Abstract
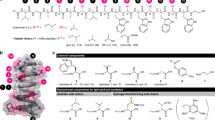
Natural products have been major sources of antibacterial agents and remain very promising. Frequent rediscoveries of known compounds hampers progress of new discoveries and demands development and utilization of new methods for rapid biological and chemical dereplication. This paper describes an efficient approach for discovery of new thiazolyl peptides by sensitive-resistant pair screening and dereplication in a time and cost-effective manner at industrial scale. A highly effective library-based dereplication of thiazolyl peptides by high resolution fourier transform liquid chromatography mass spectrometry (HRFTLCMS) has been developed, which can detect and dereplicate very low levels of thiazolyl peptides particularly when combined with miniaturized high-throughput 96-well solid-phase extraction separation, and as well can be automated. Combination of sensitive (susceptible)-resistant pair screening, diversified screening collection and miniaturized high-throughput SPE and HRFTLCMS techniques were applied for discovery of new thiazolyl peptides. The combined approach allowed for identification of over 24 thiazolyl peptides represented by three of the five structural subgroups, including three novel compounds. In addition, it is possible for the first time to mechanistically group three structural subgroups of over 24 thiazolyl peptides. Furthermore, these studies helped to understand natural frequency of distribution of these compounds and helped in discovery of new producing strains of many thiazolyl compounds.
Similar content being viewed by others
Introduction
Bacterial resistance to antibiotics continues to challenge the current treatment paradigm of bacterial infections. Methicillin-resistant Staphylococcus aureus is well publicized and infection by methicillin-resistant Staphylococcus aureus leads to high mortality in hospital.1 Infections by many other resistant bacteria such as ESKAPE pathogens are often more dangerous, leading to poor and sometimes no available treatment options.2, 3 Bacterial strains that are resistant to one antibiotic are often susceptible to others. However, treatment options are often significantly limited in the case of multi-drug-resistant bacteria. In order to treat drug-resistant bacterial infections and avoid an epidemic, it is incumbent upon the scientific community to make new treatment options available by discovering new structural chemotypes inhibiting the growth of bacteria by new modes of action (for example, platensimycin4, 5 and platencin6, 7).
Thiazolyl peptides are a class of naturally occurring antibiotics produced by filamentous prokaryotic organisms.8 Micrococcin was isolated in 1948 and the most studied member, thiostrepton, was discovered in 1954. In the last 50 years over 90 thiazolyl peptides have been reported with varying degree of activities.8 Some of the most interesting and most potent of the thiazolyl peptides are glycothiohexide α,9, 10 S54832A-I,8, 11 MJ347-81F4A and B12 and nocathiacins.13, 14 Despite highly potent in vitro antibacterial activities, these compounds could not be advanced as clinical agents due to poor physicochemical properties, most notably low-aqueous solubility. Key attributes of this class of natural products—highly potent activity and new modes of action—remain very attractive and, therefore, prompted us to start a serious reinvestigation. Chemical synthetic methods continue to evolve with development of new mild selective reactions helpful in the chemical modification of these complex compounds. Recently, structural modification of nocathiacins, leading to the semisynthesis of compounds with improved properties was reported.15, 16, 17, 18, 19, 20, 21, 22 Discovery of new natural thiazolyl peptides that are endowed with improved physical properties (for example, aqueous solubility) and spectrum would be beneficial in addressing new direction for structure activity relationship. In order to find new thiazolyl peptides we adopted two parallel approaches. In the first approach, we searched for new compounds from the known producers of thiazolyl peptides using analytical methods, particularly LCMS after systematic manipulations of the seed and growth media. This led to the discovery of thiazomycins.23, 24, 25 The second approach involved screening with sensitive-resistant pair S. aureus strains grown on agar medium in a two-plate assay format. In this assay, one plate was seeded with wild-type (sensitive) S. aureus strain and the second plate was seeded with well-characterized thiazomycin-resistant S. aureus strain.26 The thiazomycin resistance was mapped to L11 protein in which four amino acids (Pro23-Val24-Gly25-Pro26) in the rplK gene encoding L11 protein was deleted.26 This deletion resulted in greater than 500-fold increase in MIC of thiazomycin.27 The mechanistic expectation for this two-plate assay-based screening was to discover inhibitors that exert their activity by binding at the thiazomycin-binding site of L11 protein. Conceptually, these inhibitors could be thiazolyl peptides or even better novel compounds that are non-thiazolyl peptides. This assay is similar to the empiric assays that involves screening with drug-resistant strains,13, 28, 29 but with added benefit that it yields compounds with defined binding interactions, provided such interactions are known for the original resistant antibiotic. Some of the screening assays have been summarized in these reviews.30, 31
We instituted a high-throughput screening campaign using thiazomycin sensitive-resistant pair two-plate assay26 to screen for compounds that interact at thiazomycin-binding site. We thoroughly validated the selectivity of the assay by screening a series of known antibiotics that work by alternate mode of actions including several protein synthesis inhibitors (for example, see Supplementary Figure S1A in Zhang et al.32). The screening of microbial fermentation extracts led to a series of hits representing known and novel thiazolyl peptides from small volume fermentations. We herein summarize the result of the screening campaign with a clear strategy and execution of robust dereplication and high-throughput solid-phase extraction, leading to identification of a over 24 thiazolyl peptides representing three of the five structural subseries of the thiazolyl peptide class (Chart 1), including three novel thiazolyl peptides from diverse producing actinomycetes collected from geographically distinct environments.
