Abstract
Since the early 1980s, fungi have emerged as a major cause of human disease. Fungal infections are associated with high levels of morbidity and mortality, and are now recognized as an important public health problem. Gram-negative bacterial strains of genus Xenorhabdus are known to form symbiotic associations with soil-dwelling nematodes of the Steinernematidae family. We describe here the discovery of a new antifungal metabolite, cabanillasin, produced by Xenorhabdus cabanillasii. We purified this molecule by cation-exchange chromatography and reverse-phase chromatography. We then determined the chemical structure of cabanillasin by homo- and heteronuclear NMR and MS-MS. Cabanillasin was found to be active against yeasts and filamentous fungi involved in opportunistic infections.
Similar content being viewed by others
Introduction
Since the early 1980s, fungi have increasingly emerged as a major cause of human disease, particularly in immunocompromised individuals and in hospitalized patients with serious underlying diseases.1 Fungal infections are associated with high levels of morbidity and mortality, and are now recognized as an important public health problem. Most nosocomial fungal infections are caused by Candida species, with Candida albicans being the most common etiological agent of fungal bloodstream infections. Candida and Aspergillus infections together account for 90% of all nosocomial fungal infections. Cryptococcus neoformans has also become a major opportunistic pathogen in immunocompromised individuals.2 Only a small number of classes of antifungal drugs are used in clinical practice (echinocandin, polyene, azole derivatives and fluoropyrimidine),3 and the incidence of resistance to antifungal agents is increasing worldwide. New antifungal compounds must therefore be identified, to ensure the effective treatment of fungal infections in the future. In this context, bacteria remain a major resource for the discovery of new antifungal molecules.
Our work focuses on an underinvestigated bacterial genus, Xenorhabdus. The strains of this genus are Gram-negative and are symbiotically associated with soil-dwelling nematodes of the Steinernematidae family.4, 5, 6, 7 These nematodes enter insect larvae via natural openings, and then release the bacteria from their intestine into the hemocoel of the host.8 The bacteria then kill the insect host, by producing inhibitors of the insect immune system9, 10, 11 and insecticidal proteins.12 The bacteria proliferate in the dead tissues of the host, thereby favoring the reproduction of the nematode by degrading the insect biomass13 and producing antibiotics that inhibit the development of other microorganisms present in the insect cadaver (bacteria, fungi).14 Only a few families of antifungal compounds from Xenorhabdus have been characterized and described: xenorhabdins,15 indole derivatives16, 17 and peptide antimicrobial from xenorhabdus (PAX).18, 19
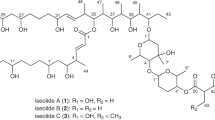
We describe here the culture conditions used for Xenorhabdus cabanillasii strain JM26, the isolation of an antifungal compound, cabanillasin (Figure 1), and the determination of its biology functions and chemical structure. An antibacterial metabolite family, nemaucins, was produced by the same Xenorhabdus species.20 These molecules are composed by a peptidic part and multiple guanidino groups. Nemaucins demonstrate strong antibacterial activities.
Results and discussion
Fermentation
Xenorhabdus cabanillasii JM26 was cultured for 48 h at 28 °C, with shaking, in a 2-l Erlenmeyer flask containing 500 ml of medium consisting of 10 g Bacto Peptone, 1 g K2HPO4, 1 g MgSO4, 7H2O, 2 g (NH4)2SO4, 2 g CaCO3 and 10 g NaCl in 1.0 l of water. The medium was inoculated with 1% (v/v) of a 24-h preculture in the same medium. The production of the antifungal compound was monitored by analytical HPLC.
Isolation
Bacterial cells were removed by low-speed centrifugation (6000 g, 10 min at 4 °C) and the supernatant was sterilized by passage through a filter with 0.22 μm pores. The sterilized supernatant was then added to an equal volume of 0.1 M NaCl and 0.02 M Tris buffer (pH 7), and the resulting mixture was subjected to cation-exchange chromatography on a Sep Pack CarboxyMethyl cartridge (Acell Plus CM; Waters, Milford, MA, USA). Unbound material was removed by washing with 0.1 M NaCl in 0.02 M Tris (pH 7) and the compound with antibiotic activity was eluted with 0.5 M NaCl in 0.02 M Tris (pH 7). This eluate was acidified by adding 0.1% (v/v) trifluoroacetic acid (TFA) and was then subjected to reverse-phase chromatography on a Sep Pack C18 cartridge (Sep-Pak plus C18; Waters). Unbound material was removed by washing with 0.1% TFA in H2O, and the antibiotic pool was eluted with acetonitrile. The eluate was lyophilized and resuspended in water. The crude extract was purified by reverse-phase HPLC on a C18 column (Waters; Symmetry Symmetry C18; 5 μm; 4.6 × 150 mm), with a linear gradient of 0.1% TFA in water–acetonitrile, increasing from 0 to 30% in 30 min, with a flow rate of 1 ml min−1, and UV detection at wavelengths of 200–400 nm, yielding pure cabanillasin at a retention time of 13.95 min.
