Abstract
Five new phenethylamine (PEA) derivatives (1–5) were isolated from the strain of Arenibacter nanhaiticus sp. nov. NH36AT derived from the marine sediment of the South China Sea by bioassay-guided fractionation. Their structures were elucidated by spectroscopic methods including UV, IR, HR-MS and NMR. Interestingly, compounds 1–4 existed as enantiomers, which were resolved by chiral liquid chromatography. The resolved configuration of each enantiomer was assigned by the Marfey’s method. Of these compounds, 5 showed weak antimicrobial activity against Staphylococcus aureus and Bacillus subtilis with MIC values of 0.50 and 0.25 mg ml−1, respectively.
Similar content being viewed by others
Introduction
Phenethylamine (PEA), the simplest endogenous aromatic amine and potential pharmacophore, provides the basic chemical structure for several classes of drugs with wide biological activity.1, 2, 3, 4 Owing to this reason, PEA derivatives have attracted intensive interests recently and a series of new ones has been obtained from natural organisms and chemical syntheses, which exhibited diverse bioactivities, including bronchiectasia, antidepressant, stimulant, etc.5, 6 Nowadays, they have emerged as a promising biologically active scaffold in the search of new agents against human diseases.
In our ongoing work to search for antimicrobial components from marine microorganisms, five new PEA derivatives (1–5) were isolated from the strain of Arenibacter nanhaiticus sp. nov. NH36AT derived from the marine sediment of the South China Sea. Here, we reported the isolation and structural elucidation of new compounds (1–5). Moreover, the chiral resolution of enantiomers and the determination of their absolute configurations were also discussed.
Results
Structure determination
Compounds 1–4 (Figure 1) gave negative reaction with ninhydrin, but were positive after hydrolysis with concentrated HCl (6.0 N), which suggested that they were cyclo-peptides.7
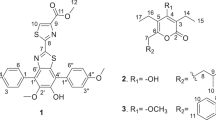
Compound 1 was obtained as a white amorphous powder. Its molecular formula was determined to be C22H25N3O3 based on the quasimolecular ion at m/z 380.1976 [M+H]+ (calcd 380.1974) in the HR-ESI-TOF MS, indicating 12 degrees of unsaturation. The 1H and 13C NMR spectra of 1 (Table 1) showed resonances for three amide N-H proton signals (δH 7.83 (1H, t, 5.2), 7.96 (1H, brs) and 8.12 (1H, brs)) and three ester/amide carbonyl carbon signals (δC 167.2, 167.3 and 171.1), which further suggested that it was a cyclopeptide. The characteristic methine resonances of α-CH at δ 3.01/52.8 and 4.12/55.4 indicated that it was a cyclodipeptide. The presence of a cluster of 10 aromatic protons at δH 7.17–7.28 in the 1H-NMR spectrum suggested that it contained two monosubstituted benzene rings. Detailed analysis of 1H_1H COSY, HSQC and HMBC data of 1 established three residues as phenylalanine (Phe), glutamic acid (Glu) and N-phenethyl group (Figure 2a). According to the HMBC correlation between H2-1′′ and C-5 (Glu), the structure of 1 was established to be cyclo (Phe-Glu) substituted with N-phenethyl, which was a new PEA derivative.
The optical rotation value of 1 was almost zero, which suggested it should be a racemate. Chiral resolution of 1 was performed by Lux Amylose-2 chiral liquid chromatography (phenomenex, φ 5 μm, 4.6 × 250 mm) using 100% CH3CN at 1.0 ml min−1. Two enantiomers were obtained from 1 (Figure 3a), namely 1a and 1b, at tR 17.11 and 24.72 min, respectively, and their relative content was nearly identical according to their relative peak areas (53.4% for 1a and 46.6% for 1b) in the HPLC chromatogram.