Results and Discussion
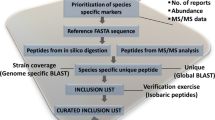
The 21 700 actinomycete strains selected for this study were isolated from diverse environmental samples including plant rhizosphere, salt marshes, mangrove sediments and soils collected from the largest diversity of ecological niches throughout the World. The strains were isolated using specific enrichment isolation approaches as described earlier.33 Strains were assembled in taxonomic groups based on morphological criteria, identifying among them 16 900 Streptomyces and 4800 strains belonging to minor genera or not identified. They were cultivated in different production media according to the preferred nutritional requirements of the actinomycetes groups. Pure microbial strains were grown in three or four media in 10 ml volume and fermentation was carried out at 28 °C and 220 r.p.m., for 7–13 days according to the taxonomic group. The fermentation whole broth was extracted with equal volume of acetone and extract was centrifuged to remove cellular debris and supernatant was concentrated to ½ volume to remove most of the acetone. Ten microliters of aqueous extracts were spotted on each of the thiazomycin sensitive and resistant plates containing a wild-type S. aureus and thiazomycin-resistant S. aureus strains, respectively, and incubated at 37 °C for 24 h. Inhibition zone sizes for each extract were scored on each plate and differential was measured. The extracts showing larger growth inhibition zone on the sensitive plate and smaller or no inhibition zone on the resistant plate were considered active hit in the assay. In less than 1 year we screened 75 000 extracts leading to 163 primary hits.
Targeted and direct dereplication of thiazolyl peptides by LCMS against known standards is relatively easy, but generally complicated by low titer of metabolites in screening broth unoptimized for metabolite production. Highly potent biological activity exhibited by thiazolyl peptides allowed for rapid and easy detection by bioassay even when titer was low. In order to gain higher sensitivity and detection accuracy, we utilized high resolution fourier transform liquid chromatography mass spectrometry (HRFTLCMS) as an analytical tool and high-throughput 96-well solid-phase extraction/purification using prepacked OASIS hydrophilic lipophilic balance reversed phase resin, which allowed for enrichment of the thiazolyl peptides and improved bioassay and MS sensitivity. An aliquot of 0.4 ml of each broth was loaded on the 96-well plate containing resin and eluted with 0.4 ml each of 5 and 50% aqueous methanol, 100% methanol, 100% acetone and finally 100% dimethyl sulfoxide (Figure 1). Fractions were collected in separate 96-well plates either by gravity elution or by a vacuum manifold. The fractions were tested in the thiazomycin sensitive-resistant two-plate assay for antibacterial activity. The activity eluted in either 100% methanol or 100% acetone or split in 100% methanol and 100% acetone fractions. Over 85% recovery of the thiazolyl peptides was achieved from the high-throughput purification technique whether conducted from extract or purified control (for example, 5–500 μg load of thiostrepton) as quantitated by bioactivity and HPLC. The processing of samples helped to clean up fractions providing enriched thiazolyl peptides as exemplified by the enrichment of philipimycin (Chart 1), a novel thiazolyl peptide produced by MA7347 shown in Figure 2. This rapid clean up and enrichment method allowed for significant improvement in detection of thiazolyl peptides by HRFTLCMS.
HPLC chromatograms of a representative example of high-throughput SPE purification in 96-well plate format. HPLC condition: Zorbax SB-phenyl, 4.6 × 75 mm, 1.0 ml min−1, 38–60% CH3CN (0.1% trifluoroacetic acid) 7 min, 60–95% acetonitrile 1 min, hold 95% 1 min, back to 38% 1 min, hold 38% 4 min, 2–7 min. A full color version of this figure is available at The Journal of Antibiotics journal online.
Active fractions were analyzed by HRFTLCMS in a batch mode and were automatically compared using Apex software against a growing library of thiazolyl peptides (Supplementary Table S1, see Supplementary Information) that consisted of calculated high-resolution mass values of M+H, M+Na and M+NH4 ions, and multiply charged species of the same adduct of each of the reported compounds. This process provided significant confidence for the identification of the compounds and was further supported by detection of a family of congeners present in the same extract as exemplified by the analysis of sulfomycin and thiostrepton families (Supplementary Table S2, Supplementary Information). This process of HRFTLCMS-based dereplication comparison does not require significant automation and can be done by searching the calculated ions in an excel table if necessary.
