Abstract
A novel actinomycete strain, designated VRC21T, was isolated from the rhizosphere of Callistemon citrinus collected from Hyderabad, India. The morphological and chemotaxonomic properties of strain VRC21T was consistent with the characteristics of members of the genus Streptosporangium, that is, the formation of sporangia on aerial mycelium, coiled unbranched hyphae within the spore vesicle, the presence of meso-diaminopimelic acid in the cell wall, and madurose and galactose as major whole-cell sugars. Diagnostic polar lipids were phosphatidylethanolamine, diphosphatidylglycerol, phosphatidylglycerol, phosphatidylinositol and phosphatidylinositol-mannosides. The predominant menaquinones were MK-9(H2) and MK-9(H4). The major cellular fatty acids were iso-C14:0, iso-C16:0, C17:0 10-methyl, C18:1w9c and C18:0 10-methyl. 16S rRNA gene sequence analyses revealed that strain VRC21T was a member of the genus Streptosporangium. The highest similarity values were observed with S. carneum DSM 44125T (98.2%) and S. fragile DSM 43847T (98.2%); the values of the remaining type strains were below 98%. The values of DNA–DNA relatedness between the strain VRC21T and the type strains of the related species were below 70%. On the basis of the polyphasic evidence, the strain VRC21T should be classified as novel species Streptosporangium terrae sp. nov. in the genus Streptosporangium. The type strain is VRC21T (=KCTC 29207T=MTCC 11724T).
Similar content being viewed by others
Introduction
The genus Streptosporangium was first described by Couch.1 The genus encompasses aerobic, Gram-stain-positive, non-acid-fast organisms with stable, unbranched, non-fragmenting substrate mycelium that carries cottony aerial mycelium, which differentiates into sporangiophores; spores are spherical, rod or oval shape and the sporangial walls are thick; organism has meso-diaminopimelic acid and madurose in the peptidoglycan; MK-9(H2, H4) as predominant menaquinones; complex mixtures of iso-, anteiso-, saturated, unsaturated and 10-methyl branched fatty acids; phospatidylethanolamine, diphosphatidylglycerol, phosphatidylglycerol, phosphatidylinositol and glucosamine-containing polar lipids as major components; and has a DNA base composition within the range 69–71 mol% GC.1 At the time of manuscript preparation, there were 17 species and 2 subspecies with validly published names (http://www.bacterio.net/streptosporangium.html): Streptosporangium roseum,1 S. amethystogenes, S. album, S. vulgare, S. amethystogenes, subsp. amethystogenes,2 and subsp. fukuiens,3 S. longisporum,4 S. nondiastaticum, S. pseudovulgare,5 S. violaceochromogenes,6 S. fragile,7 S. carneum,8 S. subroseum,9 S. purpuratum, S. yunnanense,10 S. canum,11 S. oxazolinicum,12 S. anatoliense13 and S. sandarakinum.14
During our research work entitled ‘Discovery of novel antimicrobial agents against Streptococcus pneumonia—A multidisciplinary approach’, strain VRC21T was isolated from rhizosphere of Callistemon citrinus, with the prospect that it might produce novel antimicrobial agents. Although our investigation was multidisciplinary, the main aim was to discover novel antimicrobial agents against Streptococcus pneumonia. To achieve the goal, we have isolated few active compounds (under the process of publication) from C. citrinus leaves solvent extracts. Another major source for novel antimicrobial agents is soil microbes. In this regard, soil was collected from C. citrinus plant rhizosphere, because C. citrinus roots produce leptospermone. Leptospermone is the blue print of the compound mesotrione that has been proven to be an effective herbicide.15 C. citrinus being rich in terpenoids, produces many essential oils, which give good fragrance that surrounds the plant, including its rhizosphere soil. This plant grows widely as an ornamental plant in India, having potential medicinal properties that were proved not only by literature but also by our previous research work. This formed the basis in selecting the plant rhizosphere for actinomycetes isolation, identification, characterization and screening for secondary metabolites. The present study was carried out to determine the taxonomic status of the strain VRC21T by using a polyphasic approach.
