Abstract
Many polyketide antibiotics contain macrolactones that arise from polyketide synthase chain release via thioesterase (TE) domain-catalyzed macrolactonization. The hydroxyl groups utilized in such macrolactonization reactions typically derive from reduction of β-ketothioester intermediates in polyketide chain assembly. The stambomycins are a group of novel macrolide antibiotics with promising anticancer activity that we recently discovered via rational activation of a silent polyketide biosynthetic gene cluster in Streptomyces ambofaciens. Here we report that the hydroxyl group utilized for formation of the macrolactone in the stambomycins is derived from cytochrome P450-catalyzed hydroxylation of the polyketide chain rather than keto reduction during chain assembly. This is a novel mechanism for macrolactone formation in polyketide antibiotic biosynthesis.
Similar content being viewed by others
Introduction
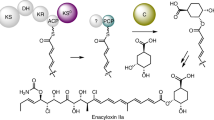
Several polyketide natural products with important applications in medicine and agriculture contain a macrolactone. Examples include the antibacterial erythromycin A,1 the antifungals amphotericin B and nystatin,2, 3 the insecticide spinosyn A4 and the avermectins5 that are used as both insecticides and anthelmintics. Such macrolactones are typically biosynthesized by thioesterase (TE) domains located at the C-terminus of type I modular polyketide synthase (PKS) assembly lines.6, 7, 8, 9, 10 The fully assembled polyketide chain is transferred from the acyl carrier protein domain within the last module of the PKS to a conserved Ser residue in the active site of the TE domain.11 The resulting ester undergoes general base-assisted intramolecular nucleophilic attack by a hydroxyl group, generated by reduction of a β-ketothioester intermediate in polyketide chain assembly.11 This leads to formation of a tetrahedral intermediate that collapses to release the product of the PKS as a macrolactone (Figure 1).11
Mechanism for polyketide chain release from type I modular polyketide synthases (PKSs) via macrolactonization. The hydroxyl group required for macrolactonization is typically generated by reduction of a β-ketothioester intermediate in polyketide chain assembly; in this example, the ketoreductase (KR) domain within module 1 of 6-deoxyerythronolide B synthase catalyzes this reaction. The fully assembled polyketide chain is transferred from the phosphopantetheine thiol of the acyl carrier protein (ACP) domain within the last module of the PKS to the active site serine residue of the thioesterase (TE) domain at the C-terminus of the last module. The TE domain then utilizes an active site His residue (B) as a general base to form the macrolactone (in this case 6-deoxyerythronolide B) via an addition–elimination mechanism.
We recently reported the discovery of the stambomycins 1–4, a remarkable family of 51-membered macrolide antibiotics with promising activity against human cancer cell lines, via rational activation of a silent gene cluster in Streptomyces ambofaciens ATCC23877 that encodes a cryptic modular PKS assembly line (Figure 2).12 Although the stambomycin PKS contains a C-terminal TE domain that is proposed to catalyze macrolactone formation,12 the C-50 hydroxyl group utilized for this process does not appear to derive from PKS-mediated keto reduction during polyketide chain assembly. Instead, we hypothesized that this hydroxyl group and the C-28 hydroxyl group are derived from molecular oxygen via cytochrome P450 (CYP)-catalyzed hydroxylation reactions (Figure 2).12 Here we report the results of experiments aimed at investigating this hypothesis.
Materials and methods
HR MS, MS/MS and UHPLC-MS were carried out using a Bruker MaXis electrospray ionization time-of-flight (ESI-TOF) MS connected, where appropriate, to a Dionex 3000 RS UHPLC fitted with an Agilent Zorbax Eclipse Plus (Agilent, Wokingham, UK) C18 column (100 × 2.1 mm, 1.8 μm). NMR spectra were recorded at 25 °C in CD3OD on a Bruker Avance II 700 MHz spectrometer (Bruker, Coventry, UK) equipped with a TCI cryoprobe. Stambomycin-related metabolites accumulated in S. ambofaciens mutants were purified by semi-preparative HPLC on an Agilent 1100 instrument fitted with a C-18 column (100 × 21 mm, 5 μm, Agilent). The strains and plasmids used in this study are listed in Table 1.