When the thiazolyl peptides were produced in higher titers they can be dereplicated without fractionation. At the later stage of the screening we employed two stage processes in which we first analyzed the two-plate screening hits directly by HRFTLCMS. The hits that showed unambiguous identification of known thiazolyl peptides were eliminated from further studies. Extracts showing ambiguous identification due to low signal intensity or showing the absence of any known thiazolyl peptides were fractionated by 96-well fractionation procedures and then reanalyzed by HRFTLCMS. A combination of 96-well solid-phase extraction fractionation, activity recovery and quantification, and HRFTLCMS-based dereplication allowed for rapid identification of known thiazolyl peptides in 125 extracts, suggesting that seven contained potentially novel compounds (Supplementary Table S2). Geographical distribution and frequency of the occurrence (compounds produced by the same strain in more than one producing media were counted only once) of thiazolyl peptides identified during this study are summarized in Table 1, including taxonomic position deduced from the phylogenetic analysis of all producing strains. These strains were isolated from a broad diversity of geographical origins and environmental ecosystems. Representative isolates were isolated from Africa (South Africa, Togo and Equatorial Guinea), Central America (Panama, Costa Rica and Puerto Rico), North America (USA, Mexico), Australasia (New Caledonia), South America (Bolivia and French Guinea), Asia (Sri Lanka) and Europe (Spain). The taxonomic position of the strains producing different active compounds was studied to evaluate their relatedness and the potential association with the production of these bioactive molecules (Figure 3). This analysis has shown that these thiazolyl peptides are wide-spread across different actinomycetes lineages represented by at least seven different genera. Whereas representative strains of the genus Streptomyces dominate the number and diversity of the compounds produced, as could be expected from the larger number of strains tested of this taxon (77.8%), thiazolyl peptides were also produced by members of the genera Nonomuraea, Actinoplanes, Micromonospora, Nocardia, Actinokineospora and Amycolatopsis, all minor taxa only representing 22% of the strains screened. All known compounds identified in this screening represented a, d and e structural subseries of thiazolyl peptides.8 This screening process allowed rapid identification of 24 of the 80 known thiazolyl peptides and three new thiazolyl peptides, one of them elucidated as philipimycin (Chart 1), produced by a strain of Actinoplanes philippinensis32 and two compounds (MW1520 and 1636) that remain under investigation. Preliminary characterization of the producer strains suggests that the compound with MW1520 is produced by unrelated members of the genus Nonomuraea and a potential new species of Streptomyces, whereas the compound with MW1636 is produced by strains of the genus Amycolatopsis (Figure 3). The compounds most frequently detected were sulfomycin I (35 times) followed by multhiomycin (28 times) and sulfomycin II (7 times), all of them produced by strains widely distributed among environmental samples recovered from four continents (Table 1). Major compound sulfomycin I was produced by more than ten different Streptomyces spp. and Nocardia spp., whereas multhiomycin was produced by strains of the genera Streptomyces and Actinoplanes. Several compounds were detected less than five times, most only once. It is interesting to mention that glycothiohexide was produced by strains of Nonomuraea spp with geographical origins as diverse as Costa Rica, Togo and New Caledonia.
During the discovery period, we also added a series of thiazomycins to the list of new thiazolyl peptides discovered by chemical screening from nocathiacin producers, which were not discovered by the two-plate screening although a nocathiacin I producer was discovered.25 All compounds discovered by the thiazomycin sensitive and resistant pair two-plate assay during this study showed differential zone of inhibition in this assay, suggesting that these compounds bind to the same binding site as thiazomycin, thus for the first time validating their binding relationship to the target. GE2270 series of thiazolyl peptides bind to elongation factor TU site and do not show activity in this two-plate assay, and, therefore, were not discovered, confirming screening specificity. It was certainly not due to lack of producing strain of GE2270 in our collection. While conceptually it is possible that a non-thiazolyl peptide can bind to the thiazomycin-binding site we failed to find such a compound. However, this screening method represents a great tool for the discovery of new members of the same class of compounds and provides a very valuable tool for the discovery of new producing strains of known compounds as demonstrated by discovery of multiple producing strains of many compounds (Table 1, and Supplementary Table S2). Despite the similar frequency of discovery of these compounds observed in both groups of strains (0.54 and 0.58%), the occurrence of novel compounds of this class of metabolites is mostly concentrated in minor actinomycetes taxa (Table 2).
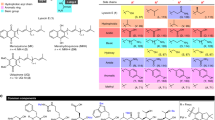
A number of compounds that were identified by HRFTLCMS were isolated and characterized by comparison of the spectroscopic data. These compounds are: Sulfomyin I, multhiomycin, thiostrepton A, thiocillin I and II. The structural identity of thiostrepton A and multhiomycin was confirmed by comparison with authentic samples. There were two objectives for these purifications: (1) Confirmation of the compounds shown by HRFTLCMS to build confidence in the dereplication process and (2) head to head comparison of activity of the compounds to correlate the binding site selectivity.
The isolation of S54832 A-1 from Micromonospora globosa, recovered from a Spanish soil sample has been reported in a patent literature with 90 MHz NMR data without assignment.11 Therefore, the isolation and structural elucidation of S54832 A-1 has been briefly described which serves as an example for the purification of other compounds described in Table 3.
