Abstract
In the course of screening for breast cancer cell migration inhibitors, we isolated two novel compounds, migracins A and B from the culture broth of Streptomyces sp. MI264-NF2. Their structures are related to those of luminacins previously isolated from Streptomyces. Migracins A and B inhibited breast cancer cell migration, monitored by wound healing assay with IC50 values of 1.31 and 1.99 μg ml−1, respectively, in human breast carcinoma MDA-MB-231 cells without showing any cytotoxicity. Migracins also inhibited the migration of human lung adenocarcinoma A549 cells and human fibrosarcoma HT-1080 cells. Therefore, migracins may become new cancer metastasis inhibitors.
Similar content being viewed by others
Introduction
Breast cancer often metastasizes to distal organs such as bone, lung and liver through migration, invasion, and blood and lymphatic vessel transport.1, 2, 3 Furthermore, metastasized breast cancer is often highly resistant to radiation and conventional chemotherapeutic agents.1, 4 Therefore, novel therapeutic agents that suppress tumor metastasis are needed to advance the efficacy of chemotherapy in human breast cancer. These inhibitors should be also useful to find out the molecular mechanism of metastasis. Mechanisms of metastasis include detachment, migration, invasion, blood and lymphatic vessel transport, and growth in the secondary sites. Among them, steps of migration and invasion are involved in all cases of cancer metastasis.5 Especially, cancer cell migration is easily measured by wound healing assay. Hence, we conducted screening of breast cancer cell migration inhibitors from microbial secondary metabolites. NF-κB promotes migration and invasion of cancer cells and suppression of NF-κB inhibits cancer cell migration.6, 7, 8, 9, 10 As (-)-dehydroxymethylepoxyquinomicin ((-)-DHMEQ), a specific NF-κB inhibitor, covalently binds to a specific cysteine of NF-κB component proteins,11, 12 it is an effective inhibitor of cancer cell migration.13, 14 Therefore, we employed (-)-DHMEQ as a positive control of breast cancer cell migration inhibitor and tried to obtain more effective inhibitors of breast cancer cell migration than (-)-DHMEQ. As a result, we isolated two novel compounds, migracins A and B, from the culture broth of Streptomyces sp. MI264-NF2. We report on the fermentation, isolation, structure elucidation and biological activities of these compounds.
Results
Screening
We have evaluated 632 culture broth samples derived from microbes. Cancer cell migration was monitored by wound healing assay with breast carcinoma MDA-MB-231 cells. We found the inhibitory activity on cancer cell migration in the culture broth derived from Streptomyces sp. MI264-NF2. This strain was isolated from a soil sample collected at Shinagawa-ku, Tokyo, Japan. Then, we tried to isolate active principles from the culture filtrate of this strain. After fermentation of the hit strain, ethanol extract from the culture media was concentrated and then re-extracted with ethyl acetate. The crude solid obtained from the ethyl acetate extract was purified by sephadex LH-20 column chromatography. Then, semipreparative HPLC gave two pure compounds that were later named as migracins A and B.
Structure elucidation
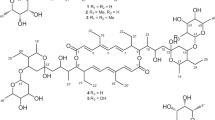
The physicochemical properties of migracins are summarized in Table 1. The IR absorptions at 3402-3389, 1634-1628 and 1094-1092 cm−1 are due to hydroxyl, carbonyl and ether moieties, respectively. Analysis of UV spectrum revealed that the absorptions of migracins are similar to those of luminacins.15, 16 The molecular formulae of migracins were both determined as C26H38O9, based on high resolution ESI-MS, indicating 8 degrees of unsaturation corresponding to the number of protons and carbons observed by NMR. The 1D and 2D NMR analyses of migracins (Tables 2 and 3) revealed the presence of seven quaternary carbons: one ketone carbonyl, five aromatic and one aliphatic (C-O) carbons; and the following proton-bearing carbons: one formyl, one aromatic CH, five CH-O (at C-1′, C-3′, C-5′, C-7′ and C-8′), two CH-C (at C-2′ and C-2′), four CH2-C (at C-4′, C-9′, C-11′ and C-12′), one ethoxyl, and four methyls. From these results, we propose that migracin have the structure as shown Figure 1. The COSY and HMBC analyses (Figures 2a and b) indicated that migracins A and B have the same planar structure with each other. Moreover, 1H and 13C NMR data of migracins A and B (Tables 2 and 3) are coincident with those of luminacins C1 and C2, respectively. The difference between migracins and luminacin Cs is that the 1′-methoxyl groups on the 5-isobutyl side chains of luminacins were replaced by the 1′-ethoxyl groups on those of migracins. The proposed structures of migracins A an B (Figure 1) will be also described in Discussion.