Chiral HPLC analytical chromatograms of compounds 1–4. (a) Chiral HPLC analytical chromatogram of compound 1; (b) Chiral HPLC analytical chromatogram of compound 2; (c) Chiral HPLC analytical chromatogram of compound 3; (d) Chiral HPLC analytical chromatogram of compound 4. A full color version of this figure is available at The Journal of Antibiotics journal online.
Marfey’s method8, 9 was applied to assign the absolute configuration of enantiomers 1a and 1b. The FDAA derivatives of the acid hydrolysate of 1a/1b and the authentic D- and L-amino acids were subjected to HPLC analysis (Cosmosil C18 column, φ 5 μm, 4.6 × 250 mm, 0.8 ml min−1) at 35 °C using the following gradient program: solvent A, H2O (0.1% HCOOH); solvent B, CH3CN; linear gradient, 30–80% of B in A over 30 min with UV detection at 340 nm. The absolute configurations of amino acid residues in 1a/1b were established by comparison of retention times of their derivatives with those of corresponding authentic D- and L-amino-acid derivatives. Therefore, absolute configurations of the Glu and Phe residues of 1a were assigned to be D-Glu and L-Phe, respectively (Figure 4a). The absolute configurations of Glu and Phe residues of 1b were assigned to be L-Glu and D-Phe, respectively (Figure 4b).
Determination of the absolute configurations of compounds 1 and 2. (a) HPLC chromatogram of derivatives of the acid hydrolysate of 1a and those of corresponding authentic amino acids; (b) for 1b and (c) for 2. A full color version of this figure is available at The Journal of Antibiotics journal online.
Compound 2 was obtained as a white amorphous powder. [α]25D −26.3 (c 0.2, MeOH). HR-ESI-TOF MS gave the molecular formula of C22H26N3O3 (m/z 380.1977 [M+H]+, calcd 380.1974), the same as that of 1. The 1H and 13C NMR data of 2 were almost identical with those of 1 (Table 1), which revealed that they had closely similar structures. However, they showed different retention behaviors in normal and reversed-phase chromatography. Interpretation of 1D and 2D NMR spectra indicated that 2 had the same planar structure to that of 1.
In view of the existence of enantiomers of 1, compound 2 was also subjected to Lux Amylose-2 chiral liquid chromatography analysis. Two enantiomers were obtained at tR 8.19 min for 2a and 10.54 min for 2b (Figure 3b), and their relative content ratio was ∼9:1 according to their relative peak areas in the HPLC chromatogram (89.3% for 2a and 10.7% for 2b).
As the relative contents of enantiomers 2a and 2b were of great disparity, and available amount of 2 was also small, the absolute configurations of compunds 2a and 2b were determined directly by the Marfey’s method without chiral resolution of 2. Results revealed that configurations of Glu and Phe residues of 2a were L-Glu and L-Phe, respectively, and the opposite to those of 2b (Figure 4c).
Compound 3 was obtained as a faint yellow amorphous powder. Its molecular formula was determined to be C19H27N3O3 based on the quasimolecular ion at m/z 346.2130 [M+H]+ (calcd 346.2131) in the HR-ESI-TOF MS, indicating 8 degrees of unsaturation. The 13C NMR spectrum of 3 (Table 1) showed three ester/amide carbonyl carbon signals (δC 167.2, 167.3 and 171.1). The characteristic methine resonances of α-CH at δ 3.94/55.1 and 4.08/55.0 indicated that it was a dipeptide. The presence of a cluster of five aromatic protons at δ 7.17–7.27 suggested the presence of one monosubstituted benzene ring in the 1H-NMR spectrum of 3. Detailed analyses of 1H_1H COSY, HSQC and HMBC data of 3 established the three residues as leucine (Leu), glutamic acid (Glu) and N-phenethyl (Figure 2b). According to the HMBC correlation between H2-1′′ and C-5 (Glu), the structure of 3 was established to be cyclo (Leu-Glu) substituted with N-phenethyl group, which was a new PEA derivative.
