Abstract
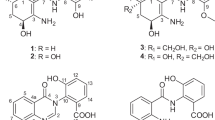
Two novel furaquinocin (FQ) analogues, I (1) and J (2), were isolated from Streptomyces reveromyceticus SN-593 strain NRM2. Their structures were elucidated by MS and NMR analyses. Similar to the previously described FQ D (3), both 1 and 2 possessed a dihydrofuran ring fused to a polyketide naphthoquinone skeleton. The main difference between 1, 2 and 3 was the type of residue attached to C-13; these were a carboxyl, a carboxamide and a methyl residue, respectively.
Similar content being viewed by others
Introduction
The majority of bioactive compounds used as drugs and bioprobes originally derive from the secondary metabolites of microorganisms. Genome sequence analyses have shown that the number of bioactive compounds isolated from Streptomyces spp. is much lower than the number of gene clusters harbored by each species.1, 2, 3 This indicates that the majority of gene clusters are either not expressed, or are only minimally expressed, under standard laboratory conditions. To discover novel natural products that are normally expressed at a very low level, we have been performing a systematic isolation of secondary metabolites. As a result, we isolated verticilactam4 from Streptomyces spiroverticillatus JC-8444, a tautomycin producer,5 and 6–dimethylallylindole-3-carbaldehyde6 from S. reveromyceticus SN-593, a reveromycin producer.7 In addition, from S. reveromyceticus SN-593, we detected metabolites showing UV spectra similar to furaquinocin (FQ), but different mass from the reported FQs. Consistent with the finding, we could annotate an FQ biosynthetic gene cluster from the genome draft sequence. However, with the quantity produced by wild type strain, it was not possible to isolate and determine the structure. So, in order to improve the production of novel FQ derivatives, we performed the expression of pathway-specific regulatory genes associated with the FQ biosynthetic gene cluster. Activation of pathway-specific positive regulatory gene is commonly used for augmentation of secondary metabolites in Streptomyces.8, 9, 10
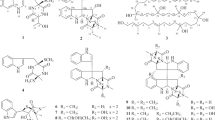
FQs, originally isolated from the culture broth of Streptomyces sp. KO-3988,11 have cytocidal activities without antibacterial activity. So far, eight analogues of these polyketide isoprenoid hybrid compounds (Figure 1)11, 12, 13 and their gene cluster14 have been identified. However, no new analogues have been described since the discovery of FQ two decades ago. Here, we report the identification of the novel FQ analogues FQ I (1) and FQ J (2) from a genetically-engineered strain of S. reveromyceticus SN-593.
Results and discussion
Enhanced FQ production on strain NRM2
The production of reveromycins was completely abolished in the strain NRM1. In addition, it accumulated a derivative of benzoxazole-4-carboxylic acid, possibly due to the changes in the metabolism of primary metabolites. On the basis of the structure, it is speculated that the benzoxazole-4-carboxylic acid derivative is not involved in FQ biosynthesis. The structure elucidation and biosynthesis of this compound will be reported elsewhere. In wild type strain, the benzoxazole-4-carboxylic acid derivative could also be detected in the early stationary phase of culture (day 3–4), whereas it diminished on the fifth day. The amount of the benzoxazole-4-carboxylic acid derivative was reduced in the NRM2 strain, and its peak showed similar UV spectrum to that of FQs on the HPLC chromatogram (Figure 2). Products corresponding to the extract's three major peaks were isolated so that their structures could be elucidated. These were identified as 1, 2 and 3, respectively (Figure 1), where 3 was the major metabolite. The production of 3 as a major metabolite is also reported by heterologous expression of fur1 to fur21 on S. lividans14 and S. albus.15 Given the findings of heterologous studies and our observations that FQ production was enhanced simply by the introduction of fur22 to the strain NRM1, Fur22 can be considered a positive regulator of FQ biosynthesis.