Biological properties
We assessed the antifungal activity of cabanillasin against a wide range of yeasts and filamentous fungi involved in nosocomial infections.
In assessments carried out after 24 h of incubation, cabanillasin was found to be highly active against Candida krusei ATCC 6258 and Candida lusitaniae CBS 6936 (Table 1). Indeed, we obtained IC50 values of 6.25 μg ml−1 for Candida krusei and 1.56 μg ml−1 for Candida lusitaniae, and an MIC of 25 μg ml−1. This molecule did not completely inhibit Candida albicans ATCC 90029 and Candida glabrata ATCC 90030, for which we observed 20–40% residual growth. The IC50 values for Candida albicans and Candida glabrata were 0.78 and 6.25 μg ml−1, respectively (Table 1). After 48 h of incubation, the level of activity against yeasts was significantly lower (Table 1). This loss of inhibitory activity over time may be due to the chemical instability of cabanillasin.
The inhibitory activity of cabanillasin against the basidiomycete yeast Cryptococcus neoformans was moderate after 48 h (IC50=12.5 μg ml−1) and was no longer observed after 72 h (IC50>50 μg ml−1; Table 1).
Cabanillasin had only weak activity against the filamentous fungi Aspergillus fumigatus and Rhizopus oryzae at 48 h, and no activity against these fungi at 72 h (Table 1). By contrast, it was more active against Fusarium oxysporum, with an IC50 of 6.25 μg ml−1 at 72 h of incubation (Table 1). The discovery of new molecules with inhibitory activity against Fusarium spp. is of particular interest, because this genus includes several animal pathogen species with multiple resistance to almost all the available systemic antifungal drugs.21
Cytotoxicity assays revealed that LD50 of cabanillasin in human prostatic carcinoma cells (PC-3) was 46.83±1.55 μg ml−1, whereas that in human normal mammary epithelial cells (hTERT HME-1) was 24.46±2.51 μg ml−1.
Physicochemical properties of cabanillasin
Cabanillasin was obtained as a colorless substance and was soluble in H2O, dimethyl sulfoxide (DMSO) and methanol. ESI-MS showed its MW to be 511 Da. Its molecular formula was determined to be C22H49N13O1 by high-resolution ESI-MS measurement (found 511.4261, cald 511.4261). The cabanillasin had a UV λmax of 214 nm (methanol).
Chemical structure elucidation
We investigated the chemical structure of cabanillasin by collecting two sets of NMR data, in both DMSO-d6 and D2O. The 1H NMR spectra recorded in DMSO-d6 contained four groups of signals. First, a sharp signal at 2.02 p.p.m. indicated the presence of an acetyl group (Table 2). There were then two multiplets of similar intensity at 3.25 and 1.50 p.p.m. were each considered to correspond to eight methylene groups. The low-field-shifted multiplet (3.25 p.p.m.) probably reflects the bonding of these methylene groups to a nitrogen atom, whereas the other multiplet, at 1.50 p.p.m., is typical of saturated methylene groups in an aliphatic chain. The fourth group of signals included several signals differing in breadth, ranging from 12.3 to 7.4 p.p.m., indicating the presence of amine or guanidine groups. The protons giving rise to this group of signals were not observed in D2O and are, thus, exchangeable, consistent with such an assignment. Furthermore, the three signal groups obtained in D2O, at 3.40–3.00 p.p.m. (8 CH2), 1.75–1.60 p.p.m. (8 CH2) and 2.23 p.p.m. (acetyl group), are consistent with the proposed assignment in DMSO-d6.
The 13C one-dimensional spectrum and the two-dimensional spectra (HSQC and HMBC) clearly confirmed the presence of the chemical groups identified above. The carbonyl and methyl signals of the acetyl group and the quaternary carbon of the guanidine groups were identified at 175.79, 23.7 and 155.72–152.93 p.p.m., respectively, in D2O.