Materials and Methods
Strain VRC21T was isolated from the rhizosphere of C. citrinus (Curtis) Skeels, from Hyderabad, India (GPS coordinates for the sampling site is 17°23′6.7374″N 78°29′11.979″E). Soil samples were collected in sterile tubes and brought to the laboratory of Osmania University, India. Samples were dried in laminar flow under aseptic conditions. Samples were serially diluted with sterilized distilled water, and up to 10−5 dilutions were made; 0.1 ml suspension from each dilution was spread on yeast extract malt extract agar (ISP medium 2)16 plates. The plates were observed intermittently during incubation. Pinpoint colonies with a clear zone of inhibition and the dominant reddish pink color colony were selected and maintained on ISP medium 2 at 4 °C and in glycerol suspensions (20% v/v) at −20 °C.
The strain VRC21T, S. fragile DSM 43847T and S. carneum DSM 44125T were cultured for 3 weeks at 30 °C and the cultural characteristics were observed on ISP (International Streptomyces Project) media 2, 3, 4, 5, 6 and 7,16 starch casein agar and nutrient agar.17 For morphological characterization, 21-day-old cultures on ISP medium 2 were taken. The cover slip technique18 was used to observe hyphae and spore chains by light microscopy (Olympus microscope BH-2, Delhi, India). Spore texture, spore-chain morphology and spore ornamentation were studied by scanning electron micrography. Specimens were prepared according to Williams and Davies,19 and stub was prepared according to our previous study.20 Finally, the sample was sputtered with gold (E-1010, Ion sputter with Gold, Model S-3700N, Hitachi, Japan). The color of substrate, aerial mycelia and soluble pigments were determined by comparison with chips from the ISCC-NBS color charts.21 A range of physiological tests such as growth at different temperatures (4, 10, 15, 20, 25, 30, 35, 40 and 45 °C), pH values (4.0–11.0) and NaCl concentrations (3, 5, 7 and 9% (w/v)) were examined on ISP medium 2.18 Assimilation of various carbohydrates as the sole carbon source was tested using ISP medium 9.22 Biochemical characteristics, H2S production and sensitivity of the strain to different antibiotics23 were determined using the methods of Korn-Wendisch et al.24 Starch hydrolysis was examined using ISP medium 4, and for nitrate reduction ISP medium 8 was used.16 Tyrosinase activity was determined using ISP medium 7.25 Enzyme activities were determined using the API ZYM system (bioMerieux, Lyon, France) according to the manufacturer’s instructions. Biomass for molecular and chemotaxonomic studies was obtained after incubation in shake flasks of trypticase soy broth medium (Hi-Media, Mumbai, India) at 30 °C for 7 days with rotary aeration (180 r.p.m.). The isomer type of diaminopimelic acid in cell wall peptidoglycan was determined by the method of Hasegawa et al.26 Whole-cell sugars were studied as described by Staneck and Roberts.27 Cellular fatty acid analysis was determined by the method of Sasser,28 using MIDI Sherlock version 6.0, MIDI database RTSBA6. Polar lipids were extracted and analyzed according to the method of Minnikin et al.,29 and Komagata and Suzuki.30 Mycolic acids were tested by the acid methanolysis method of Minnikin et al.31 Menaquinones were extracted and examined by using the method of Collins et al.32 and analyzed by HPLC.33 The N-acyl types of muramic acid were determined by the method of Uchida and Aida.34 Isolation of DNA35 and determination of DNA G+C content was carried out according to the method of Marmur and Doty.36 The levels of DNA–DNA relatedness was performed by using dot-blot hybridization method of Chung et al.37 and a simple fluorimetric method for estimation of DNA–DNA relatedness based on thermal denaturation temperatures.38 The 16S rRNA was amplified using the bacterial universal primers and sequencing was performed under contract by Macrogen Inc. (Kumchun-Ku, Seoul, South Korea) using a 3730XL DNA analyzer (Applied Biosystems, Seoul, South Korea). The 16S rRNA gene sequence was aligned with related sequences belonging to the genus Streptosporangium by using CLUSTAL W.39 Pairwise evolutionary distances were calculated using the DNADIST program with the Kimura two parameter model as developed by Kimura.40 Multiple sequence alignments with most closely related Streptosporangium species similarity were carried out using EzTaxon-e server (http://eztaxon-e.ezbiocloud.net/).41 Phylogenetic tree analysis was performed using MEGA6.42 The phylogenetic tree was constructed based on the neighbor-joining,43 maximum-parsimony44 and maximum-likelihood45 algorithms. Data were resampled 1000 bootstrap replications.46
