Abstract
Antimicrobial peptides (AMPs) are multifunctional compounds that may show antimicrobial and immunomodulatory activities. With the rapid increase in the incidence of multidrug-resistant bacteria, there is an enormous interest in AMPs as templates for the production of new antibiotics. However, there are concerns that the therapeutic administration of AMPs can select resistant strains. In order to distinguish between resistant and non-resistant strains and verify resistance specificity to AMPs, in this study a magainin I-resistant Escherichia coli model was used. First, the identity of all strains was confirmed by matrix-assisted laser desorption ionization-time of flight (MALDI-TOF)-MS, VITEK 2 and MicroScan, and the susceptible and magainin-resistant strains were successfully differentiated by MALDI-TOF-MS analysis. Furthermore, cross-resistances to a broad spectrum of antibiotics were evaluated, showing that all E. coli strains are susceptible to the drugs tested, suggesting that the resistance seems to be specific to AMPs. Finally, the specific resistance to magainin I compared with other AMPs was checked by microdilution. This experiment showed that the magainin MICs were 62 and 104 μM for susceptible and resistant strains, respectively. The other AMPs MICs were 3.4 μM to proline-arginine-rich 39-amino-acid peptide, 43 μM to porcine myeloid antimicrobial 23-amino-acid peptide-23 and 1.2 μM to cecropin P1 for all strains, demonstrating any additional resistance to peptides here evaluated, confirming that the resistance seems to be essentially specific to magainin I. In summary, the data reported here reinforce the proposal that magainin I seems not to be merely a membrane disruptor, probably showing additional molecular targets in pathogenic bacteria.
Similar content being viewed by others
Introduction
There is an emerging global health concern: while bacteria increasingly adapt and develop mechanisms to become resistant to the broad spectrum of conventional antimicrobial drugs, the discovery and development of new antibiotic drugs is clearly in decline.1, 2, 3 Infectious diseases caused by resistant bacteria are commonly associated with higher mortality and morbidity levels in hospitals worldwide.4, 5, 6 This situation is made even worse by a poor pipeline for the discovery of new antibiotics to treat health-care-associated multidrug-resistant Gram-negative infections such as in Klebsiella ssp., Enterobacter ssp. and Escherichia coli.7
For this reason, studies have been carried out to identify target molecules that could be the focus for the development of new therapeutic drugs to combat resistant pathogens and to treat severe infectious diseases.8 In this context, the antimicrobial peptides (AMPs), commonly produced with an innate immune response by microorganisms, plants and animals alike, have generated interest as promising lead compounds for new classes of antimicrobial drugs.7, 8, 9
AMPs are effective small polypeptides widespread among organisms ranging from the microorganism to humans.10, 11, 12 Their mechanisms of action are not fully elucidated,8 but in general they are correlated with direct interaction on bacterial membranes to form pores leading to membrane disturbance and cell lysis.13, 14 Additional mechanisms of action and targets include inhibition of DNA, RNA and protein synthesis, inhibition of membrane proteins and enzymes, activation of autolysins, interference in cell wall formation, formation of macropinosomes15, 16, 17, 18 and immunomodulatory activities,19, 20 that is, angiogenesis, cytokine production, chemotactic function,21 histamine and prostaglandin release and modification of cell migration and maturation.13 The mechanisms of AMP action seem to be highly complex, and these mechanisms might work together to provide a potential antimicrobial activity.
The magainins isolated from a skin secretion of the African clawed frog Xenopus laevis,22 are α-helical cationic peptides of 23 amino-acid residue length with bactericidal activity. So far, the main proposed modes of action for magainins are correlated with the random insertion of the peptide hydrophobic regions into the bacterial membrane, forming pores, disturbing membrane stability and further causing cell membrane disruption.14, 23 However, the internalization of magainin by E. coli has been observed, suggesting a possible intracellular target and perhaps a different and more complex mechanism of action than only membrane pore formation.16
Despite the complexity of their mechanisms of action, resistance to AMPs due to bacterial adaptation has been observed for Salmonella enterica with resistance to PR-39 (proline-arginine-rich 39-amino-acid peptide),24 and E. coli resistant to magainin I.25 To explore the mechanisms of action and resistance to AMPs, we sought a bacterial AMP-resistant model among susceptible and magainin I-resistant strains of E. coli. The E. coli strains were first differentiated between susceptible and resistant strains to magainin I by using matrix-assisted laser desorption ionization-time of flight (MALDI-TOF)-MS analysis and then the level of resistance specificity to magainin I was evaluated and compared with other AMPs and antibiotics.