The optical rotation value of 3 was almost zero, which suggested that it was also a racemate. Further chiral resolution was performed by Lux Amylose-2 chiral liquid chromatography (phenomenex, φ 5 μm, 4.6 × 250 mm) using 100% CH3CN at 1.0 ml min−1. Two enantiomers were obtained from compound 3, namely 3a and 3b, at tR 8.91 and 15.26 min, respectively (Figure 3c), and their relative contents were nearly identical according to their relative peak areas in the HPLC chromatogram (44.9% for 3a and 55.1% for 3b).
The absolute configurations of the Glu and Leu residues of 3a were assigned to be L-Glu and D-Leu, respectively, by Marfey’s method8, 9 (Figure 5a), and opposite to those of 3b (Figure 5b).
Determination of the absolute configuration of compounds 3 and 4. (a) HPLC chromatogram of derivatives of the acid hydrolysate of 3a and those of corresponding authentic amino acids; (b) for 3b and (c) for 4. A full color version of this figure is available at The Journal of Antibiotics journal online.
Compound 4 was obtained as a faint yellow amorphous powder. [α]25D −17.7 (c 0.5, MeOH). HR-ESI-TOF MS gave the molecular formula of C19H28N3O3 (m/z 346.2130 [M+H]+, calcd 346.2131), the same as that of 3. The 1H and 13C NMR data were almost identical with those of 3 (Table 1), which revealed that they had closely similar structures. However, they showed different retention behaviors in normal and reverse-phase chromatography. Interpretation of 1D and 2D NMR spectra indicated that 4 has the same planar structure as that of 3.
In view of the existence of enantiomers of 3, compound 4 was also subjected to Lux Amylose-2 chiral liquid chromatography analysis. Two enantiomers were obtained at tR 8.30 min for 4a and 10.90 min for 4b (Figure 3d), and their relative content ratio was ∼9:1 according to their relative peak areas in the HPLC chromatogram (89.2% for 4a and 10.8% for 4b).
As the relative contents of enantiomers 4a and 4b were of great disparity, and the available amount of 4 was also small, the absolute configurations of compunds 4a and 4b were determined directly by the Marfey’s method without chiral resolution of 4. Results revealed that configurations of Glu and Leu residues of 4a were L-Glu and L-Leu, respectively, and are opposite to those of 4b (Figure 5c).
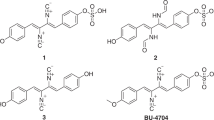
Compound 5 was obtained as a red amorphous powder. Its molecular formula was determined to be C24H19NO4 based on the quasimolecular ion at m/z 386.1386 [M+H]+ (calcd 386.1371) in the HR-ESI-TOF MS, indicating 16 degrees of unsaturation. The 1H and 13C NMR spectra of 5 (Table 2) showed two methylenes and three phenyl rings (including a monosubstituted benzene ring and two para-disubstituted benzene rings). Detailed analyses of 1H–1H COSY, HSQC and HMBC correlations further revealed the presence of an N-phenethyl group and two para-disubstituted benzene residues. In the HMBC spectra of 5, correlations between H2-1′′ (δH 3.82, 2H, t, J=7.2 Hz) and C-1 (δC 172.9) and C-1′ (δC 172.9), H-4/H-8 (δH 6.75, 2H, d, J=8.8 Hz ) and C-2 (δC 135.5), H-4′′/H-8′′ (δH 7.21–7.25, 2H, m) and C-2′ (δC 135.5) suggested that the N-phenethyl group and para-disubstituted benzene residues were linked with a 1H-pyrrole-2,5-dione ring (Figure 2c), which was further confirmed by comparison of its NMR data with that of 1-benzyl-3,4-bis(4-hydroxyphenyl)-1H-pyrrole-2,5-dione.10 Thus, compound 5 was established to be 1-(2-phenylethyl)-3,4-bis(4-hydroxyphenyl)-1H-pyrrole-2,5-dione, which was a new compound.