Deletion of samR0478 and samR0479 in S. ambofaciens
The samR0478 and samR0479 genes in S. ambofaciens ATCC/OE48412 were replaced with a spectinomycin resistance cassette using the REDIRECT system, as described previously.13 The spectinomycin resistance cassette, derived from the plasmid pIJ778,13 was used as the template for PCR reactions with the primers 5′-TGCAGACGGGCTGCTCGACGCGCGGGGAGTGACGGCATGTGTAGGCTGGAGCTGCTTC-3′ (forward)/5′-GTCCCGGGCTCGGGCCGCGCCTTTGCCGCTGCCGGACTCATTCCGGGGATCCGTCGACC-3′ (reverse) and 5′-GAGCGGCCGGGAGGCGGGCGCGCGGCGCAGTCGGCGTTCTGTAGGCTGGAGCTGCTTC-3′ (forward)/5′-CGCCCCGCAGGACACGGCAACCACCGACGTAGAGGGATGATTCCGGGGATCCGTCGACC-3′ (reverse) to create disruption cassettes (∼1400 bp) for samR0478 and samR0479, respectively. Escherichia coli BW25113/pKD20 was transformed with the bacterial artificial chromosome (BAC) BBC and then separately with each of the PCR products to replace either samR0478 or samR0479 in the BAC by homologous recombination. E. coli ET12567/pUZ8002 was separately transformed with each of the mutated BACs (BBC/Δ478::oriT-aadA and BBC/Δ479::oriT-aadA) that were subsequently transferred independently to S. ambofaciens ATCC/OE484 by conjugation. Spectinomycin-resistant colonies were picked and grown in Hickey-Tresner (HT) liquid medium.14 After several rounds of growth, cultures were spread on HT agar medium, and individual colonies were picked and analyzed by PCR and Southern hybridization to identify those in which the desired gene replacements had occurred via double crossover homologous recombination. The resulting mutants were named ATCC/OE484/Δ478 and ATCC/OE484/Δ479.
Incorporation of 18O2 into stambomycins A–D 1-4
The ATCC/OE484 strain was cultured on MP5 agar medium overlaid with a sterile cellophane membrane as described previously12 in a glove bag filled with 18O2 gas. After 4 days of incubation at 30 °C, the cellophane membrane was lifted off the plate and the mycelia were scraped into a 50 ml plastic tube. Then, 10 ml of methanol was added and the mixture was sonicated for 10 min. The mycelia were pelleted by centrifugation for 10 min at 4 °C and the supernatant was analyzed by HR LC-MS using a flow rate of 0.2 ml min−1 and water containing 0.1% formic acid (solvent A)/acetonitrile containing 0.1% formic acid (solvent B) as eluents. The following elution profile was used: 80% solvent A/20% solvent B for 5 min; gradient to 100% solvent B in 15 min; 100% solvent B for 5 min; gradient back to 80% solvent A/20% solvent B in 3 min. The MS was calibrated at the beginning of each run via loop injection of 20 μl of 10 mM sodium formate and was operated in positive ion mode with settings as follows. Full MS scan range: 50–2000 m/z; End plate offset: −500 V; capillary: −4000 V; nebulizer gas (N2): 1.6 bar; dry gas (N2): 8 l min−1; dry temperature: 180 °C.
Analysis of the production of stambomycins and related metabolites in S. ambofaciens ATCC/OE484/Δ478 and ATCC/OE484/Δ479
S. ambofaciens ATCC/OE484/Δ478 was grown and analyzed as described above for the incorporation of 18O2 into the stambomycins. S. ambofaciens ATCC/OE484/Δ479 was grown in liquid MP5 medium, as described previously.12 The culture supernatant was separated from the biomass by centrifugation and passed through a 0.4 μm filter. The filtrate was analyzed by LC-MS, as described above for the incorporation of 18O2 into the stambomycins.