Materials and methods
Bacterial strains
The magainin I susceptible and resistant strains were prepared from E. coli ATCC 8739 (American Type Culture Collection, Manassas, VA, USA) as previously described.25 Briefly, a single colony of E. coli ATCC 8739 isolated by the streak method in Luria-Bertani (LB) solid medium was cultivated in LB broth at 37 °C, shaking at 240 r.p.m. for 16 h in the absence or presence of magainin I at 0.5 × MIC in order to induce resistance.26 After 10 successive propagations under the same experimental conditions, the colonies selected were grown in LB liquid medium at 37 °C and shaken at 240 r.p.m. for 16 h; the inoculums were stored in sterile 10% glycerol at −80 °C until the experiments. The bacterial suspensions stock of the magainin-resistant colonies were named R1, R2 and R3, and the control magainin-susceptible colonies were called C1, C2 and C3; these were used in all of the ensuing experiments.
Strain identification by automated microbiological systems
The culture strains (E. coli C1, C2 and C3 controls; R1, R2 and R3 magainin-resistant strains) were propagated on LB agar plates and then sub-cultured on MacConkey agar plates at 37 °C for 16 h. After this, the bacterial inoculum for the MicroScan WalkAway system (Siemens Healthcare Diagnostics, Deerfield, IL, USA) was prepared with a wand by the Prompt Inoculation System (3M Company, St Paul, MN, USA) in a standard manufacturer’s solution for all tested strains, and the MC50 test panel wells were loaded with Renok rehydrating (Siemens Healthcare Diagnostics). The bacterial suspension for the VITEK 2 system (BioMérieux, Durham, NC, USA) was prepared with 1–3 colonies from each strain diluted in 0.9% saline. In the turbidity meter DensiChek (BioMérieux), the turbidity was adjusted to 0.5 McFarland standard according to manufacturer’s directions, and this inoculum was immediately used to load the ID-GNBcard for Gram-negative identification. For bacterial identification via automated systems, the biochemical results obtained after sample incubation were compared against the taxonomic database from the software, and a numerical probability calculation was performed to determine a numerical value for proximity. On the basis of numerical probability calculation, different confidence levels of identification were assigned, such as 96–99% of probability, excellent; 93–95%, very good; 89–92%, good; 85–88%, acceptable.
MALDI-TOF-MS strain identification
In order to carry out identification of the strains and differentiate between resistant and susceptible, MALDI-TOF-MS and BioTyper software version 3.0 (Bruker Daltonics, Fremont, CA, USA) were used. The samples for MALDI-TOF-MS were prepared as previously described.27 First, the E. coli magainin susceptible (C1, C2 and C3) and the E. coli magainin-resistant (R1, R2 and R3) strains were cultured for 12 h at 37 °C in solid LB medium. Then six colonies of each strain were individually homogenized in 1 ml of 70% ethanol. The suspensions were centrifuged at 13 000 g for 2 min and the supernatant was discarded. The pellet was resuspended in 10 μl of 70% formic acid and 100% acetonitrile in a 1:1 ratio (v/v). Following homogenization, the samples were centrifuged at 13 000 g for 2 min. Then, 1 μl of the final supernatant was applied to a MALDI plate (Bruker Daltonics) and air dried at 25 °C, and a further 1 μl of a saturated matrix solution of α-cyano-4-hydroxycinnamic acid (10 mg ml−1) in 0.3% trifluoroacetic acid and 50% acetonitrile was added. The samples were analyzed on a Microflex LRF mass spectrometer (Bruker Daltonics) using the model-based testing method in a positive linear mode. More than 24 spectra of each strain were acquired over the m/z range of 2000–20 000 for protein profile generation.