Results and Discussion
Morphological, cultural and physiological characteristics
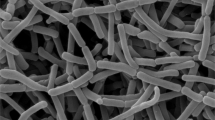
The cultural characteristics of strain VRC21T, along with those of the type strains of the closely related species, S. fragile DSM 43847T and S. carneum DSM 44125T, are given in Table 1. Strain VRC21T grew well and formed extensively branched and non-fragmented substrate mycelia on various tested agar media. The colony color of strain VRC21T was dark reddish brown with a dark reddish brown color soluble pigment when grown on various media (Table 1). The temperature and pH range for growth were 10–35 °C and pH 7–9, respectively, with optimum growth occurring at 30 °C and pH 7. Strain VRC21T tolerated up to 3% NaCl (w/v) and exhibited good growth on ISP 2, ISP 3, ISP 4, ISP 5, ISP 6 and ISP 7, sparse growth on nutrient agar and starch casein agar, and no growth on Czapek’s agar and potato dextrose agar. It produced pink colored aerial mycelia and very dark reddish brown colored substrate mycelia, and its spores were spherical, non-motile and present within sporangia; abundant sporangia formed from the aerial hyphae (Figure 1), and a very dark reddish brown diffusible pigment was observed. Casein was degraded. Starch was hydrolyzed. Gelatin was liquefied, milk was peptonized and coagulated. Nitrate was not reduced to nitrite and H2S gas was not produced. Melanin was produced. The Voges Proskauer’s reactions and methyl red tests were positive. The results of other physiological and biochemical analyses are summarized in the species description and Table 2 below.
Chemotaxonomic characteristics
Cell-wall peptidoglycan of strain VRC21T contained meso-diaminopimelic acid as the diagnostic diamino acid. Whole-cell sugars were madurose, galactose, glucose, xylose and arabinose. The acyl type of muramic acid was N-acetyl. Mycolic acids were not found. The major cellular fatty acids were (%): iso-C14:0 (9.07), C14:0 (5.06), iso-C16:1 (11.80), C16:0 (14.11), C-17:0 10-methyl (5.98), C18:1w9c (4.26), C18:0 10-methyl, TBSA (8.16). (Table 3). The predominant menaquinones were MK-9(H2) (52%) and MK-9(H4) (44%), and MK-9(H6) (4%) was detected as a minor component. Strain VRC21T contained phosphatidylethanolamine, diphosphatidylglycerol, phosphatidylglycerol, phosphatidylinositol, two phosphatidylinositol-mannosides, one unidentified glycolipid, two unidentified phosphoaminolipids, three unidentified phospholipids and nine unidentified lipids. The DNA G+C content was 69.5±1.5 mol%.
Phylogenetic analysis
The 16S rRNA gene sequence (1466 bp) of strain VRC21T showed a close relationship with members of the genus Streptosporangium, and the similarity values between strain VRC21T and the type strains in the genus Streptosporangium were 96.7–98.2%. Although strain VRC21T showed the highest similarity values to S. carneum DSM 44125T (98.2%) and S. fragile DSM 43847T (98.2%), the strain did not form a reliable cluster with any members of the genus Streptosporangium (Figure 2). The Genbank accession number of the 16S rRNA gene sequences of strain VRC21T is JX082289.
Neighbor-joining (NJ) phylogenetic dendrogram, based on 16S rRNA gene sequence analysis showing the position of strain VRC21T within the genus Streptosporangium. Only bootstrap values above 50% (percentages of 1000 replications) are indicated. Actinomadura flavalba DSM 45200T (FJ157185) was taken as an out group. L, branch also recovered in the maximum-likelihood tree; P, branch also recovered in the maimum-parsimony tree. Bar 0.005 nucleotide substitutions per position.