Isolation of 28-deoxystambomycins A–D 5–8 from S. ambofaciens ATCC/OE484/Δ478
The 28-deoxystambomycins A–D 5–8 were isolated from cultures of S. ambofaciens ATCC/OE484/Δ478 grown on solid MP5 medium, as described previously for the isolation of stambomycins A–D 1–4 from S. ambofaciens ATCC/OE484.12
Isolation of shunt metabolites 9–12 from S. ambofaciens ATCC/OE484/Δ479
Twenty 50 ml cultures of S. ambofaciens ATCC/OE484/Δ479 were grown in MP5 liquid medium, as described previously.12 The cultures were combined and centrifuged for 20 min at 4000 r.p.m. (3220 g). The supernatant was freeze-dried and the residue was re-suspended in 25 ml of 1:4 MeOH/H2O. The resulting solution was passed through a 0.4 μm syringe filter and separated by HPLC, monitoring absorbance at 240 nm. The column was eluted with water containing 0.1% formic acid (A) and acetonitrile containing 0.1% formic acid (B) using the following elution profile: 80% solvent A/20% solvent B for 10 min; gradient to 100% solvent B in 10 min; 100% solvent B for 15 min. Fractions containing 9–12 were identified using ESI-MS, combined and evaporated under reduced pressure. The residue was resuspended in 5 ml of 50% aqueous methanol and further purified by HPLC using the following elution conditions: 0 min, 50% A/50% B; 15 min, 5% A/95% B; 20 min, 100% B; 25 min, 100% B. Fractions containing an inseparable mixture of 9 and 10 were collected, combined and lyophilized, as were fractions containing an inseparable mixture of 11 and 12, yielding 4.3 and 3.2 mg, respectively.
Results
If our hypothesis for the origin of the C-28 and C-50 oxygen atoms is correct, we would expect two oxygen atoms of the stambomycins to be derived from molecular oxygen. Thus, we investigated the incorporation of 18O2 into stambomycins A–D 1–4. The previously reported stambomycin producer S. ambofaciens ATCC/OE48412 was grown on a solid medium under an 18O2 atmosphere and methanol extracts of the mycelia were analyzed by HR LC-MS. An approximately 3:1 mixture of doubly and singly 18O-labeled stambomycins A and B 1 and 2 was observed (Figure 3). Analogous 18O2 incorporation patterns were observed for stambomycins C and D 3 and 4.
Mass spectra from LC-MS analysis of the incorporation of 18O2 into stambomycins A/B 1/2. The top panel shows the mass spectrum of a mixture of singly and doubly 18O-labeled stambomycins A/B; none of the unlabeled metabolites are observed. The middle panel shows the simulated mass spectrum for a species with the molecular formula C73H134N16O2118ONa2+, corresponding to the doubly charged ion of singly labeled stambomycins A/B. The bottom panel shows the simulated mass spectrum for a species with the molecular formula C73H134N16O2018O2Na2+, corresponding to the doubly charged ion of doubly labeled stambomycins A/B.
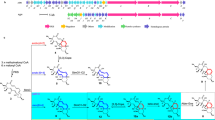
The samR0478 and samR0479 genes within the stambomycin biosynthetic gene cluster encode proteins with sequence similarity to CYPs (Figure 4a).12 Thus, we hypothesized that SamR0478 and SamR0479 catalyze the hydroxylation of C-28 and C-50 (or vice versa) of the stambomycin polyketide chain. To investigate the role played by these enzymes in stambomycin biosynthesis, we independently replaced samR0478 and samR0479 on the chromosome of S. ambofaciens ATCC/OE484 with a spectinomycin resistance cassette.
The stambomycin biosynthetic gene cluster and the structures of stambomycin-related metabolites accumulated in the samR0478 and samR0479 mutants of Streptomyces ambofaciens. (a) Organization of the stambomycin biosynthetic gene cluster. The samR0478 and samR0479 genes, which were independently deleted in this study, are black. The polyketide synthase (PKS) genes are dark gray and other genes are light gray. The samR0487 to samR0478 and samR0473-samR0468 regions are magnified 5 and 10 times, respectively, compared with the regions containing the PKS genes. (b) Structures of 28-deoxystambomycins A–D 5–8 accumulated in the samR0478 mutant. (c) Structures of the shunt metabolites 9–12 that accumulate in the samR0479 mutant showing observed COSY correlations (black lines) and key heteronuclear multiple-bond correlations (HMBC; arrows).