Biological activity
The antimicrobial activity of compounds 1–5 was evaluated by an agar dilution method.11 The results showed that compound 5 displayed antimicrobial activity against S. aureus and B. subtilis, whereas compounds 1–4 were inactive against the two strains (MIC >1.00 mg ml−1).
Methods
General experiments
Optical rotations were measured on a JASCO P-1020 digital polarimeter at room temperature (JASCO Inc., Tokyo, Japan). IR spectra were recorded on a JASCO FT/IR-480 plus Fourier transform infrared spectrometer (JASCO Inc.). UV spectra were recorded in CH3OH by using a JASCO V-550 UV/VIS spectrophotometer (JASCO Inc.). HR-ESI-MS was recorded on an Agilent 6200 ESI/TOF mass spectrometer (Agilent Technologies, Santa Clara, CA, USA). 1D NMR (1H, 13C, DEPT-135) and 2D NMR (1H–1H COSY, HSQC and HMBC) were recorded on a Bruker AVANCE 400 spectrometer at 300 K (Bruker Amazon AV400, Zurich, Switzerland). The chemical shifts were given in δ, with the solvent signals (DMSO-d6: δH 2.49/δC 39.5; CD3OD: δH 3.31/δC 49.2; Acetone-d6: δH 2.05/δC 29.9, 206.7) as an internal standard (CIL Inc., Berkeley, CA, USA). TLC was carried out on Silica gel GF254 (YanTai Chemical Inst., Yantai, China) plates. HPLC was performed on a SHIMADZU LC-6AD LC-6AD Liquid Chromatography with SPD-20A Detector by using an ODS column (Welch Materials XB-C18 column (φ 5 μm, 4.6 × 250 mm), YMC-Pack ODS-A (φ 5 μm, 10.0 × 250 mm)) (Nacalai Tesque Inc., Kyoto, Japan) at 220 and 254 nm. Chiral HPLC analysis was performed using a phenomenex Lux Amylose-2 column (φ 5 μm, 4.6 × 250 mm) (Phenomen, Torrance, CA, USA). Column chromatography (CC) was carried out on silica gel (200–300 mesh; Qingdao Marine Chemical Ltd., Qingdao, China), Sephadex LH-20 (50 μm; Amersham Pharmacia Biotech AB, Uppsala, Sweden) and ODS (60–80 mesh; YMC Ltd., Kyoto, Japan). All solvents used in CC were of analytical grade (Tianjin Damao Chemical Plant, Tianjin, China).
Strain
The strain of A. nanhaiticus sp. nov. NH36AT was isolated from a sandy sediment sample containing shell fragments collected from a water depth of 36 m in the South China Sea (19° 44.775′ N 111° 6.192′ E) in May 2007.12 16S rRNA gene sequence analysis revealed that strain NH36AT was most closely related to members of the genus Arenibacter. On the basis of phenotypic, chemotaxonomic and phylogenetic data, this organism was classified as a representative of a novel species in the genus Arenibacter. A voucher specimen was deposited at the China Center for Type Culture Collection (CCTCC AB 208315T) and at the Marine Culture Collection of China (MCCC 1A04137T).
Fermentation of strain
The producing strain was cultured and fermented as follows. The monoclonal strain growing on marine 2216E plate13 was inoculated into a 250-ml Erlenmeyer flask containing 100 ml marine 2216E culture medium and cultured at 30 °C for 24 h on a rotary shaker at 180 r.p.m. Then, 20 ml of the resultant seed culture was inoculated into 1000-ml Erlenmeyer flasks, each containing 500 ml of the above culture medium and incubated for 2 days at the same conditions. After centrifugation, the total 60 l of fermentation broth was collected.