DNA–DNA hybridization
The DNA–DNA relatedness values among strain VRC21T, S. carneum DSM 44125T and S. fragile DSM 43847T were in the range of 28–43% (Table 4). These values were below the value of 70% that was recommended by Wayne et al.47 for the assignment of strains to the same species, and these results thus confirm that strain VRC21T distinct from their closely related phylogenetic neighbors.
Conclusion
The phylogenetic analysis and morphological and chemotaxonomic properties indicated that the strain VRC21T belongs to the genus Streptosporangium. However, DNA–DNA relatedness values between strain VRC21T and the closely related type strains were below 70% (Table 4). Furthermore, colonies of strain VRC21T could also be distinguished from its closest relatives by additional phenotypic characteristics. In strain VRC21T, the spore shape was spherical, whereas in S. carneum DSM 44125T and S. fragile DSM 43847T, spores were in oval shaped. Strain VRC21T produced very dark reddish brown colored substrate mycelium, whereas S. carneum DSM 44125T produced orange to yellow brown colored and S. fragile DSM 43847T produced dark brown to black colored substrate mycelium. S. carneum DSM 44125T did not produce a soluble pigment and S. fragile DSM 43847T produced a brown colored diffusible pigment, but strain VRC21T produced a very dark reddish brown colored pigment. A comparison of the cultural and physiological characteristics and fatty acid composition of strain VRC21T with its closest relatives of the genus Streptosporangium is given in Tables 1, 2 and 3.
Furthermore, as shown in Tables 1 and 2, many differences among strain VRC21T, S. carneum DSM 44125T and S. fragile DSM 43847T were observed. On the basis of the data presented in this study, we suggest that VRC21T represents a novel species of the genus Streptosporangium, for which the name Streptosporangium terrae sp. nov. is proposed.
Description of Streptosporangium terrae sp. nov.
Streptosporangium terrae (ter’rae. L. gen. n. terrae of the earth).
It is a aerobic, Gram-positive, non motile actinomycete with extensive, non-fragmented dark reddish brown colored substrate mycelium. The sporangia contains spherical shape smooth spores. Colony appears leathery and cottony with aerial mycelium, sporangial walls are thick, and abundant big sporangiophores with spherical and non-motile spores are present. A very dark reddish brown colored diffusible pigment is observed. Colony size is 2–4 mm. The temperature and pH range for growth were 10–35 °C and pH 7–9, with optimum growth at 30 °C and pH 7.0, and tolerates 0–3% NaCl. Siderophore activity is not seen. Growth is not found in the presence of inhibitory compounds such as crystal violet (0.05% w/v), phenol (1.5% w/v), sodium azide (0.001% w/v), lysozyme (0.005% w/v), potassium tellurite (0.005% and 0.01% w/v). Acid is produced from lactose, inositol, sorbitol, mannitol and maltose. No acid is produced from arabinose, xylose, adonitol, rhamnose and glucose. It gives a positive result in tests of catalase, oxidase and urease activities. Positive results were observed for gelatin liquefaction, milk coagulation, peptonization and starch hydrolysis. Tweens 40, 80 and aesculin are not hydrolysed. It is able to degrade casein, starch, urea, xanthine and hypoxanthine. It does not degrade adenine, chitin, elastin and tyrosine. Nitrate is not reduced to nitrite and H2S gas is not produced. Enzyme activities of the API ZYM system are positive for esterase (C4), valine allylamidase, α-chymotrypsin, β-galactosidase, β-glucuronidase, α-glucosidase, β-glucosidase, N-acetyl-β-glucosaminidase and α-mannosidase, weakly