Bruker MALDI BioTyper spectral analysis
The acquired spectra of each strain were smoothed; the baseline was corrected and compared against MALDI Biotyper database version 3.0, which contains 3995 reference microorganisms. The standard parameters of the pattern-matching algorithm were applied. The MALDI Biotyper break points consist of a log (score) in the range 0–3.0 by comparing the peak list for an unknown strain with the reference database, and a score (log) near 2.0 is considered a good correlation within species level.28
MSP dendrogram
The 24 mass spectra acquired by MALDI-TOF-MS for each strain were used to construct a main spectrum profile (MSP) to allow bacterial differentiation between the susceptible and resistant E. coli strains to magainin I. The MSP dendrogram was performed using the standard parameters of the flexAnalysis software (Bruker Daltonics) considering the mass range from 3000 to 15 000 Da with a resolution set to one, the factor of compression set to 10, tolerance of distance of 300, tolerance of peak 25, intensity correction function 0.25, the baseline subtracted and intensity smoothed by the Savitzky Golay algorithm. For the differentiation between resistant or susceptible E. coli strains, the MSP of the E. coli strains were compared with each other and they were clustered in an analysis tree.
Antimicrobial susceptibility assay via MicroScan and VITEK 2 systems
The bacterial suspensions of all susceptible and magainin I-resistant strains were prepared according to the manufacturer’s directions, as described above in strain identification by automated microbiological systems. The antimicrobial susceptibility testing was performed using the MC50 test panel and the AST-N105 test card for the MicroScan WalkAway system (Siemens) and VITEK 2 system (BioMérieux), respectively. The broad spectrum of antibiotic resistance was tested for amikacin, ampicillin, amoxicilin/clavulanate, aztreonam, cefepime, cefotaxime, cefoxitin, cefazolin, ceftazidime, ceftriaxone, cefuroxime, ciprofloxacin, gentamicin, levofloxacin, meropenem, imipenem, ertapenem, piperacillin, piperacillin/tazobactam, tetracycline, ticarcillin/clavulanate, tobramycin and trimethoprim/sulfamethoxazole via the MicroScan system and for amikacin, ampicillin, ampicillin/sulbactam, piperacillin/tazobactam, aztreonam, cephalothin, cefepime, cefotaxime, cefoxitin, ceftazidime, ciprofloxacin, colistin, gentamicin, meropenem, imipenem, ertapenem and tigecycline via the VITEK 2 system. After incubation, the automated systems calculated the MIC for each antimicrobial tested, and the strain was classified as susceptible (S), intermediate (I) and resistant (R) according to the interpretative break points for MIC described in the Clinical and Laboratory Standards Institute document M100-S22.29
Evaluation of AMP susceptibility
Magainin I (Peptide 2.0, Chantilly, VA, USA), cecropin P1, PMAP-23 (porcine myeloid antimicrobial 23-amino-acid peptide) and PR-39 (Shanghai Hanhong Chemical, Shanghai, China) were synthesized by the Fmoc (9-fluorenylmethyl carbamate) method. These peptides were chosen according to two main characteristics. First, they are from the same family of magainin I, the cathelicidins, and second, cecropin P1 and PMAP-23 have a similar magainin I mode of action. In contrast, PR-39 has a completely different mechanism of action, allowing the other AMPs and magainin I to be compared. The lyophilized powder of these peptides was reconstituted in ultrapure sterile water and stored at−80 °C until use. The concentration and purity (>95% purity) were assessed by MS analyses using an Autoflex Speed mass spectrometer (Bruker Daltonics) by the model-based testing method in a positive linear mode over the range of 500–5000 Da.28 In order to determine the peptide’s MICs, all the E. coli strains were tested in a 96-well polypropylene microplate by the broth microdilution method according to the Clinical and Laboratory Standards Institute document M07-A9.30 The bacterial suspension was inoculated to a final concentration of 5 × 105 CFU ml−1 in LB broth containing increasing concentrations of the peptides at 1.2–124 μM. Milli-Q water (EMD Millipore, Billerica, MA, USA) and LB without peptides were used as negative controls and chloramphenicol 99 μM was applied as a positive control. The plates were incubated at 37 °C for 12 h with stirring at 40 r.p.m. and the end point of bacterial growth was measured at 595 nm. The experiment was performed in triplicate.
Results
Strain identification by automated systems and MALDI-TOF-MS
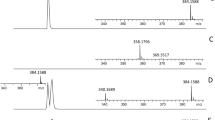
The VITEK and MicroScan systems identified the magainin I-resistant and -susceptible E. coli strains with confidence identification levels of 95% and 99.99%, respectively (data not shown). The Bruker Biotyper MALDI-TOF-MS analyses matched all magainin I-resistant and -susceptible E. coli strains present in the database consistent with a log pattern-algorithm score of 1.93, which is considered a reliable correlation within species levels (Figure 1).