Together, the MS (Figure 2) and NMR data support the multimeric chemical structure for cabanillasin displayed in Figure 1, generated from four units of the amino-1 guanidino-4 butane moiety. Cabanillasin thus has a free amino group for the first unit and an acetylated guanidine group at the end of the fourth unit. Cabanillasin is markedly cationic (one amino and four guanidine functional groups), but also has hydrophobic properties (four methylene groups), resulting in an amphipathic molecule. Interestingly, its antifungal activity appears to predominate. Cabanillasin and nemaucins20 have a common structural part (four units of the amino-1 guanidino-4 butane moiety) and the same bacterial producer. Common genes may be involved in the biosynthesis of these molecules.
Conclusion
We describe here the discovery of an antifungal molecule, cabanillasin, produced by Xenorhabdus cabanillasii strain JM26 and purified by cation-exchange chromatography and reverse-phase HPLC. The chemical structure of cabanillasin was determined by homo- and heteronuclear NMR and MS-MS. Cabanillasin was found to be active against yeasts and filamentous fungi responsible for opportunistic infections in immunocompromised patients.
Methods
The producing organism
Xenorhabdus cabanillasii JM26 (CNCM I-4418) was grown in Luria–Bertani medium (10 g Bacto Tryptone, 5 g yeast extract and 10 g NaCl in 1.0 l of water) for liquid culture and on Luria–Bertani agar for solid cultures. The phase status (I or II) of this strain was determined by culture on nutrient bromothymol blue agar medium (31 g nutrient agar, 25 mg bromothymol blue and 40 mg of 1% 2, 3, 5-triphenyl tetrazolium chloride in 1.0 l water)12 and the measurement of antibacterial activity against Micrococcus luteus. When cultured in vitro, Xenorhabdus produces two colony forms or variants. Variant I is blue, due to the adsorption of bromothymol blue, whereas variant II is red. Variants I and II are indicated by suffixes (/1 and/2, respectively) attached to strain designations. This strain was maintained at 15 °C on nutrient bromothymol blue agar medium.
Antifungal susceptibility tests
We assessed the activity of cabanillasin against various fungal pathogens of humans: ascomycetes yeasts (Candida albicans ATCC 90029, Candida glabrata ATCC 90030, Candida krusei ATCC 6258 and Candida lusitaniae CBS 6936), basidiomycete yeasts (Cryptococcus neoformans, clinical strain), ascomycete filamentous fungi (Aspergilus fumigatus, clinical strain; Fusarium oxysporum, clinical strain) and zygomycete filamentous fungi (Rhizopus oryzae, environmental strain). The CLSI M27-A2 broth microdilution method and the CLSI M38-A microdilution method with RPMI 1640 medium (recommended in the Clinical and Laboratory Standards Institute M23-A document)22 were used for the testing of yeasts and filamentous fungi, respectively.23, 24
Inocula of yeasts and filamentous fungi were prepared on YPD medium (10 g yeast extract, 10 g peptone and 20 g glucose in 1.0 l tap water) and malt medium (20 g malt extract in 1.0 l tap water), respectively. The density of each inoculum was adjusted by dilution in RPMI 1640 broth to between 5 × 102 and 5 × 103 cfu ml−1 for yeast isolates and 5 × 103 and 5 × 104 cfu ml−1 for filamentous fungi. We added 10 μl of cabanillasin solution to 190 μl of fungal inoculum in microdilution trays (96 U-bottomed plates; final concentration of 50–0.098 μg ml−1 for all fungi). The inoculated microdilution trays were incubated at 35 °C in ambient air. The absorbance of the culture was measured at 450 nm, after 24 and 48 h of incubation for the yeasts and after 48 and 72 h of incubation for filamentous fungi and Cryptococcus neoformans. Activity was assessed by determining the percentage growth inhibition ((1−A450 nm culture with cabanillasin)/A450 nm culture without cabanillasin) × 100.
The IC50 is the concentration at which the growth observed is 50% of that observed in the absence of the antifungal compound (that is, the concentration inhibiting fungal growth by 50%). The MICs for Candida species were determined at 24 and 48 h, and corresponded to 100% growth inhibition. Amphotericin B was used as a standard (final concentration of 16–0.03 μg ml−1 for all fungi). Experiments were carried out in duplicate.