Extraction and purification
The fermentation broth was extracted repeatedly with EtOAc, and the organic solvent was evaporated under reduced pressure to afford the crude extract (38.5 g). The extract was subjected to silica gel CC using CHCl3-CH3OH gradient elution to obtain 10 fractions (F1–F10). The fraction F4 (1.82 g) was subjected to ODS medium-pressure liquid chromatography eluted with a gradient of MeOH in H2O (30, 50, 70 and 100%) to afford six subfractions (F4-1–F4-6). F4-6 (0.27 g) was subjected to a Sephadex LH-20 CC eluted with 1:1 CHCl3-MeOH to yield three subfractions (F4-6-a–F4-6-c). F4-6-c (35.5 mg) was purified by reversed-phase HPLC on an YMC-Pack ODS-A column (5 μm, 10.0 × 250 mm, 4.0 ml min−1), with CH3CN and H2O (50:50, v/v) as the eluents to yield compound 5 (3.0 mg, tR=16.5 min).
The fraction F7 (0.78 g) was subjected to ODS medium-pressure liquid chromatography eluted with a gradient of MeOH in H2O (30, 50, 70 and 100%) to afford five subfractions (F7-1–F7-5). F7-5 (0.28 g) was purified by reversed-phase HPLC on an YMC-Pack ODS-A column (5 μm, 10.0 × 250 mm, 4.0 ml min−1) with CH3OH and H2O (40: 60, v/v) as the eluent to yield compounds 1 (5.0 mg, tR=27.2 min), 2 (2.4 mg, tR=38.1 min), 3 (4.1 mg, tR=24.5 min) and 4 (3.8 mg, tR=32.0 min).
Physico-chemical properties
Table 3 summarized the physico-chemical properties of compounds 1–5.
Determination of absolute configuration by the Marfey’s method
Marfey’s method8, 9 was applied to assign the absolute configuration of amino-acid residues of each enantiomer. Compound 1a (0.5 mg) in 6.0 N HCl (1.0 ml) was heated at 155 °C for 2 h. Upon removal of excess HCl under vacuum, the hydrolysates were placed in a 1-ml reaction vial and treated with 1% solution of 1-fluoro-2, 4-dinitrophenyl-5-L-alanine amide (FDAA; 150 μl) in acetone, followed by 1.0 N NaHCO3 (40 μl). The reaction mixtures were heated at 45 °C for 1.5 h, cooled to room temperature and acidified with 2.0 N HCl (20 μl). Similarly, the standard L- and D-amino acids were derivatized according to the method mentioned above. The derivatives of the hydrolysates and the standard amino acids were subjected to HPLC analysis (Kromasil C18 column; φ 5 μm, 4.6 × 250 mm; 1.0 ml min−1) at 35 °C using the following gradient program: solvent A, H2O (0.1% HCOOH); solvent B, CH3CN; linear gradient, 30–80% of B in A over 30 min with UV detection at 340 nm. Absolute configurations of compounds 1b, 2, 3a, 3b and 4 were also analyzed and determined by the Marfey’s method, just like that of 1a.
Antimicrobial assays
The antimicrobial activities of compounds 1–5 against S. aureus and B. subtilis were evaluated by an agar dilution method.11 The tested strains were cultivated on MH agar plates for bacteria at 35 °C. Compounds 1–5 and positive controls (Tobramycin) were dissolved in EtOH at different concentrations from 1000 to 0.25 μg ml−1 by the continuous two-fold dilution methods. A 10-μl quantity of test solution was absorbed by a paper disk (5 mm diameter) and placed on the assay plates. After 20 h incubation, zones of inhibition (mm in diameter) were recorded. The MICs were defined as the lowest concentration at which no microbial growth could be observed. Experimental results showed that compound 5 displayed a weak antimicrobial activity against S. aureus and B. subtilis, with MIC values of 0.50 and 0.25 mg ml−1, respectively (Tobramycin-displayed MIC values of 0.30 μg ml−1 and 0.01 mg ml−1 against the two strains, respectively), whereas compounds 1–4 were inactive against the two strains (MIC >1.00 mg ml−1).