positive for esterase lipase (C8) and negative for trypsin, acid phosaphatase, napthol-AS-BI-phosphohydrase. The Voges Proskauer’s reactions and methyl red are positive; lysine, ornithine, citrate and malonate utilization, phenylalanine deamination, and indole, inulin, sodium gluconate, glycerol, salicin, dulcitol, arbitol, erythritol and alpha-methyl-D-glucoside are positive. It produces melanoid pigments. It utilizes a variety of organic compounds as sole carbon sources, including inulin, arabinose, dextrin, dextrose, fructose, glycerol, galactose, mannose, maltose, starch, xylose, raffinose, rhamnose, lactose and sucrose. But sodium gluconate, cellobiose, melibiose, saccharose and trehalose are not utilized. It utilizes arginine, asparagine, histidine, methionine, proline and tyrosin. It is resistant to ofloxacin (1 μg l−1), penicillin G (1 μg l−1), cephalothin (1 μg l−1), gentamycin (1 μg l−1) and vancomycin (1 μg l−1), but susceptible to co-trimoxazole (1 μg l−1), clindamycin (1 μg l−1), erythromycin (1 μg l−1), kanamycin (25 μg l−1) and amphicillin (10 μg l−1). The major cellular fatty acids are iso-C16:1, C16:0 and C18:0 10-methyl, TBSA. Cell wall peptidoglycan contains meso-diaminopimelic acid. Whole-cell sugars are madurose, galactose, glucose, xylose and arabinose. Major diagnostic polar lipids are phosphatidylethanolamine, diphosphatidylglycerol, phosphatidylglycerol, phosphatidylinositol and two phosphatidylinositol-mannosides. The predominant menaquinones are MK-9 (H2) and MK-9 (H4). The G+C content of the genomic DNA of the type strain is 69.5±1.5 mol%. Inhibitory activity is showed against Staphylococcus aureus (MTCC 7443). The type strain, VRC21T (=KCTC 29207T=MTCC 11724T), was isolated from the rhizosphere of Callistemon citrinus collected from Hyderabad, India.
References
Couch, J. N. A new genus and family of the Actinomycetales, with a revision of the genus Actinoplanes. J. Elisha Mitchell Sci. Soc. 71, 148–155 (1955).
Nonomura, H. & Ohara, Y. Distribution of the actinomycetes in soil. V. The isolation and classification of the genus Streptosporangium. J. Ferment. Technol. 38, 405–409 (1960).
Iinuma, S., Yokota, A. & Kanamura, T. New subspecies of the genus Streptosporangium Streptosporangium amethystogenes subsp. fukuiense subsp. nov. Actinomycetologica 10, 35–42 (1996).
Schafer, D. Eine neue. Streptosporangium-Art aus turkischer Steppenerde. Arch. Mikrobiol. 66, 365–373 (in German) (1969).
Nonomura, H. & Ohara, Y. Distribution of the actinomycetes in soil. VII. A culture method effective for both preferential isolation and enumeration of Microbispora and Streptosporangium strains in soil. Part 2. Classification of the isolates. J. Ferment. Technol. 47, 701–709 (1969).
Kawamoto, I. et al. A new antibiotic victomycin (XK 49-1-B-2). I. Taxonomy and production of the producing organism. J. Antibiot. (Tokyo) 28, 358–365 (1975).
Shearer, M. C., Colman, P. M. & Nash, C. H. III Streptosporangium fragile sp. nov. Int. J. Syst. Bacteriol. 33, 364–368 (1983).
Mertz, F. P. & Yao, R. C. Streptosporangium carneum sp. nov. isolated from soil. Int. J. Syst. Bacteriol. 40, 247–253 (1990).
Zhang, L., Jiang, C. & Chen, W. Streptosporangium subroseum sp. nov., an actinomycete with an unusual phospholipid pattern. Int. J. Syst. Evol. Microbiol. 52, 1235–1238 (2002).
Zhang, L. P., Jiang, C. L. & Chen, W. X. Streptosporangium yunnanense sp. nov. and Streptosporangium purpuratum sp. nov., from soil in China. Int. J. Syst. Evol. Microbiol. 55, 719–724 (2005).
Zhang, L. P., Zhang, L.M. & Zhang, X. M. Streptosporangium canum sp. nov., isolated from soil. Int. J. Syst. Evol. Microbiol. 59, 1715–1719 (2009).