Metabolite profiling of the ethyl acetate extract from wild type strain S. reveromyceticus SN-593, reveromycin polyketide synthase disruption mutant NRM1, and FQ-producing strain NRM2. The pictured chromatogram was produced on a PDA detector (Waters, Milford, MA, USA) at a wavelength of 254 nm. The marked peaks indicate 1, 2 and 3. (x) indicates benzoxazole-4-carboxylic acid derivative that accumulated in the NRM1 strain.
The total amount of FQs produced by strain NRM2 was calculated on the basis of a calibration curve in which the quantity of 3 was plotted against the peak area obtained on the HPLC chromatogram. These values were highly correlated, (R2=0.999), and the curve indicated that 705 mg of FQs were produced per liter of culture broth. Although FQs were major metabolites compared with the benzoxazole-4-carboxylic acid derivative in the strain NRM2, peak area determination for quantification of FQs was carried out at a wavelength of 261 nm, where the benzoxazole-4-carboxylic acid derivative had lowest UV absorption.
Physico-chemical properties and structure elucidation of 1 and 2
HR–MS (m/z: 415.13634 [M-H]− calcd. for C22H23O8 415.13929) indicated that the molecular formula of compound 1 was C22H24O8. The UV absorption spectrum showed λmax (MeOH) values at 221, 268, 294 and 407 nm (Table 1). These values were identical with those of 3 and FQ A,12 suggesting that 1 was an FQ derivative possessing a naphthoquinone skeleton. Similarly, results of the 13C NMR spectrum were identical for 1, 3 and FQ A, with signals at 9.3, 61.1, 109.7, 110.4, 126.0, 133.9, 135.1, 158.4, 163.3, 181.9 and 185.3 ppm (Table 2). The naphthoquinone skeleton was confirmed by HMBCs of H-5 to C-3a, C-4, C-5a, C-6 and of C-9a, and C-8-Me to C-7, C-8 and C-9 (Figure 3a). A methoxy group was assigned at C-7 by an HMBC of C-7-Me to C-7. A dihydrofuran ring and its fusion to the naphthoquinone were confirmed by HMBCs of H-2 to C-3a and C-9b, C-2-Me to C-3, and C-3-Me to C-3 and C-3a. The structure of the side chain at C-3 was determined by DQF-COSY, which revealed connectivity between C-10 and C-12, and by an HMBC spectrum showing the following correlations: H-12 to C-13 and C-14, and H-15 to C-12, C-13 and C-14. This confirmed the attachment of a carboxyl group to C-13. The side chain was assigned at C-3 by HMBCs of H-10 and H-11 to C-3. The 13C NMR chemical shift of C-15 (12.8 ppm) suggested that the double bond at C-12 was in the E-configuration; this was confirmed by 1D NOE difference spectra, which showed an NOE at H-11 by an irradiation to H-15 (Figure 3b). Results from the 1D NOE difference spectra also indicated that both the C-2-Me and C-3-Me had a trans-configuration, as an NOE at H-2 was observed by an irradiation to C-3-Me. Given the structure of compound 1, we determined it was a novel FQ, which we named FQ I. A comparison of optical rotation values between 1, 3 and FQ A (1: −51.2° (c 0.15, CHCl3), 3: −85.6° (c 0.147, CHCl3) and FQ A13: −46° (c 0.58, CHCl3)) indicated that the three compounds possessed identical absolute stereochemistry, including at C-10. The complete NMR spectra of 1 are presented in Supplementary Figures 1–8.