Cytotoxicity test
We used 100 μl of a suspension of PC-3 (human prostatic carcinoma) or hTERT HME-1 (human normal mammary epithelium) cells prepared in RPMI 1640+10% fetal calf serum+1% glutamine to inoculate in 96-well plates. We used 3500 cells per well for PC-3, and 7500 cells per well for hTERT HME-1 cells. The microplates were incubated at 37 °C for 24 h under an atmosphere containing 5% CO2. After 24 h of culture, the medium was removed by aspiration and we added 100 μl of RPMI 1640+10% fetal calf serum+1% glutamine, supplemented with eight concentrations of cabanillasin (from 0.78 to 100 μg ml−1), to each well. Microplates were incubated at 37 °C for 48 h under an atmosphere containing 5% CO2. After 48 h of culture, the medium was removed by aspiration and we added 100 μl of RPMI 1640+thiazolyl blue tetrazolium bromide ( 0.5 mg ml−1) to each well. Microplates were incubated at 37 °C for 150 min. We then removed all the medium by inverting and tapping the plate, and added 100 μl of DMSO to each plate. Spectrophotometric absorbance was then measured at 550 nm.
The lethal dose 50 was calculated as LD50=((50-(Y2-((Y1-Y2)/(X1-X2))*X2))/((Y1-Y2)/(X1-X2))). X1 is a concentration at which viability exceeded 50%, X2 is a concentration at which viability was below 50%, Y1 is the percentage viability at concentration X1 and Y2 is the percentage viability at concentration X2. Experiments were done in triplicate.
NMR and MS analysis
The purified compound was analyzed by MS and NMR to determine its chemical structure.
We first carried out LC-MS to determine the m/z value of the protonated molecule of cabanillasin. We then subjected cabinallasin to MS-MS fragmentation. ESI-LC-MS data were obtained in the positive mode, on a Waters alliance LC-MS system (Waters ZQ mass detector, Waters photodiode array detector 2696, Waters alliance HPLC systems 2790). We used a C18 column (Waters Symmetry C18 5 μm 4.6 × 150 mm) for HPLC. The solvents used were (A) water+0.1% TFA and (B) acetonitrile+0.1% TFA, and the flow rate was 1 ml min−1. The mobile phase composition was 100% A at 0 min, ramped to 30% B at 30 min. Samples were dissolved in solvent A (100 μl). The sample injection volume was 10 μl. Detection in the UV-visible range was carried out by determining absorbance at 200–400 nm. Solvent flow to the MS was diverted to waste for the first 5 min to minimize salt build-up. MS-MS fragmentation data were obtained on a Waters Micromass Q-ToF micromass spectrometer.
NMR spectra were recorded for two samples of 5 mg of cabanillasin dissolved in 750 μl of either D2O or DMSO-d6. All spectra were recorded at 298 °K.
The 1H–1H homonuclear (COSY, TOCSY) and 1H–13C heteronuclear (HSQC and HMBC) experiments in D2O were recorded on a Bruker Avance spectrometer (Billerica, MA, USA) equipped with a cryoprobe at 500.13 MHz for 1H and 125.76 MHz for 13C. With the DMSO-d6 sample, a similar set was recorded on a Varian Gemini 300-BB spectrometer (Palo Alto, CA, USA). The full assignment was obtained from the combined analysis of each data set.
References
Pfaller, M. A. & Diekema, D. J. Epidemiology of invasive candidiasis: a persistent public health problem. Clin. Microbiol. Rev. 1, 133–163 (2007).
Perea, S. & Patterson, T. F. Antifungal resistance in pathogenic fungi. Clin. Infect. Dis. 35, 1073–1080 (2002).
Stevens, D. A. Advances in systemic antifungal therapy. Clin. Dermatol. 30, 657–661 (2012).
Poinar, G. O. J. The presence of Achromobacter nematophilus in the infective stage of a Neoaplectana sp. (Steinernematidae: Nematoda). Nematologica 12, 105–108 (1966).
Akhurst, R. J. Neoaplectana species: specificity of association with bacteria of the genus Xenorhabdus. Exp. Parasitol. 55, 258–263 (1983).
Herbert, E. E. & Goodrich-Blair, H. Friend and foe: the two faces of Xenorhabdus nematophila. Nat. Rev. Microbiol. 5, 634–646 (2007).
Tailliez, P., Pagès, S., Ginibre, N. & Boemare, N. New insight into diversity in the genus Xenorhabdus, including the description of ten novel species. Int. J. Syst. Evol. Microbiol. 56, 2805–2818 (2006).
Sicard, M. et al. Stages of infection during the tripartite interaction between Xenorhabdus nematophila, its nematode vector, and insect hosts. Appl. Environ. Microbiol. 70, 6473–6480 (2004).
Park, Y. & Kim, Y. Eicosanoids rescue Spodoptera exigua infected with Xenorhabdus nematophilus, the symbiotic bacteria to the entomopathogenic nematode Steinernema carpocapsae. J. Insect. Physiol. 46, 1469–1476 (2000).