Matrix-assisted laser desorption ionization-time of flight mass spectra of E. coli strains. C1–3, magainin I-susceptible control strains; R1–3, magainin I-resistant strains. The proteins were analyzed on a Microflex LRF mass spectrometer in positive linear mode using the model-based testing method. Axis ‘y’ shows the intensity and axis ‘x’ shows mass/charge.
The MALDI-TOF-MS analysis in positive linear mode from 2000 to 20 000 Da demonstrated a similar, but not identical, protein profile among all magainin I-resistant and -susceptible E. coli strains (Figure 1). An MSP dendogram constructed using flexAnalysis software was used to generate an MSP dendogram to analyze all 24 acquired spectra from each strain, and the results were compared in a cluster tree. The MSP dendrogram showed a clear group cluster difference between magainin I-resistant and -susceptible strains owing to subtle modifications in molecular masses between magainin I-resistant and -susceptible strains (Figure 2).
Susceptibility to antimicrobial drugs
With the aim of analyzing the resistance specificity to magainin I, the magainin-resistant and -susceptible E. coli strains, we used the Vitek and MicroScan automated expert systems to test bacterial susceptibility to 27 antibiotics used in clinical practice. These represent 11 distinct antimicrobial classes with distinct mechanisms of action correlated with bacterial metabolism and the physiologic process, such as DNA replication, translation and biosynthesis of the bacterial cell wall.8 The antimicrobial susceptibility testing results, interpreted according to the Clinical and Laboratory Standards Institute29 parameters, showed that all of the magainin-resistant and -susceptible strains were susceptible to all of the antibiotics tested (Tables 1 and 2), confirming that AMP resistance is very specific and distinct to that observed for antibiotics tested.
Evaluation of magainin I-resistant E. coli strains for susceptibility/resistance to AMPs
The three cationic AMPs magainin I, cecropin P1 and PMAP-23, with a mechanism of action related to cell lysis mediated by pores in bacterial membranes,13, 14 and PR-39, with a mechanism of action related to interruption of DNA and protein synthesis24 (Table 3), allowed a comparison between magainin I and other AMPs.
Susceptibility to AMPs was also tested by the broth microdilution method. The magainin I MICs for magainin I-susceptible, and -resistant strains were 62 μM and 104 μM, respectively (Table 4). There were no differences in MICs between magainin I-susceptible and -resistant strains for the other cationic AMPs. The MICs were 3.4, 43 and 1.2 μM for PR-39, PMAP-23 and cecropin P1, respectively (Table 4).
According to this, no cross-resistance was developed by the magainin-resistant strains for the AMPs tested, and this shows that the resistance observed in R1, R2 and R3 strains seems to be essentially specific to magainin I. The resistance specificity to magainin, evaluated by antimicrobial susceptibility testing and the microdilution method, demonstrates that this AMP seems not to be merely a membrane disruptor, probably having diverse other targets among pathogenic bacteria. The same could be concluded for the other peptides evaluated here.
Discussion
The prevalence of multidrug-resistant bacterial pathogens has inspired a widespread search for new antibacterial agents. In this regard, AMPs may be promising because it is apparently difficult for bacteria to develop resistance to these peptides.9, 31, 32
In this work, the specificity of resistance to AMPs was investigated by using a magainin I-resistant bacterial model to contribute to a better understanding of magainin’s mode of action and resistance. This model was provided by successive propagations with E. coli ATCC 8739 at sub-inhibitory concentration of magainin I in accordance with Maria-Neto et al.25 to promote magainin I-resistant strains. Multiple mechanisms of action are involved in the antimicrobial activities observed with various magainin classes. In turn, magainin-resistant bacteria appear to have several mechanisms of action likely the result of a complex defense system, much of which has not yet been described in the literature.25
The automated expert systems, that is, VITEK 2, BD Phoenix and MicroScan WalkAway33 are often applied in microbiology analysis and allow bacterial identification through a comparison between the colorimetric reaction and fluorescence quantification results obtained from classical biochemical analysis and the database available from the equipment software.34 Then, all the strains were identified as E. coli, as expected, with a 95% score, which is considered very high accuracy for Vitek software, and a 99.99% score, considered excellent confidence for MicroScan analysis.