Compound 2 was obtained as a yellow powder. Its UV absorption spectrum, with λmax (MeOH) values of 221, 268, 292 and 407 nm, was identical to that of 1, suggesting that 2 also had a naphthoquinone skeleton and was an FQ derivative. An ESI–MS showed a quasi-molecular ion peak at m/z 414 [M-H]−, indicating that 2 contained a nitrogen atom. HR–MS (m/z: 414.15819 [M-H]− calcd. for C22H24NO7 414.15528) determined that the molecular formula of the compound was C22H25NO7. 1H and 13C NMR spectra were identical with those of 1 except for the signals associated with the side chain. These suggested that the hydroxyl group present in 1 was replaced with an amino group in 2. The FQ skeleton was determined using 2D NMR correlations (DQF-COSY and an HMBC spectrum) and a comparison of NMR chemical shifts between 1 and 2. The structure of the side chain was determined by DQF-COSY, which revealed the connectivity between C-10 and C-12 and HMBC, which showed the following correlations: H-10 to C-3, C-3a and C12; H-11 to C-12 and C-13; and H-15 to C-12, C-13 and C-14 (Figure 4a). The low C-14 field shift and the similarity in C-10 chemical shifts between the 13C NMR spectra for 1 and 2, both indicated that the amino group was attached to C-14. The attachment of the side chain to C-3 was confirmed by HMBC correlation from H-10 to C-3. The double bond at C-12 was determined to be in the E-configuration, as indicated by the 13C NMR chemical shift at C-15 and the NOE correlation between H-11 and H-15 in the NOESY spectrum (Figure 4b). The NOESY spectrum also showed an NOE between H-2 and C-3-Me, suggesting they were in the trans-configuration. According to the optical rotation value (−39.9° (c 0.48, CHCl3)), this compound had the same absolute stereochemistry as 1. Thus, we determined that compound 2 was a novel FQ, which we have named FQ J. The complete NMR spectra of 2 are presented in Supplementary Figures 9–15.
Biosynthesis of 1 and 2
FQs are polyketide isorpenoid hybrid compounds biosynthesized by prenylation of a 1, 3, 6, 8—tetrahydronaphthalene derivative (polyketide) with a geranyl diphosphate (isoprenoid), followed by a series of reactions.14, 15 By taking advantage of known FQ gene cluster from original producer strain S. sp. KO-3988,14 we could find a highly homologous gene cluster from S. reveromyceticus SN-593. On the basis of the structures of 1, 2 and 3, and gene annotations (Supplementary Table 1), it is expected that three rounds of oxidation at C-14 of 3 will lead to the formation of carboxyl group (1) by a P450 (Fur8), whereas carboxamide formation from 1 by an aminotransferase (Fur3) will lead to 2. Although the FQ gene cluster includes candidate genes of P450 and aminotransferase, it is too early to conclude that these genes are responsible for oxidation and transfer of amino group in FQs, without genetic and biochemical characterization.
Biological activity of 1 and 2
In order to investigate potential cytocidal activities of 1 and 2, the compounds were added to the culture medium of HeLa cells. No significant cytocidal activity was detected at concentrations of 0.1–30 μg ml−1. Among FQs, FQ H displays the strongest cytocidal activity, with an IC50 value of 0.08 and 0.22 μg ml−1 against B16 melanoma cells and HeLa S3 cells, respectively.12 The difference in activity level between FQ H and the novel compounds reported here may have resulted from the structural modification at C-14. Future research should investigate whether 1 and 2 display bioactivity in other assay systems. To facilitate these and other related studies, 1 and 2 will be deposited to RIKEN NPDepo (http://www.npd.riken.jp), where they will be made available for use by other researchers.
In summary, we not only enhanced the production of FQs from S. reveromyceticus SN-593, but also isolated two novel FQ analogues. Our results suggest that a systematic isolation and the engineering of pathway specific regulatory genes might be a promising approach to identify metabolites hidden inside Streptomyces.
Methods
General
All chemicals and reagents were of analytical grade and were purchased from Wako (Osaka, Japan), Nacalai Tesque (Kyoto, Japan) or Sigma (St Louis, MO, USA). UV spectra were measured with a JASCO 660 UV-vis spectrometer (JASCO International, Tokyo, Japan), whereas optical rotations were measured with a HORIBA SEPA-300 high sensitive polarimeter (Horiba, Kyoto, Japan). HR–MS were measured with a JEOL AccuTOF JMS-T700 LCK mass spectrometer (JEOL, Tokyo, Japan). 1H, 13C and 2D NMR spectra of 1 and 2 were measured in CD3OD, whereas FQ D (3) was measured in CDCl3 on a JEOL ECP 500 FT-NMR spectrometer (JEOL) (500 MHz for 1H and 125 MHz for 13C and 2D). Infrared spectra were recorded on a Fourier transform infrared spectrometer (FT-720, Horiba, Kyoto, Japan).