Inhibition of cancer cell migration by migracins A and B
Human breast carcinoma MDA-MB-231 cells and human fibrosarcoma HT-1080 cells showed high migration potency without stimulation. Treatment of the cells with migracins A or B inhibited the cell migration in a dose-dependent manner at the concentration that was not cytotoxic, as shown in Figures 3a and b. In one hand, migration of human lung adenocarcinoma A549 cells were highly stimulated when treated with tumor necrosis factor-α (TNF-α) and transforming growth factor-β 1 (TGF-β). Then, treatment of the stimulated cells with migracin A or B inhibited the cell migration in a dose-dependent manner and more effectively than the positive control, SB431542 that inhibited the activity of TGF-β1 superfamily activin receptor-like kinase receptors as shown in Figure 3c. The IC50 values of migracins A and B are 1.31 and 1.99 μg ml−1 in MDA-MB-231 cells, 0.135 μg ml−1 and <0.1 μg ml−1 in HT-1080 cells and 0.236 and 0.249 μg ml−1 in A549 cells, without showing any cytotoxicity.
Inhibition of cell migration by migracins A and B in indicated cell lines. (a) An artificial wound was made by a pipette tip. The MDA-MB-231 cells were treated with indicated concentrations of migracin A or B. After 24 h, photographs were taken in phase-contrast microscopy to quantitate the distance of wound (column). For the toxicity evaluation, the effect of migracin A or B on the cell number was also examined (line). (b) The HT-1080 cells were treated with indicated concentrations of migracin A or B. After 24 h, the cell migration and cell number were analyzed as in (a). (c) The TNF-α and TGF-β stimulated-A549 cells (opened columns) or unstimulated-A549 cells (closed columns) were treated with migracin A or B for 24 h. Then, the cell migration and cell number were analyzed as in (a).
Cytotoxicity of migracins A and B
The cytotoxicity values of migracins A and B were evaluated against the MDA-MB-231, A549 and HT-1080 cell lines using MTT method. The concentrations to reduce viability by 50% (ED50) of migracin A on these three cancer cell lines are 16.7 μg ml−1 in MDA-MB-231 cells, 17.8 μg ml−1 in A549 cells and 5.0 μg ml−1 in HT-1080 cells. The corresponding values of of migracin B on these three cancer cell lines are 26.4 μg ml−1 in MDA-MB-231 cells, 26.7 μg ml−1 in A549 cells and 15.0 μg ml−1 in HT-1080 cells.
Effects of migracins A and B on NF-κB activity in MDA-MB-231 cells
(-)-DHMEQ inhibited the cell migration in MDA-MB-231 cells via NF-κB inhibition. Therefore, we evaluated the effect of migracins A and B on NF-κB activity in MDA-MB-231 cells. As shown in Figure 4, migracins A and B did not inhibit NF-κB activity.
Discussion
In the course of screening of breast cancer cell migration inhibitors, we found strong inhibitory activity in the culture broth of Streptomyces sp. MI264-NF2 and isolated two novel compounds, migracins A and B. The structures of migracins are related to those of luminacins C1 and C2. Moreover, migracin A has the same planar structure as migracin B in the same way that luminacin C1 has the same planar structure as luminacin C2. In comparison of the NMR data and optical rotation values of migracins A and B with those of luminacins C1 and C2, it is suggested that they have the same stereochemistry at C-1, respectively. Tatsuta et al.17 confirmed absolute structures of luminacins C1 ([α]D26 -98o (c 0.28, CHCl3) and C2 ([α]D26 +54o (c 0.13, CHCl3) by the total synthesis to be (1′S,2′S)- and (1′′R,2′S)-configurations, respectively. Based on these data, the stereostructures of migracins A (1′′S) and B (1′R) are proposed as shown in Figure 1.