In our studies, the VITEK 2, BD Phoenix and MicroScan WalkAway automated expert systems both successfully identified all of the E. coli strains in about 24–48 h. In contrast, the MALDI-TOF-MS for rapid strain identification, had comparable accuracy but was capable of precisely identifying many strains in a single analysis within a few minutes. We propose therefore, that this tool in the clinical laboratory would allow rapid and precise identification of clinical pathogens at reduced cost and time, resulting in more rapid initiation of appropriate chemotherapy,35, 36 and probably fewer therapeutic failures owing to the use of ineffective antibiotics.3, 4
In addition, MALDI-TOF-MS analyses allowed the distinction of susceptible and magainin I-resistant bacteria (Figure 2), which as far as we know is described here for the first time in the literature. This result demonstrated that the difference between the degree of distance from susceptible and resistant strains could be correlated with subtle physiological differences, allowing molecular mass modifications and difference in ions’ intensity, which can be easily detected by MALDI-TOF-MS analysis.37, 38
MALDI-TOF-MS has been used to rapidly discriminate bacterial strains resistant to a single antibiotic or multiple antibiotics, such as Streptococcus pneumoniae strains resistance to ciprofloxacin, ofloxacin, levofloxacin or moxifloxacin,39 methicillin-resistant Staphylococcus aureus strains38, 40 and antibiotic-resistant E. coli.38, 41
Our data demonstrate that magainin resistance was specific and there does not appear to be cross-resistance to antibiotic resistance. A recent study by our group, which examined comparative proteomics of magainin I-resistant E. coli strains, showed similar growth profiles between susceptible and resistant strains. However, the most important protein differences in resistant strains were directed to improve cell energy maintenance (∼49% of differential identified cytosolic proteins), suggesting an intense energy metabolism in the magainin-resistant strains.25 In summary, data reported here introduce a new understanding of magainin-specific bacterial resistance.
The AMPs tested in this study, PMAP-23, cecropin P1 and PR-39 are all members of the cathelicidin family of magainin peptides. Both the cathelicidins and the defensins, belong to a large group of primarily cationic, amphipathic AMPs active at the microbial membrane.21, 42 Cathelicidins are found in myeloid (also named myeloid AMPs) and epithelial tissues, isolated from diverse organisms21, 43 that is, the cecropins from insect tissues,44 the magainins from frog skin22 and cecropin P1 from a porcine nematode.45
The cathelicidin family members vary in amino-acid sequence (Table 3), structure and size.42, 46 They have complex mechanisms of action, usually associated with their charge and hydrophobicity pattern, which enable bacterial membrane binding and control subsequent antimicrobial activity.47 In order to verify the specificity of resistance to magainin I, all magainin-resistant and -susceptible strains were tested by microdilution with the cathelicidin family members, PR-39, PMAP-23, cecropin P1 and also magainin I. PMAP-23, identified from cDNA cloning encoding a putative AMP precursor of porcine bone marrow48, 49 and cecropin P1, first isolated from the porcine small intestine nematode Ascaris suum,45, 50 are amphipathic α-helical peptides, composed of 23 and 31 amino-acid residues, respectively. The PR-39, originally isolated from the porcine small intestine,51 is a linear AMP with 39 amino-acid residues and an unusual high proline content (∼49%) and arginine (∼26%) residues (Table 3).43 PR-39 AMP mediates bacterial killing via a mechanism that is not involved in pore formation and cell lysis, mainly through stereospecific interaction with bacterial cells, resulting in the interruption of DNA and protein synthesis,24, 43, 52 whereas the antimicrobial activity of PMAP-23 and cecropin P1 is related to the amphipathic helix interaction and penetration into the lipid bilayer bacterial membrane, forming pores and causing its disruption in a manner similar to that proposed for magainins.43, 53, 54
Every peptide’s mechanism of action described above, similarly to magainin mechanism of action,49, 55, 56 has been associated with the peptide structural conformation, allowing interaction with a bacterial cell either with or without translocation across biological membranes.24, 43, 52 However, the microdilution results demonstrated that the magainin-resistant strains have not developed cross-resistance to the other AMPs tested (Table 4). These data suggest that the resistance seems to be specific to magainin I in all the resistant strains (R1, R2 and R3), as it was only reported for this AMP (Table 4).
Magainin I-specific resistance reported here, indicates that the previously reported magainin mechanism of action, formation of toroidal pores in the bacterial membrane,56 is probably not as simple as previously thought. In contrast, these data suggest that the AMPs have a much more complex mode of action, and in accordance with previous reports, is not the result of a single molecular mechanism.25 This suggests that there may well be other cathelicidins effective against magainin-resistant bacteria.