Bacterial strains and culture conditions
Escherichia coli (E. coli) DH5α was utilized for general gene cloning and E. coli GM2929 hsdS::Tn10 (pUB307::Tn7)16 was used for conjugal gene transfer. Each strain was cultured in Luria-Bertani broth (Nacalai Tesque) supplemented with appropriate antibiotics (ampicillin 100 μg ml−1, kanamycin 50 μg ml−1, streptomycin 50 μg ml−1, spectinomycin 100 μg ml−1, chloramphenicol 30 μg ml−1) as required.
Spores of different S. reveromyceticus SN-593 strains were prepared on an mannitol soya flour plate (2% soya flour, 2% D (−)-mannitol and 2% agar) containing 10 mM MgCl2 and were inoculated on soluble starch yeast extract medium (70 ml: 0.1% yeast extract, 0.1% NZ-amine and 1% soluble starch) contained in a 500-ml cylindrical flask rotating at 150 rpm for 2 days at 28 °C. After seed cultures (1 ml) were inoculated into a 500-ml cylindrical flask containing 70 ml of production medium,6 they were cultured for an additional 5 days under the same conditions.
DNA manipulation and construction of the FQ-producing strain
DNA manipulation including genomic DNA isolation was done as described in standard protocols.17, 18 An 822-bp DNA fragment of fur22, a gene belonging to Streptomyces antibiotic regulatory protein family, was amplified from genomic DNA using sets of primers Bam-fur22-F: CGCGGATCCATGAAGATTCAGCTGCTTGGC and Hind-fur22-R: CCCAAGCTTTCACGCGGCGTAGAGGTCGAA (restriction site underlined), using a DNA Engine thermal cycler (Bio-Rad, Hercules, CA, USA). The DNA fragment was isolated and ligated to BamHI and HindIII sites of pTYM1919 containing an aphII promoter. The resulting plasmid (pTYM-Fur22) was transformed to E. coli GM2929 hsdS::Tn10 (pUB307::Tn7) and then to the NRM1 strain20 by conjugal gene transfer to construct FQ-producing strain (NRM2) (Scheme 1). Selection of exconjugants was performed on an mannitol soya flour agar plate containing 25 μg ml−1 thiostreptone and 5 μg ml−1 carumonam, as described.6
Cell culture and cell proliferation assay
HeLa S3 cells were cultured in Dulbecco's modified Eagle medium, supplemented with 10% fetal bovine serum and antibiotics. For the cell viability assay, 4 × 103 cells per well were seeded in a 96-well culture plate. FQs were added to the culture medium at various concentrations from 0.1–30 μg ml−1. After 48 h of incubation, we counted the cells by measuring the absorbance at 450 nm, using a Cell Count Reagent SF kit (Nacalai Tesque).21
Extraction and analysis of secondary metabolites
The culture broth (4 ml) of different strains of S. reveromyceticus SN-593 was extracted with an equal volume of acetone and centrifuged at 5000 g for 10 min. The supernatant was concentrated in vacuo to remove acetone. The pH of the aqueous extract was adjusted to 4.0 by adding acetic acid, and the extract was extracted twice with an equal volume of ethyl acetate. The organic layer was dried in vacuo and the residue was dissolved in 1.2 ml of methanol. An ESI-MS analysis was performed using a Waters Alliance HPLC system equipped with a mass spectrometer (Q-Trap; Applied Biosystems, Foster City, CA, USA). The HPLC used an XTerraMSC18 (Waters, Milford, MA, USA) 5 μm (2.1 × 150 mm) column and a flow rate of 0.2 ml min−1. Solvent A was 0.05% aqueous formic acid and solvent B was acetonitrile. After the column was equilibrated with 30% solvent B, 1 μl of extracted sample was injected into the column. The column was then developed with a linear gradient from 30 to 100% of solvent B over 20 min and was kept at 100% of solvent B for 20 min. Mass spectra were collected in the ESI-negative mode.