We report here the production of S54832 A-1 by two new strains of the genus Micromonospora. Strain P-000503611 was isolated from a rhizosphere soil collected in Tenerife Island, Spain, and the second strain P-000507284 was isolated from a moss collected in South Africa. Furthermore, the phylogenetic analysis shows that the strain P-000503611 is associated with the strain Micromonospora auratinigra TT1-11T, a relationship highly supported by the bootstrapping value (100%) and permitting to propose the strain as a new member of the species Micromonospora auratinigra (Figure 3). Similarly, the second strain of Micromonospora (P-000507284) producing S54832 A-1 was also found to cluster with the same species in the phylogenetic analysis. In spite of their relatedness, the compounds were produced in each case in different production media with different incubation periods. In the case of the strain P-000503611, the compound was produced after 13 days fermentation, whereas the same molecule was produced by P-000507284 after 7 days of cultivation in oatmeal-based medium corn lactose ardamine (CLA).34
The fermentation broth of P-000503611 was extracted with acetone and chromatographed on an Amberchrome column eluting with aqueous methanol. The antibiotic active fractions were chromatographed on Sephadex LH20 to afford S54832 A-1 as an amorphous powder. Most of the other compounds were isolated with a similar two to three step process involving Amberchrome, LH20 and reversed phase HPLC. The HRESIFTMS analysis of the S54832 A-1 showed a molecular formula C59H55N13O19S5. The 1H and 13C NMR spectra of S54832 A-1 were very similar to the corresponding spectra of thiazomycin except for the absence of the olefinic methoxyl, N-methyl, two methyls and oxazolidine groups and the presence of an olefinic quartet, olefin coupled methyl, two methoxyl and a hydroxy methyl groups. The structure was elucidated by full suites of one-dimensional and two-dimensional NMR spectra including COSY, HMQC and HMBC experiments in CDCl3. Full assignment is presented in Figure 4.
The list of purified compounds and their antibacterial activities and spectrum is presented in Table 3. These compounds represent structural subseries a, d and e class of thiazolyl peptides.8 The hydroxy-pyridine class (subseries e) of the thiazolyl peptides represented by multhiomycin, S54832 A-1 and thiazomycin showed most potent activities. The glycosylated hydroxy-pyridine thiazolyl peptide, philipimycin, was slightly less active than the other three of the subseries e compounds. Thiostrepton A, the series a thiazolyl peptide showed intermediate potency and the subseries d, the pyridine class of thiazolyl peptides, represented by sulfomycin I, thiocillin I and thiocillin II showed lease potent activity (Table 3). Regardless of their overall potency, all compounds showed cross-resistance to thiazomycin-resistant strain, suggesting for the first time that they all bind to L11-binding site and confirming that they all share the common binding site.
In addition, S54832 A-1 was tested in a disseminated murine model of S. aureus infection.27 It showed over 5.5 log reduction of CFU of S. aureus burden in kidney at 2.5 mg kg−1 (ED99 of 0.29 mgkg−1 by s.c. administration (dosing vehicle dimethyl sulfoxide).
Sensitive-resistant pair-based screening for antibacterial agents provides a great opportunity for discovery of novel antibacterial agents with defined mode of action. The actives from such screening are amenable for easy dereplication by HRFTLCMS particularly when combined, if necessary, with high-throughput small-scale fractionation and component concentration. This approach allows for rapid discovery of novel compounds, new congeners (for example, philipimycin), as well as new strains for the production of various known thiazolyl peptides. The library with list of compounds with calculated accurate masses represents a great tool that can be used in an automated or manual fashion with HRFTLCMS for rapid and efficient dereplication of natural products.
Experimental section
General procedure
All reagents were obtained from Sigma-Aldrich (St Louis, MO, USA) and were used without further purification. Multhiomycin and thiostrepton A were purchased from Sigma-Aldrich. The NMR spectra were recorded on a Varian Inova 500 or 600 MHz spectrometers operating at 500 or 600 MHz for 1H and 125 or 150 MHz for 13C nuclei. Residual solvent signal was used as a reference. Optical rotations were obtained on a Perkin-Elmer 241 Polarimeter (Perkin-Elmer, Waltham, MA, USA), and IR spectral data were obtained on a Perkin-Elmer Spectrum One spectrometer. High-resolution mass spectra were obtained on a Thermo Finnigan Linear Trap Quadrupole-Fourier Transform (LTQ-FT) with the standard Ion Max Atmospheric Pressure Ionization (API) source (without the sweep cone) and ESI probe. Three scan events were used. The ion trap was scanned from 150–2000 first in negative ion mode and then in positive ion mode. The FT was scanned from 200–2000 in the positive ion mode only. In all cases, the source induced dissociation (SID) was set to 18 V to try to reduce multiple ion clusters. Apex software was used for the data analysis with the following standard conditions; peak width/resolution was set to 100 000 resolution at mass 400, MS search tolerance set between 0.008 and 0.003 Da. Apex uses a proprietary algorithm to determine the baseline and compute the peak centroids of continuum mass spectral data. The primary objective of the algorithm is to accurately determine peak centroid m/z values to facilitate use in accurate mass applications. Apex targets compounds in acquired mass spectral data sets by computing theoretical isotope clusters of all library compounds and exhaustively comparing each cluster to each spectrum in the data set, taking into account elevated mass accuracy. Apex is designed for high-throughput application, with imported sample list and tabular results files in XML format. Apex provides a viewer for batch analysis of hits.