The precise AMP mechanism of action still remains to be elucidated, and the understanding of AMPs properties can be of extreme importance for clinical development of peptide-based therapies. The study reported here describes the specificity of but one of a large family of AMPs. Future research using different bacterial resistance models are warranted to broaden our knowledge of AMPs in an effort to identify potential candidate peptides with a high potential for clinical application as novel antimicrobial agents to treat emergent multidrug-resistant bacteria.
References
Fischbach, M. A. & Walsh, C. T. Antibiotics for emerging pathogens. Science 325, 1089–1093 (2009).
Conly, J. M. Antimicrobial resistance programs in Canada 1995–2010. Antimicrob. Resist. Infect. Control 1, 10–15 (2012).
Rosenthal, V. D. Health-care-associated infections in developing countries. Lancet 377, 186–188 (2011).
Park, S. H. et al. Emergence of extended-spectrum β-lactamase-producing Escherichia coli as a cause of community-onset bacteremia in South Korea: risk factors and clinical outcomes. Microb. Drug Resist. 17, 537–544 (2011).
World Health Organization.. Report on the Burden of Endemic Health Care-Associated Infection Worldwide, WHO: Geneva, Switzerland, (2011).
Karakoc, C., Tekin, R., Yesilbag, Z. & Cagatay, A. Risk factors for mortality in patients with nosocomial Gram-negative rod bacteremia. Eur. Rev. Med. Pharmacol. Sci. 17, 951–957 (2013).
Arias, C. A. & Murray, B. E. Antibiotic-resistant bugs in the 21st century-a clinical super-challenge. New Engl. J. Med. 360, 439–443 (2009).
Lima, T. B. et al. Bacterial resistance mechanism: what proteomics can elucidate. FASEB J. 27, 1291–1303 (2013).
Bocchinfuso, G., Bobone, S., Mazzuca, C., Palleschi, A. & Stella, L. Fluorescence spectroscopy and molecular dynamics simulations in studies on the mechanism of membrane destabilization by antimicrobial peptides. Cell. Mol. Life Sci. 68, 2281–2301 (2011).
Hancock, R. E. & Chapple, D. S. Peptide antibiotics. Antimicrob. Agents Chemother. 43, 1317–1323 (1999).
Candido, E. S. et al. Plant storage proteins with antimicrobial activity: novel insights into plant defense mechanisms. FASEB J. 25, 3290–3305 (2011).
Fulmer, P. A., Lundin, J. G. & Wynne, J. H. Development of antimicrobial peptides (AMPs) for use in self-decontaminating coatings. ACS Appl. Mater. Interfaces 4, 1266–1270 (2010).
Wiesner, J. & Vilcinskas, A. The ancient arm of the human immune system. Virulence 1, 440–464 (2010).
Huang, Y., Huang, J. & Chen, Y. Alpha-helical cationic antimicrobial peptides: relationships of structure and function. Protein Cell 1, 143–152 (2010).
Brogden, K. A. Antimicrobial peptides: pore formers or metabolic inhibitors in bacteria? Nat. Rev. Microbiol. 3, 238–250 (2005).
Splith, K. & Neundorf, I. Antimicrobial peptides with cell-penetrating peptide properties and vice versa. Eur. Biophys. J. 40, 387–397 (2011).
Nguyen, L. T., Haney, E. F. & Vogel, H. J. The expanding scope of antimicrobial peptide structures and their modes of action. Trends Biotechnol. 29, 464–472 (2011).
Pushpanathan, M., Gunasekaran, P. & Rajendhran, J. Antimicrobial peptides: versatile biological properties. Int. J. Pept. 13, 1–15 (2013).
Steinstraesser, L. et al. Innate defense regulator peptide 1018 in wound healing and wound infection. PLoS ONE 7, 1–7 (2012).
Pena, O. M. et al. Synthetic cationic peptide IDR-1018 modulates human macrophage differentiation. PLoS ONE 8, 1–10 (2013).
Steinstraesser, L., Kraneburg, U., Jacobsen, F. & Al-Benna, S. Host defense peptides and their antimicrobial-immunomodulatory duality. Immunobiology 216, 322–333 (2011).