Isolation of 1, 2 and 3
The NRM2 strain was cultured as described above. An equal volume of acetone (770 ml) was added to the total culture broth and filtered to remove mycelia. The filtrate was evaporated in vacuo to remove acetone; the pH of the water layer (770 ml) was adjusted to 4.0 by adding acetic acid. Two extractions were performed using the same volume (770 ml) of ethyl acetate. The organic layer was dried in vacuo to obtain 410 mg of yellow–brown residue. This material was applied to a silica gel column and stepwise elution with CHCl3/MeOH (250 ml each of 100/0, 100/1, 100/2, 100/5, 100/10, 100/20, 100/50, 100/100 and 0/100) was carried out to yield nine fractions.
The CHCl3/MeOH 100/5 fraction (185 mg) was purified with a reversed-phase HPLC column PEGASIL ODS (Senshu Scientific, Tokyo, Japan) (i. d. 20 × 250 mm, SSC) equipped with a 2996 Photodiode Array Detector (Waters). Elution proceeded at a flow rate of 9 ml min−1 with 40% CH3CN, yielding 9 mg of 1 and 15 mg of 2 at 17 and 12 min, respectively. The CHCl3 100% fraction (153 mg) was applied to the same HPLC system and eluted with 60% CH3CN-0.05% formic acid to yield 39 mg of 3, in the form of a yellow powder (Figure 1).

Construction of S. reveromyceticus SN-593 NRM2 by conjugal gene transfer.
References
Bentley, S. D. et al. Complete genome sequence of the model actinomycete Streptomyces coelicolor A3(2). Nature 417, 141–147 (2002).
Ikeda, H. et al. Complete genome sequence and comparative analysis of the industrial microorganism Streptomyces avermitilis. Nat. Biotech. 21, 526–531 (2003).
Ohnishi, Y. et al. Genome sequence of the streptomycin-producing microorganism Streptomyces griseus IFO 13350. J. Bacteriol. 190, 4050–4060 (2008).
Nogawa, T. et al. Verticilactam, a new macrolactam isolated from a microbial metabolite fraction library. Org. Lett. 12, 4564–4567 (2010).
Cheng, X.- C. et al. A new antibiotic, tautomycin. J. Antibiot. 40, 907–909 (1987).
Takahashi, S. et al. Biochemical characterization of a novel indole prenyltransferase from Streptomyces sp. SN-593. J. Bacteriol. 192, 2839–2851 (2010).
Osada, H., Koshino, H., Isono, K., Takahashi, H. & Kawanishi, G. Reveromycin A, a new antibiotic which inhibits the mitogenic activity of epidermal growth factor. J. Antibiot. 44, 259–261 (1991).
Chen, Y., Wendt-Pienkowski, E. & Shen, B. Identification and utility of FdmR1 as a Streptomyces antibiotic regulatory protein activator for fredericamycin production in Streptomyces griseus ATCC 49344 and heterologous hosts. J. Bacteriol. 190, 5587–5596 (2008).
Komatsu, M., Uchiyama, T., Omura, S., Cane, D. E. & Ikeda, H. Genome-minimized Streptomyces host for the heterologous expression of secondary metabolism. Proc. Natl. Acad. Sci. USA 107, 2646–2651 (2010).
Stutzman-Engwall, K. J., Otten, S. L. & Hutchinson, C. R. Regulation of secondary metabolism in Streptomyces spp. and overproduction of daunorubicin in Streptomyces peucetius. J. Bacteriol. 174, 144–154 (1992).