HR LC-MS analysis of methanol extracts of the samR0478 mutant showed that the production of stambomycins A–D 1–4 was abolished and identified new metabolites with molecular formulae corresponding to stambomycin derivatives lacking an oxygen atom (calculated for C73H134NO21Na2+: 691.9668, found: 691.9682; calculated for C72H132NO21Na2+: 684.9589, found: 684.9601) (Supplementary Information). The new metabolites were purified from extracts of large-scale cultures of the samR0478 mutant by semipreparative HPLC. 1H and 13C NMR data for the purified metabolites were very similar to those for stambomycins A–D 1–4 (Supplementary Information). Only the signals due to C-28 differed significantly (δH=3.47 p.p.m. and δC=74.2 p.p.m. for stambomycins A/B; δH=1.30, 1.58 p.p.m. and δC=35.8 p.p.m. for the corresponding novel metabolites), leading us to conclude that the novel metabolites are 28-deoxystambomycins A–D 5–8 (Figure 4b and Supplementary Information).
The production of stambomycins A–D 1–4 was also found to be abolished in the samR0479 mutant, which accumulated new metabolites with the molecular formulae C65H120O19 (calculated for [M+H]+: 1205.8497, found: 1205.8488) and C64H118O19 (calculated for [M+H]+: 1191.8340, found: 1191.8332) (Supplementary Information). A neutral loss of 44 Da, corresponding to CO2, was observed in negative ion mode MS/MS analyses of the purified metabolites (Supplementary Information). This indicated that they are the shunt metabolites 9–12, resulting from hydrolytic release and C-28 hydroxylation of the fully assembled polyketide chains from the stambomycin PKS. 1H, 13C, COSY, TOCSY, heteronuclear single quantum correlation and heteronuclear multiple-bond correlation NMR spectroscopic analyses confirmed this hypothesis and showed that these metabolites possess the C-28 hydroxyl group, but lack the C-50 hydroxyl group required for macrolide formation (Figure 4c and Supplementary Information). Coupling constants of 15.0 and 15.2 Hz for H-12/H-13 and H-48/H-49, respectively, confirmed the E configurations of the C-12/C-13 and C-48/C-49 double bonds in 9 and 10. Similarly, correlations between the C-10 methyl group and H-12, and the C-24 methyl group and H-26 in the NOESY spectrum of 9 and 10, were consistent with E configurations for the C-10/C-11 and C-24/C-25 double bonds (Supplementary Information).
Discussion
The incorporation of two atoms of oxygen from 18O2 into stambomycins A–D 1–4 is consistent with our hypothesis that the C-28 and C-50 oxygen atoms of these metabolites derive from hydroxylation of the polyketide chain. The accumulation of 28-deoxystambomycins A–D 5–8 in the samR0478 mutant of S. ambofaciens implies that the putative CYP encoded by samR0478 is responsible for introducing the C-28 hydroxyl group into the stambomycins. Whether this hydroxylation reaction occurs during or after polyketide chain assembly is unclear. The observation that the shunt metabolites accumulated in the samR0479 mutant have a hydroxyl group at C-28 tempts us to speculate that C-28 hydroxylation occurs during polyketide chain assembly (Figure 5). Several examples of analogous CYP-catalyzed in trans oxidations have been reported for intermediates bound to the carrier proteins of nonribosomal peptide synthetases.15
Proposed roles of the putative cytochromes P450 (CYPs) encoded by samR0478 and samR0479 in stambomycin biosynthesis, and mechanisms for the formation of 28-deoxystambomycins A–D 5–8 and shunt metabolites 9–12 (R=CH2CH(CH3)CH2CH3, CH2CH2CH(CH3)CH3, CH2CH(CH3)CH3, or CH2CH2CH2CH3) in the samR0478 and samR0479 mutants, respectively, of Streptomyces ambofaciens. SamR0478 and SamR0479 are hypothesized to catalyze hydroxylation of C-28 and C-50, respectively, of the fully assembled polyketide chains attached to the acyl carrier protein (ACP) domain within the last polyketide synthase (PKS) module. Alternatively, these enzymes may catalyze hydroxylation of earlier intermediates in polyketide chain assembly. In the samR0478 mutant, the ACP-bound intermediates in which X=H and Y=OH are transferred to the active site Ser residue of the thioesterase (TE) domain that catalyzes chain release via macrolactonization utilizing the C-50 hydroxyl group introduced by SamR0479. The resulting 28-deoxystambomycin A–D algycones 13–16 undergo SamR0481-mediated mycaminosylation, yielding 28-deoxystambomycins A–D 5–8. In contrast, the ACP-bound intermediates in which X=OH and Y=H accumulate in the samR0479 mutant. These intermediates are transferred to the active site Ser residue of the TE domain, but cannot be offloaded via macrolactonization because the C-50 hydroxyl group is absent. Instead, the TE domain catalyzes hydrolysis of the enzyme-bound intermediates, yielding shunt metabolites 9–12.