General procedure of isolation of cultures
The collection of actinomycetes used in this screening were selected from isolation plates obtained following multiple strategies that combine the use of specific isolation methods with selective and diluted nutritional media favoring the occurrence of slow growing species. Isolation methods ranged from the serial dilution plating, differential centrifugation and enrichment of some taxa by pretreatment.
General procedures for DNA extraction and phylogenetic analysis
Total genomic DNA from the different actinomycetes used in this study was recovered and purified as previously described35 from actinomycete cultures grown in ATCC-2 liquid medium (0.5% yeast extract (Difco, Franklin Lakes, NJ, USA), 0.3% beef extract (Difco), 0.5% peptone (Difco), 0.1% dextrose (Difco), 0.2% starch from potato (Panreac, Barcelona, Spain), 0.1% CaCO3 (E. Merck, Darmstadt, Germany) and 0.5% NZ amine E (Sigma, St Louis, MO, USA)).36 DNA preparations were used as template DNA for Taq Polymerase. PCR primers fD1 and rP2 were used for amplifying the 16S ribosomal RNA genes of the strain.36 Reactions were performed in a final volume of 50 μl containing 0.4 μM of each primer, 0.2 mM of each of the four deoxyribonucleotide triphosphates (Roche, Indianapolis, IN, USA), 5 μl of extracted DNA, 1 U Taq polymerase (Appligene, Watford, UK) with its recommended reaction buffer. PCR amplifications were performed in a Peltier Thermal Cycler PTC-200, according to the following profile: 5 min at 95 °C and 40 cycles of 30 s at 94 °C, 30 s at 52 °C for and 1 min at 72 °C, followed by 10 min at 72 °C. The amplification products were analyzed by electrophoresis in 2% (w/v) pre-cast agarose gels stained with ethidium bromide (E-gel 2%, 48 wells, Invitrogen, Carlsbad, CA, USA). PCR products were sent to Secugen (http://www.secugen.es/) for sequencing, and were purified and used as a template in sequencing reactions using the primers fD1 and rP2, and 1100R and 926F.37 Amplified DNA fragments were sequenced using the ABI PRISMDYE Terminator Cycle sequencing kit and fragments were resolved using the ABI3130 genetic analyzer (Applied Biosystems, Foster City, CA, USA). Partial sequences were assembled and edited using the Assembler contig editor component of Bionumerics (ver 5.10) analysis software (Applied Maths NV, Sint-Martens-Latem, Belgium).
The identification of phylogenetic neighbors was initially carried out by the BLAST38 and mega BLAST39 programs against the database of type strains with validly published prokaryotic names.40 The 50 sequences with the highest scores were then selected for the calculation of pairwise sequence similarity using global alignment algorithm, which was implemented at the EzTaxon server (http://www.eztaxon.org).40
Multiple alignment was carried out using CLUSTALX,41 integrated in the software MEGA version 5.05,42 followed by manual editing as recommended by Tindall et al.43 Phylogenetic analysis was also conducted using MEGA version 5.05. The distance matrix was calculated using pairwise comparisons of sequences corrected with Jukes and Cantor algorithm44 and the phylogenetic tree was built using the Neighbor-Joining method.45 The robustness of the phylogenetic tree was estimated using the bootstrap method with 1000 pseudo-replicates.46
General procedure of high-throughput fermentation in 50 ml vials
Strains were inoculated as fresh agar plugs in 10 ml ATCC-2 seed tubes (15 × 2.5 cm) and incubated at 28 °C, 220 r.p.m. and 70% humidity during 4–7 days. Seeds were used to inoculate 40 ml Environmental Protection Agency (EPA) vials containing 10 ml of fermentation medium (inoculation with 5% v/v). Fermentations were carried out at 28 °C, 220 r.p.m., 70% humidity for 7–13 days. Strains were grouped in taxonomic groups and cultivated in three different production media according to the preferred nutritional requirements of the actinomycetes groups tested.
General procedure of extraction of metabolites for primary screening
Ten milliliter (equal volume of broth) acetone was added to each fermentation broth and was shaken in a Kuhner for 15 min at room temperature. Two milliliter broth supernatant was transferred into deep-well plates and the extract was dried down in a Genevac to 500 μl and transferred into ABgene plates. An aliquot was spotted to the two-plate screening assay and remainder was saved as is for dereplication and identification of antibacterial active compounds.
Thiazomycin sensitive-resistant two-plate assay
The details of the assay have been previously described.26
High-throughput 96-well fractionation
A quantity 0.4 ml aqueous fermentation broth that showed antibacterial activity in the two-plate sensitive-resistant pair assay was charged on to a 96-well plate Waters Oasis hydrophilic lipophilic balance plate (30 mg resin consisting of a balanced ratio of lipophilic divinylbenzene and hydrophilic N-vinylpyrrolidone resins, Waters, Milford, MA, USA). The spent was collected in a 96-well elution plate and the hydrophilic lipophilic balance plate was sequentially eluted with 0.4 ml each of 5 and 50% aqueous methanol, 100% methanol, 100% acetone and 100% dimethyl sulfoxide in separate 96-well elution plates. The 10 μl each fraction generated from these fractionations including spent was tested in the thiazomycin sensitive-resistant two-plate assay. The active fraction(s) were evaluated by HRFTLCMS.