It is possible both migracins and luminacins may be artifact compounds, and one of the possible natural precursor would be 1′′,2′′-olefinic or 1′′-carbonyl compound, although both groups did not isolate this compound. However, it is still difficult to exclude the possibility that migracins and luminacins are natural compounds. We carried out additional experiments in which each migracin was dissolved in either MeOH or EtOH, then its MS and biological activity was measured. As a result, there was no change after the EtOH treatment. Therefore, it is likely that the compounds are stable in alcohol.
In biological experiments, we have carefully selected the concentration range to inhibit migration without prominent growth inhibition. Growth inhibition is modest at 1–3 μg per ml of migracins, whereas clear inhibition of migration is observed at these concentrations. Migracin inhibited the migration of human breast carcinoma MDA-MB-231 and fibrosarcoma HT1080 cells, in which those cells were not activated by ligands. Human lung adenocarcinoma A549 cells were found to migrate sufficiently only when they were treated with TNF-α and TGF-β. Migracins inhibited the migration of human lung adenocarcinoma A549 cells induced by TNF-α and TGF-β (Figure 3c). In this assay system, (-)-DHMEQ did not inhibit the migration. Therefore, migracins should have different mechanism of inhibition. (-)-DHMEQ is an NF-κB inhibitor, but migracins did not inhibit NF-κB (Figure 4). Possibly, (-)-DHMEQ did not inhibit the TGF-β-induced activity in which NF-κB is not essentially included.
Wakabayashi et al.16 reported that luminacins C1 and C2 have antiangiogenesis activity inhibiting the tube formation of HUVEC. Cell movement is essential for the VEGF-induced tube formation in cultured HUVEC. As luminacins C1 and C2 have the similar structure as migracins, they are likely to inhibit cell migration. In one hand, based on our observation, it is very likely that migracins would also inhibit tube formation inhibiting the movement of HUVEC. This activity will also assist the antimetastatic activity.
Recently, there are many reports that small G-proteins may be involved in cancer cell migration. Small G protein is classified into six families; Ras, Rho, Rab, Arf, Ran and RGK. Ras, Rho and Ran families promote cell migration, and it has been reported that inhibition of Rac, one of the Rho family proteins, leads to the suppression of cell migration in MDA-MB-435 cells.18 It is also possible that small G protein is involved in the mechanism of inhibition by migracin. Thus, we found two new compounds named migracins A and B that may have antimetastatic activity from the culture of Streptomyces sp.
Materials and methods
General experimental procedures
The UV spectra were recorded on a Hitachi U-3310 spectrophotometer (Hitachi Ltd., Tokyo, Japan). NMR spectra were recorded on a JEOL JNM-LA500 (JEOL, Tokyo, Japan) at 500 MHz. The 1H and 13C chemical shifts were referenced to the solvent signal (δH 7.26 and δC 77.16 in CDCl3). ESI-MS was measured on a LCT Premier XE equipped with ACQUITY UPLC (Waters Corporation, Milford, MA, USA). Optical rotations were measured on a JASCO P-2200 polarimeter (JASCO, Tokyo, Japan). IR spectra were recorded on a JASCO FTIR-4200 (JASCO).