The accumulation of stambomycin shunt metabolites 9–12, lacking the C-50 oxygen atom of the macrolide and the mycaminose residue, in the samR0479 mutant indicates that the putative CYP encoded by samR0479 catalyzes hydroxylation of C-50 during (or possibly even before) polyketide chain assembly (Figure 5). It appears that the TE domain of the PKS is unable to catalyze chain release via macrolactonization if the C-50 hydroxyl group is absent from the polyketide chains, despite the availability of several alternative hydroxyl groups resulting from keto reduction during chain assembly. Thus, the fully assembled polyketide chains appear to be offloaded from the PKS in the samR0479 mutant via hydrolysis (Figure 5). In contrast to the 28-deoxystambomycin A–D aglycones 13–16, 9–12 are evidently not substrates for the mycaminosyl transferase encoded by samR048112 (Figure 5).
In conclusion, we have shown that putative CYPs encoded by the samR0478 and samR0479 genes are involved in the introduction of the C-28 hydroxyl group and the C-50 oxygen atom, respectively, into the stambomycins. Hydroxylation at C-50 is required for offloading of fully assembled polyketide chains from the stambomycin PKS via macrolactonization. Further insight into the roles played by SamR0478 and SamR0479 in stambomycin biosynthesis could be obtained by investigating the catalytic properties of the purified recombinant proteins. However, this may prove challenging, given the structural complexity of likely substrates for these enzymes.
References
Harris, D. R., McGeachin, S. G. & Mills, H. H. The structure and stereochemistry of erythromycin A. Tetrahedron Lett. 679–685 (1965).
Ganis, P., Avitabile, G., Mechlinski, W. & Schaffner, C. P. Polyene macrolide antibiotic amphotericin B. Crystal structure of the N-iodoacetyl derivative. J. Am. Chem. Soc. 93, 4560–4564 (1971).
Chong, C. N. & Rickards, R. W. Macrolide antibiotic studies. XVI. The structure of nystatin. Tetrahedron Lett. 5145–5148 (1970).
Springer, J. P., Arison, B. H., Hirshfield, J. M. & Hoogsteen, K. The absolute stereochemistry and conformation of avermectin B2a aglycone and avermectin Bla. J. Am. Chem. Soc. 103, 4221–4224 (1981).
Kirst, H. A. et al. A83543A-D, unique fermentation derived tetracyclic macrolides. Tetrahedron Lett. 32, 4839–4842 (1991).
Donadio, S. & Katz, L. Organization of the enzymatic domains in the multifunctional polyketide synthase involved in erythromycin formation in Saccharopolyspora erythraea. Gene 111, 51–60 (1992).
Caffrey, P., Lynch, S., Flood, E., Finnan, S. & Oliynyk, M. Amphotericin biosynthesis in Streptomyces nodosus: deductions from analysis of polyketide synthase and late genes. Chem. Biol. 8, 713–723 (2001).