Inahashi, Y., Matsumoto, A., Omura, S. & Takahashi, Y. Streptosporangium oxazolinicum sp. nov., a novel endophytic actinomycete producing new antitrypanosomal antibiotics, spoxazomicins. J. Antibiot. 64, 297–302 (2011).
Anil, S. et al. Streptosporangium anatoliense sp. nov., isolated from soil in Turkey. Antonie Van Leeuwenhoek 102, 269–276 (2012).
Kämpfer, P., Glaeser, S. P., Grun-Wollny, I. & Busse, H. J. Streptosporangium sandarakinum sp. nov. Int. J. Syst. Evol. Microbiol. 63, 2484–2489 (2013).
Derek, C. Callisto: a very successful maize herbicide inspired by allelochemistry. Fourth World Congress on Alleopathy. (2005) (http://www.regional.org.au/au/allelopathy/2005/2/7/2636_cornesd.htm).
Shirling, E. B. & Gottlieb, D. Methods for characterization of Streptomyces species. Int. J. Syst. Bacteriol. 16, 313–340 (1966).
Waksman S. A. (ed.) in: The Actinomycetes Vol. 2, Williams and wilkins Co.: Baltimore, (1961).
Zhou, Z. H., Liu, Z. H., Qian, Y. D., Kim, S. B. & Goodfellow, M. Saccharopolyspora spinosporotrichia sp. nov., a novel actinomycete from soil. Int. J. Syst. Bacteriol. 48, 53–58 (1998).
Williams, S. T. & Davies, F. L. Use of scanning electron microscope for the examination of actinomycetes. J. Gen. Microbiol. 48, 171–177 (1967).
Vaddavalli, R. et al. Saccharopolyspora indica sp. nov., an actinomycete isolated from the rhizosphere of Callistemon citrinus (Curtis). Int. J. Syst. Evol. Microbiol. 64, 1559–1565 (2014).
Kelly, K. L. Inter-Society Color Council-National Bureau of Standards Color Name Charts Illustrated with Centroid Colors, US Government Printing Office: Washington, DC, (1964).
Pridham, T. G. & Gottlieb, D. The utilization of carbon compounds by some Actinomycetales as an aid for species determination. J. Bacteriol. 56, 107–114 (1948).
Radha, V., Sneha, P., Srilekha., Y. K. & Venkateswar, R. L. Nocardia bhagyanesis sp. nov., a novel actinomycete isolated from the rhizosphere of Callistemon citrinus (Curtis), India. Antonie Van Leeuwenhoek 105, 443–450 (2014).
Korn-Wendisch, F., Kempf, A., Grund, E., Kroppenstedt, R. M. & Kutzner, H. J. Transfer of Faenia rectivirgula Kurup and Agre 1983 to the genus Saccharopolyspora Lacey and Goodfellow 1975, elevation of Saccharopolyspora hirsute subsp. taberi Labeda 1987 to species level, and emended description of the genus Saccharopolyspora. Int. J. Syst. Bacteriol. 39, 430–441 (1989).
Dastager, S. G. et al. Seperation, identification and analysis of pigment (melanin) production in Streptomyces. Afr. J. Bio. 8, 1131–1134 (2006).
Hasegawa, T., Takizawa, M. & Tanida, S. A rapid analysis for chemical grouping of aerobic actinomycetes. J. Gen. Appl. Microbiol 29, 319–322 (1983).
Staneck, J. L. & Roberts, G. D. Simplified approach to identification of aerobic actinomycetes by thin layer chromatography. Appl. Microbiol 28, 226–231 (1974).
Sasser, M. Identification of bacteria by gas chromatography of cellular fatty acids, MIDI Technical Note 101, MIDI Inc: Newyark, DE, (1990).
Minnikin, D. E. et al. An integrated procedure for the extraction of bacterial isoprenoid quinines and polar lipids. J. Microbiol. Methods 2, 233–241 (1984).
Komagata, K. & Suzuki, K. Lipids and cell-wall analysis inbacterial systematics. Methods Microbiol 19, 161–203 (1987).