Zasloff, M. Magainins, a class of antimicrobial peptides from Xenopus skin: isolation, characterization of two active forms and partial cDNA sequence of a precursor. Proc. Natl Acad. Sci. USA 84, 5449–5453 (1987).
Imura, Y., Choda, N. & Matsuzaki, K. Magainin 2 in action: distinct modes of membrane permeabilization in living bacterial and mammalian cells. Biophys. J. 95, 5757–5765 (2008).
Pranting, M., Negrea, A., Rhen, M. & Andersson, D. I. Mechanism and fitness costs of PR-39 resistance in Salmonella enterica serovar Typhimurium LT2. Antimicrob. Agents Chemother. 52, 2734–2741 (2008).
Maria-Neto, S. et al. Deciphering the magainin resistance process of Escherichia coli strains in light of the cytosolic proteome. Antimicrob. Agents Chemother. 56, 1714–1724 (2012).
Peng, X. Proteomic analysis of the sarcosine-insoluble outer membrane fraction of Pseudomonas aeruginosa responding to ampicillin, kanamycin and tetracycline resistance. J. Proteome Res. 4, 2257–2265 (2005).
Zautner, A. E. et al. Discrimination of multilocus sequence typing-based Campylobacter jejuni sub-groups by MALDI-TOF mass spectrometry. BMC Microbiol. 13, 247 (2013).
Mellmann, A. et al. High interlaboratory reproducibility of matrix-assisted laser desorption ionization-time of flight mass spectrometry-based species identification of nonfermenting bacteria. J. Clin. Microbiol. 47, 3732–3734 (2009).
Clinical and Laboratory Standards Institute.. Performance Standards for Antimicrobial Susceptibility Testing. 22nd informational supplement. CLSI document M100-S22. (CLSI, Wayne, PA, USA, 2012).
Clinical and Laboratory Standards Institute.. Methods for Dilution Antimicrobial Susceptibility Tests for Bacteria that Grow Aerobically 9th edn. Approved standard. CLSI document M07-A9. (CLSI, Wayne, PA, USA, 2012).
Epand, R. F. et al. Dual mechanism of bacterial lethality for a cationic sequence-random copolymer that mimics host-defense antimicrobial peptides. J. Mol. Biol. 379, 38–50 (2008).
Mckenna, M. The last resort: Health officials are watching in horror as bacteria become resistant to powerful carbapenem antibiotics—one of the last drugs on the shelf. Nature 499, 394–396 (2013).
Winstanley, T. & Courvalin, P. Expert systems in clinical microbiology. Clin. Microbiol. Rev. 24, 515–556 (2011).
Zbinden, A., Böttger, E. C., Bosshard, P. P. & Zbinden, R. Evaluation of the colorimetric Vitek 2 card for identification of Gram-negative nonfermentative rods: comparison to 16S rRNA gene sequencing. J. Clin. Microbiol. 45, 2270–2273 (2007).
Tan, K. E. et al. Prospective evaluation of a matrix-assisted laser desorption ionization-time of flight mass spectrometry system in a hospital clinical microbiology laboratory for identification of bacteria and yeasts: a bench-by-bench study for assessing the impact on time to identification and cost-effectiveness. J. Clin. Microbiol. 50, 3301–3308 (2012).
Van Veen, S. Q., Claas, E. C. J. & Kuijper, E. J. High-throughput identification of bacteria and yeast by matrix-assisted laser desorption ionization-time of flight mass spectrometry in conventional medical microbiology laboratories. J. Clin. Microbiol. 48, 900–907 (2010).
Arnold, R. J. & Reilly, J. P. Observation of Escherichia coli ribosomal proteins and their posttranslational modifications by mass spectrometry. Anal. Biochem. 269, 105–112 (1999).
Muroi, M., Shima, K., Igarashi, M., Nakagawa, Y. & Tanamoto, K. Application of matrix-assisted laser desorption ionization-time off light mass spectrometry for discrimination of laboratory-derived antibiotic-resistant bacteria. Biol. Pharm. Bull. 35, 1841–1845 (2012).
Malakhova, M. V. et al. MALDI-TOF mass-spectrometry in analysis of genetically determined resistance of Streptococcus pneumoniae to fluoroquinolones. Antibiotic Khimioter 52, 7–10 (2007).
Charyulu, E. M., Gnanamani, A. & Mandal, A. B. Identification and discrimination of merhicillin resistant Staphylococcus aureus strains isolated burn wound sites using PCR and authentication with MALDI-TOF-MS. Indian J. Microbiol. 52, 337–345 (2012).