HRFTLCMS-based dereplication and classification of thiazolyl peptides
The fractions that showed activity were evaluated by HRFTLCMS using following conditions. LC-conditions: Column: Zorbax SB-C8, 2.1 × 30 mm; Temp: 40 °C; Flow rate 300 μl min−1; Solvents: A=10% acetonitrile and 90% water containing 1.3 mM trifluoroacetic acid and ammonium formate, B=90% acetonitrile and 10% water containing 1.3 mM trifluoroacetic acid and ammonium formate; Gradient 10% B to 100% B in 6 min, hold 2 min, initialize 2 min.
Description of producing organism of S54832 A1 (P-000503611)-isolation conditions and growth characteristics
The producing strain (P-00503611) was obtained from the rhizosphere soil associated with a specimen of Ranunculus cortusifolius in Tenerife, Canary Islands, Spain. The strain was isolated after pretreatment of the soil with dry heat at 100 °C for 1 h and plating on NZ amine-based agar supplemented with 20 μg ml−1 nalidixic acid. The strain grows well on a range of agar media such as yeast malt extract, oatmeal, glycerol asparagine, inorganic salts starch and tyrosine agars at 28 °C. The gross colonial morphology was typical of Micromonospora and its growth characteristics, including spore-mass color, substrate mycelial pigmentation and the production of different pigments were recorded in different agar media. The colony developed on Yeast Malt Extract Agar (ISP2), a substrate mycelium initially brownish orange in young colonies that turned orange brown after 21 days of incubation and was gradually covered by a dark-brown mucoid spore layer; no aerial mycelium was present. The substrate mycelium was extensively branched and single spores were formed on substrate mycelium, a characteristic of members of the genus Micromonospora. Spores tend to coalesce in a dark mucous mass in older cultures. Sporulation was observed in most of the test media but with different degrees of coalescence.47
Chemotaxonomic analysis and physiological properties
The analysis of cell wall composition shows that whole-cell hydrolyzates from strain P-000503611 contain meso-diaminopimelic acid, and glucose and arabinose as major cell wall sugars, chemotaxonomic markers in agreement with the morphological characteristics that associate the strain to members of the genus Micromonospora.48 Growth of the compounds was determined after 7, 14 and 21 days of growth. Strain P-000503611 moderately utilized D-glucose, rhamnose and D-xylose, and did not use sucrose, D-fructose, raffinose, I-inositol, D-mannitol, L-arabinose and cellulose.
16S ribosomal DNA sequence and phylogenetic analysis
The complete 16S ribosomal DNA sequence has been determined for strain P-000503611and identification of phylogenetic neighbors was performed as described above for all the strains in the study.
Fermentation of P-000503611
Large-scale confirmation of the production of the bioactive compound was performed using first a seed culture of the strain by inoculating frozen agar plugs of the strain into 10 ml of seed medium ATCC-2 in a 50-ml tube and incubating the tube at 28 °C with shaking at 220 r.p.m. for about 96 h. A second seed culture was prepared by inoculating 50 ml of seed medium in a 250-ml flask with 2 ml of the first seed. For production conditions, 4 ml of seed culture was transferred to a 500-ml flask containing 100 ml of production medium FR23 (glucose 5 g l−1, soluble starch 30 g l−1, cane molasses 20 g l−1 and pharmamedia 20 g l−1). Each flask was incubated at 28 °C with 220 r.p.m. agitation for 13 days before extraction.
Extraction and purification of S54832 A-1
Initial purification was performed from a 1-liter regrowth by following procedure. One liter broth was extracted by 1 liter acetone by shaking for 1 h. The broth was filtered on a bed of celite and the filtrate was concentrated under reduced pressure to remove most of the acetone. The aqueous layer was charged on to a 50-ml Amberchrome column and eluted with a 100-min linear gradient of 5–100% aqueous methanol. The activity eluted in 90–100% methanol. The active fractions were pooled and concentrated under reduced pressure to dryness affording 13.7 mg material, which was then chromatographed on a 10-ml Sephadex LH20 and eluted with methanol. The fractions eluting in 1.5–2 column volumes were combined and concentrated to dryness to furnish 2.2 mg of powder of S54832 A-1. An additional 18 mg of S54832 A-1 was purified from 5 liter regrowth by extraction, Sephadex LH20 followed by reversed phase HPLC on Zorbax C8 eluting with aqueous acetonitrile. HRESIFTMS (m/z) 1410.2422 (M+H, calcd for C59H55N13O19S5+H, 1410.2419), 1236.1528 (M-glycoside+2H, calcd for C51H42N13O15S5, 1236.1527). For 1H and 13C NMR assignment, see Figure 4.
References
Klevens, R. M. et al. Invasive methicillin-resistant Staphylococcus aureus infections in the United States. J. Am. Med. Assoc. 298, 1763–1771 (2007).