Brautaset, T. et al. Biosynthesis of the polyene antifungal antibiotic nystatin in Streptomyces noursei ATCC 11455: analysis of the gene cluster and deduction of the biosynthetic pathway. Chem. Biol. 7, 395–403 (2000).
Waldron, C. et al. Cloning and analysis of the spinosad biosynthetic gene cluster of Saccharopolyspora spinosa. Chem. Biol. 8, 487–499 (2001).
Ikeda, H., Nonomiya, T., Usami, M., Ohta, T. & Omura, S. Organization of the biosynthetic gene cluster for the polyketide anthelmintic macrolide avermectin in Streptomyces avermitilis. Proc. Natl Acad. Sci. USA 96, 9509–9514 (1999).
Tsai, S.-C. et al. Crystal structure of the macrocycle-forming thioesterase domain of the erythromycin polyketide synthase: versatility from a unique substrate channel. Proc. Natl Acad. Sci. USA 98, 14808–14813 (2001).
Laureti, L. et al. Identification of a bioactive 51-membered macrolide complex by activation of a silent polyketide synthase in Streptomyces ambofaciens. Proc. Natl Acad. Sci. USA 108, 6258–6263 (2011).
Gust, B., Challis, G. L., Fowler, K., Kieser, T. & Chater, K. F. PCR-targeted Streptomyces gene replacement identifies a protein domain needed for biosynthesis of the sesquiterpene soil odor geosmin. Proc. Natl Acad. Sci. USA 100, 1541–1546 (2003).
Kieser, T., Bibb, M. J., Buttner, M. J., Chater, K. F. & Hopwood, D. A. Practical Streptomyces Genetics 407, John Innes Foundation: Norwich, (2000).
Cryle, M. J. Carrier protein substrates in cytochrome P450-catalysed oxidation. Metallomics 3, 323–326 (2011).
Pinnert-Sindico, S. Une nouvelle espèce de Streptomyces productrice d'antibiotiques: Streptomyces ambofaciens n. sp. caractères culturaux. Ann. Inst. Pasteur. (Paris) 87, 702–707 (1954).
Hanahan, D. Studies on transformation of Escherichia coli with plasmids. J. Mol. Biol. 166, 557–580 (1983).
MacNeil, D. J. et al. Analysis of Streptomyces avermitilis genes required for avermectin biosynthesis utilizing a novel integration vector. Gene 111, 61–68 (1992).
Datsenko, K. A. & Wanner, B. L. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl Acad. Sci. USA 97, 6640–6645 (2000).
Choulet, F. et al. Evolution of the terminal regions of the Streptomyces linear chromosome. Mol. Biol. Evol. 23, 2361–2369 (2006).
Acknowledgements
This research was supported by the European Commission under the 6th framework program (integrated project Actinogen; FP6-5224).The Bruker MaXis mass spectrometer used in this research was obtained through Birmingham Science City: Innovative Uses for Advanced Materials in the Modern World (West Midlands Centre for Advanced Materials Project 2), with support from Advantage West Midlands and was partly funded by the European Regional Development Fund.
Author information
Authors and Affiliations
Corresponding author
Additional information
Supplementary Information accompanies the paper on The Journal of Antibiotics website
Supplementary information
Rights and permissions
About this article
Cite this article
Song, L., Laureti, L., Corre, C. et al. Cytochrome P450-mediated hydroxylation is required for polyketide macrolactonization in stambomycin biosynthesis. J Antibiot 67, 71–76 (2014). https://doi.org/10.1038/ja.2013.119
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ja.2013.119
Keywords
This article is cited by
-
Biosynthetic pathway of peucemycin and identification of its derivative from Streptomyces peucetius
Applied Microbiology and Biotechnology (2023)
-
Engineering the stambomycin modular polyketide synthase yields 37-membered mini-stambomycins
Nature Communications (2022)
-
Hydroxylation of diverse flavonoids by CYP450 BM3 variants: biosynthesis of eriodictyol from naringenin in whole cells and its biological activities
Microbial Cell Factories (2016)
-
A crotonyl-CoA reductase-carboxylase independent pathway for assembly of unusual alkylmalonyl-CoA polyketide synthase extender units
Nature Communications (2016)