Minnikin, D. E., Hutchinson, I. G., Caldicott, A. B. & Goodfellow, M. Thinlayer chromatography of methanolysates of mycolic acid containing bacteria. J. Chromatogr. 188, 221–233 (1980).
Collins, M. D., Pirouz, T., Goodfellow, M. & Minnikin, D. E. Distribution of menaquinones in actinomycetes and corynebacteria. J. Gen. Microbiol. 100, 221–230 (1977).
Groth, I. et al. Demetria terragena gen. nov., sp. nov., a new genus of actinomycetes isolated from compost soil. Int. J. Syst. Bacteriol. 47, 1129–1133 (1997).
Uchida, K. & Aida, K. Acyl type of bacterial cell wall: Its simple identification by calorimetric method. J. Gen Appl. Microbiol 23, 249–260 (1977).
Marmur, J. A procedure for the isolation of deoxyribonucleic acid from microorganisms. J. Mol. Biol. 3, 208–218 (1961).
Marmur, J. & Doty, P. Determination of the base composition of deoxyribonucleic acid from its thermal denaturation temperature. J. Mol. Biol. 5, 109–118 (1962).
Chung, Y. R. et al. Kitasatospora cheerisanensis sp. nov., a new species of the genus Kitasatospora that produces an antifungal agent. Int. J. Syst. Bacteriol. 49, 753–758 (1999).
Gonzalez, J. M. & Saiz-Jimenez, C A. Simple fluorimetric method for the estimation of DNA-DNA relatedness between closely related microorganisms by thermal denaturation temperatures. Extremophiles 9, 75–79 (2005).
Larkin, M. A. et al. Clustal W and Clustal X version 2.0. Bioinformatics 23, 2947–2948 (2007).
Kimura, M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 16, 111–120 (1980).
Kim, O. S. et al. Introducing EzTaxon-e: a prokaryotic 16S rRNA Gene sequence database with phylotypes that represent uncultured species. Int. J. Syst. Evol. Microbiol. 62, 716–721 (2012).
Tamura, K., Stecher, G., Peterson, D., Filipski, A. & Kumar, S. MEGA6: Molecular Evolutionary Genetics Analysis Version 6.0. Mol. Biol. Evol. 30, 2725–2729 (2013).
Saitou, N. & Nei, M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4, 406–425 (1987).
Kluge, A. G. & Farris, F. S. Quantitative phyletics and the evolution of anurans. Syst. Zool. 18, 1–32 (1969).
Felsenstein, J. Evolutionary trees from DNA sequences: a maximum likelihood approach. J. Mol. Evol. 17, 368–376 (1981).
Felsenstein, J. Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39, 783–791 (1985).
Wayne, L. G. et al. International Committee on Systematic Bacteriology. Report of the ad hoc committee on reconciliation of approaches to bacterial systematics. Int. J. Syst. Bacteriol. 37, 463–464 (1987).
Acknowledgements
We are grateful to Dr Ch. Mohan Rao, Director (Center for Cellular and Molecular Biology, Hyderabad) for providing facilities to do key experiments, Dr R.B.N. Prasad, Head, Lipid Science & Technology (Indian Institute of Chemical Technology, Hyderabad) for providing facilities for chemotaxonomic studies. We thank Professor Jean Paul Marie Euzeby for his help with the nomenclature. We are thankful to Dr K. Suresh, Scientist, and Mr Pradeep Kumar Singh (Institute of Microbial Technology, Chandigarh) for helping in phospholipid analysis.
Author information
Authors and Affiliations
Corresponding author
Additional information
Supplementary Information accompanies the paper on The Journal of Antibiotics website
Supplementary information
Rights and permissions
About this article
Cite this article
Vaddavalli, R., Gaddam, B. & Linga, V. Streptosporangium terrae sp. nov., a novel actinomycete isolated from the rhizosphere of Callistemon citrinus (Curtis), India. J Antibiot 68, 425–430 (2015). https://doi.org/10.1038/ja.2015.5
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ja.2015.5