Park, Y., Kim, Y. & Stanley, D. The bacterium Xenorhabdus nematophila inhibits phospholipases A2 from insect, prokaryote, and vertebrate sources. Naturwissenschaften 91, 371–373 (2004).
Vigneux, F. et al. The xaxAB genes encoding a new apoptotic toxin from the insect pathogen Xenorhabdus nematophila are present in plant and human pathogens. J. Biol. Chem. 282, 9571–9580 (2007).
Brown, S. E. et al. Txp40, a ubiquitous insecticidal toxin protein from Xenorhabdus and Photorhabdus bacteria. Appl. Environ. Microbiol. 72, 1653–1662 (2006).
Forst, S., Dowds, B., Boemare, N. & Stackebrandt, E. Xenorhabdus and Photorhabdus spp.: bugs that kill bugs. Annu. Rev. Microbiol. 51, 47–72 (1997).
Webster, J. M., Chen, G., Hu, K. & Li, J. Bacterial metabolites In Entomopathogenic Nematology (ed. Gangler R99–114 CABI Publishing: New York, (2002).
McInerney, B. V., Taylor, W. C., Lacey, M. J., Akhurst, R. J. & Gregson, R. P. Biologically active metabolites from Xenorhabdus spp. Part 2. Benzopyran-1-one derivatives with gastroprotective activity. J. Nat. Prod. 54, 785–795 (1991).
Li, J., Chen, G., Webster, J. M. & Czyzewska, E. Antimicrobial metabolites from a bacterial symbiont. J. Nat. Prod. 58, 1081–1086 (1995).
Li, J., Chen, G. & Webster, J. M. Nematophin, a novel antimicrobial substance produced by Xenorhabdus nematophilus (Enterobactereaceae). Can. J. Microbiol. 43, 770–773 (1997).
Gualtieri, M., Aumelas, A. & Thaler, J. O. Identification of a new antimicrobial lysine-rich cyclolipopeptide family from Xenorhabdus nematophila. J. Antibiot. 62, 295–302 (2009).
Fuchs, S. W., Proschak, A., Jaskolla, T. W., Karas, M. & Bode, H. B. Structure elucidation and biosynthesis of lysine-rich cyclic peptides in Xenorhabdus nematophila. Org. Biomol. Chem. 9, 3130–3132 (2011).
Gualtieri, M., Villain-Guillot, P., Givaudan, A. & Pagès, S. Nemaucin, an antibiotic produced by entomopathogenic Xenorhabdus cabanillasii WO/2012/085177, 28 June (2012).
Spader, T. B. et al. In vitro interactions between amphotericin B and other antifungal agents and rifampin against Fusarium spp. Mycoses 54, 131–136 (2011).
Clinical and Laboratory Standards Institute. Development of in Vitro Susceptibility Testing Criteria And Quality Control Parameters: Approved Guideline 2nd edn CLSI document M23–A2 Clinical and Laboratory Standards Institute: Wayne, PA, (2002).
Clinical and Laboratory Standards Institute. Reference Method for Broth Dilution Antifungal Susceptibility Testing of Filamentous Fungi: Approved Standard 2nd edn CLSI Document M38–A2 Clinical and Laboratory Standards Institute: Wayne, PA, (2008).
Clinical and Laboratory Standards Institute. Reference Method for Broth Dilution Antifungal Susceptibility Testing of Yeasts: Approved Standard 2nd edn CLSI document M27–A2 Clinical and Laboratory Standards Institute: Wayne, PA, (2002).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Houard, J., Aumelas, A., Noël, T. et al. Cabanillasin, a new antifungal metabolite, produced by entomopathogenic Xenorhabdus cabanillasii JM26. J Antibiot 66, 617–620 (2013). https://doi.org/10.1038/ja.2013.58
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ja.2013.58
Keywords
This article is cited by
-
The use of Phasmarhabditis nematodes and metabolites of Xenorhabdus bacteria in slug control
Applied Microbiology and Biotechnology (2024)
-
Endosymbiotic microbes from entomopathogenic nematode (EPNs) and their applications as biocontrol agents for agro-environmental sustainability
Egyptian Journal of Biological Pest Control (2022)
-
Antiprotozoal activity of different Xenorhabdus and Photorhabdus bacterial secondary metabolites and identification of bioactive compounds using the easyPACId approach
Scientific Reports (2022)
-
Natural products from Photorhabdus and Xenorhabdus: mechanisms and impacts
Applied Microbiology and Biotechnology (2022)
-
Relative potency of a novel acaricidal compound from Xenorhabdus, a bacterial genus mutualistically associated with entomopathogenic nematodes
Scientific Reports (2021)