Taxonomy of the producing strain
The strain MI264-NF2 was isolated from a soil sample collected at Shinagawa-ku, Tokyo, Japan. Strain MI264-NF2 formed well-branched substrate mycelia. This strain formed aerial mycelia that bore spirals of 2–7 turns. Mature spore chains consisted of 15–40 or more spores. The spore was oval with smooth surface and 0.5 × 0.9–0.8 × 1.2 μm in size. The substrate mycelia were pale yellow to yellowish brown. The aerial mycelia were light brownish gray to brownish gray. These characteristics were observed on yeast extract-malt extract agar (ISP medium No.2), inorganic salts-starch agar (ISP medium No.4) and glycerol-asparagine agar (ISP medium No.5). The type of diaminopimelic acid isomers in whole-cell hydrolysates of the strain MI264-NF2 was determined to be the LL-form by the method of Staneck and Roberts.19 The partial 16 S ribosomal RNA gene sequence (1482 bp) was determined. The GenBank/EMBL/DDBJ accession number for the 16 S rRNA gene sequence of MI264-NF2 is AB695175. The strain showed high similarity with those of the genus Streptomyces such as Streptomyces sviceus (NBRC 13980T, 1455/1482 bp, T: Type strain, 98.1%) and S. canus (NRRL B-1989T, 1453/1483 bp, 97.9%). These phenotypic and genotypic data suggested that the strain MI264-NF2 belongs to genus Streptomyces. Therefore, the strain was tentatively designated as Streptomyces sp. MI264-NF2. Detailed taxonomic studies of the strain MI264-NF2 are under investigation.
Fermentation, extraction and isolation
The producing organism Streptomyces sp. MI264-NF2 was grown on the seed medium, which consisted of 2.0 galactose, 2.0 dextrin, 1.0 soypeptone, 0.5% corn steep liquor, 0.2% (NH4)2SO4 and 0.2% CaCO3 for 2 days to obtain the seed culture. The barley medium was prepared by autoclaving the pressed barley after adding water in the ratio of 25 ml per 15 g of barley. The seed culture (7 ml) was inoculated on the barley medium, then incubated at 30 °C for 14 days under stationary conditions. After 14 days of incubation, EtOH (40 ml) was added to the medium and extraction was done overnight at room temperature. The extract (1400 ml in total) was evaporated under reduced pressure to about a quarter of the original volume, and then extracted twice with 350 ml EtOAc (pH 7). The EtOAc extract was concentrated under reduced pressure to give a crude extract (1.3 g). The crude extract was chromatographed on a silica gel column (Silica gel 60, 125 g, 0.063–0.200 mm, Merck KGeA, Darmstadt, Germany, column size: 37 × 250 mm) into 88 fractions (1–88, each 20 ml), using a stepwise elution of CHCl3/MeOH (100:0 in fractions 1–40, 100:1 in fractions 41–88). The eluate (168.0 mg) in fractions 60–80 was subjected on a column (27 × 200 mm) of sephadex LH-20 (GE Healthcare, Uppsala, Sweden) ,eluted with MeOH (100%) and cut into 55 fractions (1–55, each 4 ml). The eluate (123.0 mg) in fractions 38–45 was further purified by semipreparative HPLC (a Pegasil ODS C18 column, 5 μm, Senshu Scientific Co. Ltd., Tokyo, Japan, column size: 20 × 250 mm, CH3CN-H2O=80:20, 8 ml min−1) to obtain pure migracin A (2.0 mg, Rt 20.4 min) and migracin B (2.0 mg, Rt 21.0 min).
Materials for biological assays
Dulbecco's modified Eagle's medium was purchased from Nissui (Tokyo, Japan). SB431542 and MTT were purchased from Sigma (St Louis, MO, USA). Recombinant human TNF-α and recombinant TGF-β1 were purchased from Peprotech (London, UK) and Wako (Osaka, Japan), respectively. (-)-DHMEQ was synthesized in our laboratory as described before.12
Cell culture
Human breast carcinoma MDA-MB-231 cells, human lung adenocarcinoma A549 cells and human fibrosarcoma HT-1080 cells were cultured in Dulbecco's modified Eagle's medium containing 10% fetal bovine serum, 100 units ml−1 penicillin G, 200 mg l−1 kanamycin, 600 mg l−1 L-glutamine and 2.25 g l−1 NaHCO3, under a humidified atmosphere with 5% CO2 and 95% air at 37°C.