Camara, J. E. & Hays, F. A. Discrimination between wild-type and ampicillin-resistant Escherichia coli by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. Anal. Bioanal. Chem. 389, 1633–1638 (2007).
Kosciuczuk, E. M. et al. Cathelicidins: family of antimicrobial peptides. Mol. Biol. Rep. 39, 10957–10970 (2012).
Sang, Y. & Blecha, F. Porcine host defense peptides: expanding repertoire and functions. Dev. Comp. Immunol. 33, 334–343 (2009).
Steiner, H., Hultmark, D., Engstrom, A., Bennich, H. & Boman, H. G. Sequence and specificity of two antibacterial proteins involved in insect immunity. Nature 292, 246–248 (1981).
Lee, J. Y. et al. Antibacterial peptides from pig intestine: isolation of a mammalian cecropin. Proc. Natl Acad. Sci. USA 86, 9159–9162 (1989).
Epand, R. M. & Epand, R. F. Bacterial membrane lipids in the action of antimicrobial agents. J. Pept. Sci. 17, 298–305 (2011).
Strauss, J., Kadilak, A., Cronin, C., Mello, C. M. & Camesano, T. A. Binding, inactivation and adhesion forces between antimicrobial peptides cecropin P1 and pathogenic E. coli. Colloids Surf. B Biointerfaces 75, 156–164 (2010).
Zanetti, M., Storici, P., Tossi, A., Scocchi, M. & Gennaro, R. Molecular cloning and chemical synthesis of a novel antimicrobial peptide derived from pig myeloid cells. J. Biol. Chem. 269, 7855–7858 (1994).
Kim, J. Y., Park, S. C., Yoon, M. Y., Hahm, K. S. & Park, Y. C-terminal amidation of PMAP-23: translocation to the inner membrane of Gram-negative bacteria. Amino Acids 40, 183–195 (2011).
Pillai, A., Ueno, S., Zhang, H., Lee, J. M. & Kato, Y. Cecropin P1 and novel nematode cecropins: a bacteria-inducible antimicrobial peptide family in the nematode Ascaris suum. Biochem. J. 390, 207–214 (2005).
Agerberth, B. et al. Amino acids sequence of PR-39. Isolation from pig intestine of a new member of the family of proline-arginine-rich antibacterial peptides. Eur. J. Biochem. 202, 849–854 (1991).
Hennig-Pauka, I. et al. PR-39, a porcine host defense peptide, is prominent in mucosa and lymphatic tissue of the respiratory tract in healthy pigs and pigs infected with Actinobacillus pleuropneumoniaie. BMC Res. Notes 5, 539–545 (2012).
Boman, H. G., Agerberth, B. & Boman, A. Mechanisms of action on Escherichia coli of cecropin P1 and PR-39, two antibacterial peptides from pig intestine. Infect. Immun. 61, 2978–2984 (1993).
Arcidiacono, S., Soares, J. W., Meehan, A. M., Marek, P. & Kirby, R. Membrane permeability and antimicrobial kinetics of cecropin P1 against Escherichia coli. J. Pept. Sci. 15, 398–403 (2009).
Lohner, K. New strategies for novel antibiotics: peptides targeting bacterial cell membranes. Gen. Physiol. Biophys. 28, 105–116 (2009).
Amiche, M. & Galanth, C. Dermaseptins as models for the elucidation of membrane-acting helical amphipathis antimicrobial peptides. Curr. Pharm. Biotechnol. 12, 1184–1193 (2011).
Acknowledgements
This study was supported by grants from CNPq, CAPES, FAPDF and UCB.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
de Almeida, K., Lima, T., Motta, D. et al. Investigating specific bacterial resistance to AMPs by using a magainin I-resistant Escherichia coli model. J Antibiot 67, 681–687 (2014). https://doi.org/10.1038/ja.2014.48
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ja.2014.48
This article is cited by
-
Comparative NanoUPLC-MSE analysis between magainin I-susceptible and -resistant Escherichia coli strains
Scientific Reports (2017)
-
Identification of bacterial invasion in necrotizing enterocolitis specimens using fluorescent in situ hybridization
Journal of Perinatology (2017)
-
An anti-infective synthetic peptide with dual antimicrobial and immunomodulatory activities
Scientific Reports (2016)