Boucher, H. W. et al. Bad bugs, no drugs: no ESKAPE! An update from the Infectious Diseases Society of America. Clin. Infect. Dis. 48, 1–12 (2009).
Rice, L. B. Progress and challenges in implementing the research on ESKAPE pathogens. Infect. Control. Hosp. Epidemiol. 31 (Suppl 1), S7–10 (2010).
Singh, S. B. et al. Isolation, structure, and absolute stereochemistry of platensimycin, a broad spectrum antibiotic discovered using an antisense differential sensitivity strategy. J. Am. Chem. Soc. 128, 11916–11920 15547 (2006).
Wang, J. et al. Platensimycin is a selective FabF inhibitor with potent antibiotic properties. Nature 441, 358–361 (2006).
Wang, J. et al. Platencin is a dual FabF and FabH inhibitor with potent in vivo antibiotic properties. Proc. Natl Acad. Sci. (USA) 104, 7612–7616 (2007).
Jayasuriya, H. et al. Isolation and structure of platencin: a novel FabH and FabF Dual inhibitor with potent broad spectrum antibiotic activity produced by Streptomyces platensis MA7339. Angew. Chem. Int. Ed. Engl. 46, 4684–4688 (2007).
Bagley, M. C., Dale, J. W., Merritt, E. A. & Xiong, X. Thiopeptide antibiotics. Chem. Rev. 105, 685–714 (2005).
Northcote, P. T., Siegel, M., Borders, D. B. & Lee, M. D. Glycothiohexide alpha, a novel antibiotic produced by ‘Sebekia’ sp., LL-14E605. III. Structural elucidation. J. Antibiot. (Tokyo) 47, 901–908 (1994).
Northcote, P. T. et al. Glycothiohexide alpha, a novel antibiotic produced by ‘Sebekia’ sp., LL-14E605. II. Isolation and physical-chemical characterization. J. Antibiot. (Tokyo) 47, 894–900 (1994).
Keller-Juslen, C., Kuhn, M. & King, H. D. Antibiotics, pharmaceutical compositions and method of use USA patent US 4,478,831 (1984).
Sasaki, T. et al. MJ347-81F4 A & B, novel antibiotics from Amycolatopsis sp.: taxonomic characteristics, fermentation, and antimicrobial activity. J. Antibiot. (Tokyo) 51, 715–721 (1998).
Li, W. et al. Nocathiacins, new thiazolyl peptide antibiotics from Nocardia sp. I. Taxonomy, fermentation and biological activities. J. Antibiot. (Tokyo) 56, 226–231 (2003).
Constantine, K. L. et al. Conformation and absolute configuration of nocathiacin I determined by NMR spectroscopy and chiral capillary electrophoresis. J. Am. Chem. Soc. 124, 7284–7285 (2002).
Naidu, B. N. et al. Nocathiacin I analogues: synthesis, in vitro and in vivo biological activity of novel semi-synthetic thiazolyl peptide antibiotics. Bioorg. Med. Chem. Lett. 14, 5573–5577 (2004).
Naidu, B. N. et al. Synthesis and antibacterial activity of O-substituted nocathiacin I derivatives. Bioorg. Med. Chem. Lett. 14, 3743–3746 (2004).
Regueiro-Ren, A. et al. Novel semi-synthetic nocathiacin antibiotics: synthesis and antibacterial activity of bis- and mono-O-alkylated derivatives. Bioorg. Med. Chem. Lett. 14, 171–175 (2004).
Connolly, T. P. et al. Chemical conversion of nocathiacin I to nocathiacin II and a lactone analogue of glycothiohexide alpha. J. Nat. Prod. 68, 550–553 (2005).
Naidu, B. N. et al. Synthesis, in vitro, and in vivo antibacterial activity of nocathiacin I thiol-Michael adducts. Bioorg. Med. Chem. Lett. 15, 2069–2072 (2005).
Naidu, B. N. et al. Synthesis and antibacterial activity of nocathiacin I analogues. Bioorg. Med. Chem. Lett. 16, 3545–3549 (2006).
Xu, L., Farthing, A. K., Shi, Y. J., Meinke, P. T. & Liu, K. Conversion of nocathiacin I to nocathiacin acid by a mild and selective cleavage of dehydroalanine. J. Org. Chem. 72, 7447–7450 (2007).
Xu, L. et al. Nocathiacin analogs: synthesis and antibacterial activity of novel water-soluble amides. Bioorg. Med. Chem. Lett. 19, 3531–3535 (2009).
Jayasuriya, H. et al. Isolation and structure elucidation of thiazomycin- a potent thiazolyl peptide antibiotic from Amycolatopsis fastidiosa. J. Antibiot. (Tokyo) 60, 554–564 (2007).
Zhang, C. et al. Isolation, structure, and antibacterial activity of thiazomycin A, a potent thiazolyl peptide antibiotic from Amycolatopsis fastidiosa. Bioorg. Med. Chem. 16, 8818–8823 (2008).
Zhang, C. et al. Thiazomycins, thiazolyl peptide antibiotics from Amycolatopsis fastidiosa. J. Nat. Prod. 72, 841–847 (2009).