Komiyama, K. et al. Novel antibioitcs, furaquinocins A and B: taxonomy, fermentation, isolation and physico-chemical and biological characteristics. J. Antibiot. 43, 247–252 (1990).
Ishibashi, M., Funayama, S., Anraku, Y., Komiyama, K. & Omura, S. Novel antibiotics, furaquinocins C, D, E, F, G and H. J. Antibiot. 44, 390–395 (1991).
Funayama, S., Ishibashi, M., Anraku, Y., Komiyama, K. & Omura, S. Structures of novel antibiotics furaquinocins A and B. Tetrahedron Lett. 30, 7427–7430 (1989).
Kawasaki, T. et al. Biosynthesis of a natural polyketide-isoprenoid hybrid compound, furaquinocin A: identification and heterologous expression of the gene cluster. J. Bacteriol. 188, 1236–1244 (2006).
Kumano, T., Tomita, T., Nishiyama, M. & Kuzuyama, T. Functional characterization of the promiscuous prenyltransferase responsible for furaquinocin biosynthesis: identification of a physiological polyketide substrate and its prenylated reaction products. J. Biol. Chem. 285, 39663–39671 (2010).
Jiang, J. et al. Genome mining in Streptomyces avermitilis: a biochemical Baeyer-Villiger reaction and discovery of a new branch of the pentalenolactone family tree. Biochemistry 48, 6431–6440 (2009).
Sambrook, J. & Russell, D. W. Molecular Cloning: A Laboratory Manual. 3rd edn. (Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, 2001).
Kieser, T.,, Bibb, M. J., Buttner, M. J., Chater, K. F. & Hopwood, D. A. Practical Streptomyces Genetics (The John Innes Foundation: Norwich, United Kingdom, 2000).
Onaka, H., Taniguchi, S.-i., Ikeda, H., Igarashi, Y. & Furumai, T. pTOYAMAcos, pTYM18, and pTYM19, actinomycete-Escherichia coli integrating vectors for heterologous gene expression. J. Antibiot. 56, 950–956 (2003).
Takahashi, S. et al. Reveromycin A biosynthesis uses RevG and RevJ for stereospecific spiroacetal formation. Nat. Chem. Biol. (in press).
Muroi, M. et al. Application of proteomic profiling based on 2D-DIGE for classification of compounds according to the mechanism of action. Chem. Biol. 17, 460–470 (2010).
Acknowledgements
We are grateful to Dr Y Futamura for cytoxicity assays. This work was supported in part by a Grant-in-Aid for Creative Scientific Research from the Ministry of Education, Culture, Sports, Science and Technology of Japan.
Author information
Authors and Affiliations
Corresponding author
Additional information
Supplementary Information accompanies the paper on The Journal of Antibiotics website
Supplementary information
Rights and permissions
About this article
Cite this article
Panthee, S., Takahashi, S., Takagi, H. et al. Furaquinocins I and J: novel polyketide isoprenoid hybrid compounds from Streptomyces reveromyceticus SN-593. J Antibiot 64, 509–513 (2011). https://doi.org/10.1038/ja.2011.41
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ja.2011.41
Keywords
This article is cited by
-
Studies on Streptomyces sp. SN-593: reveromycin biosynthesis, β-carboline biomediator activating LuxR family regulator, and construction of terpenoid biosynthetic platform
The Journal of Antibiotics (2022)
-
β-carboline chemical signals induce reveromycin production through a LuxR family regulator in Streptomyces sp. SN-593
Scientific Reports (2020)
-
β-carboline biomediators induce reveromycin production in Streptomyces sp. SN-593
Scientific Reports (2019)
-
Kinanthraquinone, a new anthraquinone carboxamide isolated from Streptomyces reveromyceticus SN-593-44
The Journal of Antibiotics (2018)
-
Lysobacter species: a potential source of novel antibiotics
Archives of Microbiology (2016)