Wound healing assay
Cells in 48-well plates coated with collagen I (BD, Bedford, MA, USA) were allowed to reach confluence before the surface was uniformly scratched across the center of each well with a pipette tip. The wells were then rinsed twice with serum-free media to remove floating cells and growth media, after which the cells were replenished with serum-free media and cultured for 24 h. The initial wound area and movement of the cells into the scratched area were recorded. Experiments were performed in triplicate in three independent experiments.
Cytotoxicity assays
Cytotoxicity was evaluated by the 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl-2H-tetrazolium bromide (MTT) assay using MDA-MB-231, A549 and HT-1080 cell lines. Cell suspensions (200 μl) at a density of 1 × 104 cells per ml were plated in 96-well microtiter plates and incubated for 24 h. Then, the test compound solutions (2 μl in MeOH) at different concentrations were added to each well and further incubated for 48 h under the same conditions. MTT solution (20 μl of 5 mg ml−1 in Ca2+, Mg2+-free phosphate buffered saline) was added to each well and incubated for 2 h. Then, an old medium (150 μl) containing MTT was gently replaced by DMSO and pipetted to dissolve any formazan crystals formed. Absorbance was then determined on a Microplate Reader (Bio-Rad Laboratories, Inc., CA, USA) at 570 nm.
Electrophoresis mobility shift assay (EMSA)
MDA-MB-231 cells (5 × 105 cells) were seeded in 8 ml of complete medium into 100-mm dishes. On the next day, the volume of the culture medium was adjusted to 5 ml. The cells were treated with 3 μg ml−1 migracins A and B for indicated time and 10 μg ml−1 (-)-DHMEQ for 1 h. Nuclear extract preparation and EMSA were carried out as described previously.20, 21
References
Ya-Ling, H. et al. 4-Shogaol, an active constituent of dietary ginger, inhibits metastasis of MDA-MB-231 human breast adenocarcinoma cells by decreasing the repression of NF-κB/snail on RKIP. J. Agric. Food Chem. 60, 852–861 (2012).
Jemal, A. et al. Cancer statistics. CA Cancer J. Clin. 59, 225–249 (2009).
Sathya, S., Sudhagar, S., Vidhya Priya, M., Bharathi Raja, R. et al. 3β-Hydroxylup-20(29)-ene-27,28-dioic acid dimethyl ester, a novel natural product from Plumbago zeylanica inhibits the proliferation and migration of MDA-MB-231 cells. Chem. Biol. Interact 88, 412–420 (2010).
Feuer, E. J. et al. The lifetime risk of developing breast cancer. J. Natl. Cancer Inst. 85, 892–897 (1993).
Pan, X. et al. Nitidine chloride inhibits breast cancer cells migration and invasion by suppressing c-Src/FAK associated signaling pathway. Cancer Lett. 313, 181–191 (2011).
Zhou, Y., Eppenberger-Castori, S., Eppenberger, U. & Benz, C. C. The NFκB pathway and endocrine-resistant breast cancer. Endocr. Relat. Cancer. 12 ((Suppl.1),), S37–S46 (2005).
Mineva, N. D. et al. Inhibition of RelB by 1,25-dihydroxyvitamin D3 promotes sensitivity of breast cancer cells to radiation. J. Cell Physiol. 220, 593–599 (2009).
Fritz, R. D. & Radziwill, G. CNK1 promotes invasion of cancer cells through NF-κB-dependent signaling. Mol. Cancer Res. 8, 395–406 (2010).
Yodkeeree, S., Ampasavate, C., Sung, B., Aggarwal, B. B. & Limtrakul, P. Demethoxycurcumin suppresses migration and invasion of MDA-MB-231 human breast cancer cell line. Eur. J. Pharmacol. 627, 8–15 (2010).
Helbig, G. et al. NF-kappaB promotes breast cancer cell migration and metastasis by inducing the expression of the chemokine receptor CXCR4. J. Biol. Chem. 278, 21631–21638 (2003).