Zhang, C. et al. Isolation, structure, and antibacterial activity of philipimycin, a thiazolyl peptide discovered from Actinoplanes philippinensis MA7347. J. Am. Chem. Soc. 130, 12102–12110 (2008).
Singh, S. B. et al. Antibacterial evaluations of thiazomycin- a potent thiazolyl peptide antibiotic from Amycolatopsis fastidiosa. J. Antibiot. (Tokyo) 60, 565–571 (2007).
Baltz, R. H. Antimicrobials from actinomycetes: back to the future. Microbe 2, 125–131 (2007).
Haste, N. M. et al. Activity of the streptogramin antibiotic etamycin against methicillin-resistant Staphylococcus aureus. J. Antibiot. (Tokyo) 63, 219–224 (2010).
Singh, S. B., Young, K. & Miesel, L. Screening strategies for discovery of antibacterial natural products. Expert Rev. Anti. Infect. Ther. 9, 589–613 (2011).
Silver, L. L. in Antibiotic Discovery and Development Vol. I (eds) Dougherty T. J Pucci M. J)(Ch. 2), 33–75 Springer Science: New York, (2012).
Zhang, C. et al. Isolation, structure, and antibacterial activity of philipimycin, A thiazolyl peptide discovered from actinoplanes philippinensis MA7347. J. Am. Chem. Soc. 130, 12102–12110 (2008).
Genilloud, O. et al. Current approaches to exploit actinomycetes as a source of novel natural products. J. Ind. Microbiol. Biotechnol. 38, 375–389 (2011).
Singh, S. B. et al. Isolation, structure, and antibacterial activities of lucensimycins D-G, discovered from Streptomyces lucensis MA7349 using an antisense strategy. J. Nat. Prod. 72, 345–352 (2009).
Innis, M. A., Gelfand, D. H., Sninsky, J. J. & White, T. J. PCR Protocols. A Guide to Methods and Amplifications, Academic Press, (1990).
Weisburg, W. G., Barns, S. M., Pelletier, D. A. & Lane, D. J. 16S ribosomal DNA amplification for phylogenetic study. J. Bacteriol. 173, 697–703 (1991).
Lane, D. J. in Nucleic Acid Techniques in Bacterial Systematics (eds Stackebrandt E Goodfellow M)115–175 Wiley: New York, (1991).
Altschul, S. F. et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic. Acids. Res. 25, 3389–3402 (1997).
Zhang, Z., Schwartz, S., Wagner, L. & Miller, W. A greedy algorithm for aligning DNA sequences. J. Comput. Biol. 7, 203–214 (2000).
Chun, J. et al. EzTaxon: a web-based tool for the identification of prokaryotes based on 16S ribosomal RNA gene sequences. Int. J. Syst. Evol. Microbiol. 57, 2259–2261 (2007).
Thompson, J. D., Gibson, T. J., Plewniak, F., Jeanmougin, F. & Higgins, D. G. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic. Acids. Res. 25, 4876–4882 (1997).
Tamura, K. et al. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28, 2731–2739 (2011).
Tindall, B. J., Rossello-Mora, R., Busse, H. J., Ludwig, W. & Kampfer, P. Notes on the characterization of prokaryote strains for taxonomic purposes. Int. J. Syst. Evol. Microbiol. 60, 249–266 (2010).
Jukes, T. H. & Cantor, C. Evolution of Protein Molecules, in Mammalian Protein Metabolism, Academic Press, (1969).
Saitou, N. & Nei, M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4, 406–425 (1987).
Felsenstein, J. Evolutionary trees from DNA sequences: a maximum likelihood approach. J. Mol. Evol. 17, 368–376 (1981).
Shirling, E. B. & Gottlieb, D. Methods for characterization of Streptomyces species. Int. J. Syst. Bacteriol. 16, 313–340 (1966).
Lechevalier, H. A. & Lechevalier, M. P. in In Actinomycete Taxonomy (Society for Industrial Microbiology Special Publication no. 6) (eds Dietz A., Thayer D. W.)277–284 Society for Industrial Microbiology, (1980).
Author information
Authors and Affiliations
Corresponding author
Additional information
Supplementary Information accompanies the paper on The Journal of Antibiotics website
Supplementary information
Rights and permissions
About this article
Cite this article
Singh, S., Zhang, C., Zink, D. et al. Occurrence, distribution, dereplication and efficient discovery of thiazolyl peptides by sensitive-resistant pair screening. J Antibiot 66, 599–607 (2013). https://doi.org/10.1038/ja.2013.54
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ja.2013.54
Keywords
This article is cited by
-
Thiazomycin, nocathiacin and analogs show strong activity against clinical strains of drug-resistant Mycobacterium tuberculosis
The Journal of Antibiotics (2017)
-
Dereplication strategies in natural product research: How many tools and methodologies behind the same concept?
Phytochemistry Reviews (2017)
-
Winners of the 2014 JA Medals for excellence
The Journal of Antibiotics (2015)
-
Dereplication of microbial extracts and related analytical technologies
The Journal of Antibiotics (2014)