H. Humic acid-vitamin agar, a new medium for selective isolation of soil actinomycetes. J. Ferment. Technol. 65, 501–509 (1987).
Hayakawa, M. et al. Application of a method incorporating differential centrifugation for selective isolation of motile actinomycetes in soil in plant litter. Antonie van Leeuwenhoek 78, 171–185 (2000).
Shirling, E. B. & Gottlieb, D. Methods for characterization of Streptomyces species. Int. J. Syst. Bacteriol. 16, 313–340 (1966).
Rayner, R. W. A Mycological Colour Chart, (Commonwealth Mycological Institute and British Mycological Society, Kew, UK, (1970).
Hamada, M. et al. Luteimicrobium album sp. nov., a novel actinobacterium isolated from a lichen collected in Japan, and emended description of the genus Luteimicrobium. J. Antibiot. 65, 427–431 (2012).
Minnikin, D. E. et al. An integrated procedure for the extraction of bacterial isoprenoid quinones and polar lipids. J. Microbiol. Methods 2, 233–241 (1984).
Sasser, M. Identification of Bacteria by Gas Chromatography of Cellular Fatty Acids, MIDI Technical Note 101, (MIDI: Newark, NJ, USA, (1990).
Kim, O. S. et al. Introducing EzTaxon-e: a prokaryotic 16S rRNA gene sequence database with phylotypes that represent uncultured species. Int. J. Syst. Evol. Microbiol. 62, 716–721 (2012).
Thompson, J.D. et al. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25, 4876–4882 (1997).
Saitou, N. & Nei, M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. J. Mol. Biol. Evol. 4, 406–425 (1987).
Felsenstein, J. Evolutionary trees from DNA sequences: a maximum likelihood approach. J. Mol. Evol. 17, 368–376 (1981).
Fitch, W. M. Toward defining the course of evolution: minimum change for a specific tree topology. Syst. Zool. 20, 406–416 (1971).
Felsenstein, J. Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39, 738–791 (1985).
Tamaoka, J. & Komagata, K. Determination of DNA base composition by reversed-phase high-performance liquid chromatography. FEMS Microbiol. Lett. 25, 125–128 (1984).
Ezaki, T., Hashimoto, Y. & Yabuuchi, E. Fluorometric deoxyribonucleic acid deoxyribonucleic acid hybridization in microdilution wells as an alternative to membrane filter hybridization in which radioisotopes are used to determine genetic relatedness among bacterial strains. Int. J. Syst. Bacteriol. 39, 224–229 (1989).
Lechevalier, M. P. & Lechevalier, H. Chemical composition as a criterion in the classification of aerobic actinomycetes. Int. J Syst. Bacteriol. 20, 435–443 (1970).
Wayne, L. G. et al. International Committee on Systematic Bacteriology. Report of the ad hoc committee on reconciliation of approaches to bacterial systematics. Int. J. Syst. Bacteriol. 37, 463–464 (1987).
Acknowledgements
This work was supported by Science and Technology Research Partnership for Sustainable Development (SATREPS), which is a research program in collaboration with the Japan Science and Technology Agency (JST) and the Japan International Cooperation Agency (JICA).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies the paper on The Journal of Antibiotics website
Supplementary information
Rights and permissions
About this article
Cite this article
Nurkanto, A., Lisdiyanti, P., Hamada, M. et al. Actinoplanes bogoriensis sp. nov., a novel actinomycete isolated from leaf litter. J Antibiot 69, 26–30 (2016). https://doi.org/10.1038/ja.2015.81
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ja.2015.81