Yamamoto, M., Horie, R., Takeiri, M., Kozawa, I. & Umezawa, K. Inactivation of NF-kappaB components by covalent binding of (-)-dehydroxymethylepoxyquinomicin to specific cysteine residues. J. Med. Chem. 51, 5780–5788 (2008).
Suzuki, N., Sugiyama, C., Ohno, O. & Umezawa, K. Preparation and biological activities of optically active dehydroxymethylepoxyquinomicin, a novel NF-κB inhibitor. Tetrahedron 60, 7061–7066 (2004).
Wong, J. H., Lui, V. W., Umezawa, K., Ho, Y., Wong, E. Y. et al. A small molecule inhibitor of NF-kappaB, dehydroxymethylepoxyquinomicin (DHMEQ), suppresses growth and invasion of nasopharyngeal carcinoma (NPC) cells. Cancer Lett. 287, 23–32 (2010).
Matsumoto, G. et al. Targeting of NF-κB pathways by dehydroxymethylepoxyquinomicin, a novel inhibitor of breast carcinomas: anti-tumor and anti-angiogenic activity in vivo. Clin. Cancer Res. 11, 1287–1293 (2005).
Naruse, N., Kageyama-Kawase, R., Funahashi, Y. & Wakabayashi, T. Luminacins: a family of capillary tube formation inhibitors from Streptomyces sp. I. Taxomony, fermentation, isolation, physico-chemical properties and structure elucidation. J. Antibiot. 53, 579–590 (2000).
Wakabayashi, T., Kageyama-Kawase, R., Naruse, N., Funahashi, Y. & Yoshimatsu, K. Luminacins: a family of capillary tube formation inhibitors from Streptomyces sp. II. Biological activities. J. Antibiot. 53, 591–596 (2000).
Tatsuta, K., Nakano, S., Narazaki, F. & Nakamura, Y. The first total synthesis and establishment of absolute structure of luminacins C1 and C2 . Tetrahedron Lett. 42, 7625–7628 (2001).
Montalvo-Ortiz, B. L. et al. Characterization of EHop-016, novel small molecule inhibitor of Rac GTPase. J. Biol. Chem. 287, 13228–13238 (2012).
Staneck, J. L. & Roberts, G. D. Simplified approach to identification of aerobic actinomycetes by thin-layer chromatography. Appl. Microbiol. 28, 226–231 (1974).
Dignam, J. D., Lebovitz, R. M. & Roeder, R. G. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 11, 1475–1489 (1983).
Andrews, N. C. & Faller, D. V. A rapid micropreparation technique for extraction of DNA-binding proteins from limiting numbers of mammalian cells. Nucleic Acids Res. 19, 2499 (1991).
Acknowledgements
We wish to thank Dr Shinichi Kondo, Bioscience Associates, Tokyo, for valuable suggestions. The work described in this report was supported in part by grants from the programs Grants-in-Aid for Scientific Research (B) and the Keio Gijuku Fukuzawa Memorial Fund for the Advancement of Education and Research. This work was also supported in part by MEXT-Supported Program for the Strategic Research Foundation at Private Universities, Aichi Medical University 2011-2015 (S1101027).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interst.
Rights and permissions
About this article
Cite this article
Arai, Y., Iinuma, H., Ikeda, Y. et al. Migracins A and B, new inhibitors of cancer cell migration, produced by Streptomyces sp.. J Antibiot 66, 225–230 (2013). https://doi.org/10.1038/ja.2012.112
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ja.2012.112
Keywords
This article is cited by
-
Induction of apoptosis and cell cycle arrest in colorectal cancer cells by novel anticancer metabolites of Streptomyces sp. 801
Cancer Cell International (2022)
-
Isolation of ketomycin from Actinomycetes as an inhibitor of 2D and 3D cancer cell invasion
The Journal of Antibiotics (2019)
-
Novel p-terphenyl glycoside with a rare 2,6-dideoxyhexopyranose moiety from Actinomycete strain SF2911 that inhibits cancer cell migration
The Journal of Antibiotics (2017)