References
Mosnaim, A. D., Callaghan, O. H., Hudzik, T. & Wolf, M. E. Rat brain-uptake index for phenylethylamine and various monomethylated derivatives. Neurochem. Res. 38, 842–846 (2013).
Ma, G. Y. et al. Pharmacological effects of ephedrine alkaloids on human α1- and α2-adrenergic receptor subtypes. J. Pharmacol. Exp. Ther. 322, 214–221 (2007).
Reszka, K. J. et al. Airway peroxidases catalyze nitration of the β2-agonist salbutamol and decrease its pharmacological activity. J. Pharmacol. Exp. Ther 336, 440–449 (2011).
Joy, M. S. et al. Use of enantiomeric bupropion and hydroxybupropion to assess CYP2B6 activity in glomerular kidney diseases. J. Clin. Pharmacol. 50, 714–720 (2010).
Nakazato, A. et al. Synthesis and SAR of 1-alkyl-2-phenylethylamine derivatives designed from N, N-dipropyl-4-methoxy-3-(2-phenylethoxy) phenylethylamine to discover σ1 ligands. J. Med. Chem. 42, 3965–3970 (1999).
Malloy, K. L. et al. Credneramides A and B: neuromodulatory phenethylamine and isopentylamine derivatives of a vinyl chloride-containing fatty acid from cf. Trichodesmium sp. nov. J. Nat. Prod. 75, 60–66 (2012).
Zhou, J. & Tan, N. H. Application of a new TLC chemical method for detection of cyclopeptides in plants. Chin. Sci. Bull. 45, 1825–1831 (2000).
Marfey, P. Determination of D-amino acids. II. Use of a bifunctional reagent, 1,5-difluoro-2, 4-dinitrobenzene. Carlsberg. Res. Commun. 49, 591–596 (1984).
Bhushan, R. & Brückner, H. Marfey’s reagent for chiral amino acid analysis: a review. Amino Acids 27, 231–247 (2004).
Xie, H. D., Ho, L. A., Truelove, M. S., Corry, B. & Stewart, S. G. Fluorescent triphenyl substituted maleimide derivatives: synthesis, spectroscopy and quantum chemical calculations. J. Fluoresc. 20, 1077–1085 (2010).
Zaika, L. L. Spices and herbs: their antimicrobial activity and its determination. J. Food Safety. 9, 97–118 (1988).
Sun, F. Q. et al. Arenibacter nanhaiticus sp. nov., isolated from marine sediment of the South China Sea. Int. J. Syst. Evol. Microbiol. 60, 78–83 (2010).
Bai, X. Y., Zhang, Z. T., Bai, M. D., Yang, B. & Bai, M. B. Killing of invasive species of ship’s ballast water in 20 t/h system using hydroxyl radicals. Plasma Chem. Plasma Process. 25, 41–54 (2005).
Acknowledgements
This work was supported by grants from the Ministry of Science and Technology of China (No. 2009CB522300 and No. 2012ZX09301002-003), by Guangdong Natural Science Funds for Distinguished Young Scholar (No. S2013050014287), by the program for New Century Excellent Talents in University (No. NCET-10-0120) and Fok Ying Tung Education Foundation (121039) from the Ministry of Education of China.
Author information
Authors and Affiliations
Corresponding authors
Rights and permissions
About this article
Cite this article
Chen, Y., Tang, J., Tang, X. et al. New phenethylamine derivatives from Arenibacter nanhaiticus sp. nov. NH36AT and their antimicrobial activity. J Antibiot 66, 655–661 (2013). https://doi.org/10.1038/ja.2013.65
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ja.2013.65
Keywords
This article is cited by
-
New pyrones and their analogs from the marine mangrove-derived Aspergillus sp. DM94 with antibacterial activity against Helicobacter pylori
Applied Microbiology and Biotechnology (2020)
-
Elucidation of anti-Vibrio factors associated with green alga Picochlorum sp. strain S1b
Journal of Applied Phycology (2015)