Abstract
The purpose of this study was to investigate the phenotypic and molecular characterization of a novel plasmid-mediated AmpC-type β-lactamase in Klebsiella pneumoniae E701 isolated from Anhui province in China. In comparison with the ACT-1, sequence analysis revealed that there were 43 point mutations in the coding gene, and 10 of which led to amino-acid substitution. Resistance could be transferred by conjugation or transformation with plasmid DNA into E. coli JM109, which was due to the production of a β-lactamase with an isoelectric point of 8.4 named ACT-6. Cloning, expression, purification and kinetics were carried out to study the characterization of the novel AmpC-type β-lactamase. The results of MIC determinations and substrate profiles showed there was no significant difference in the activities of the novel enzyme and ACT-1. Moreover, the class 1 integron and the whole open reading frame of the novel AmpC-type β-lactamase from K.pneumoniae E701 were detectable in the same size plasmid. This is the first report on the emergence of the novel ACT-6 type β-lactamases in K. pneumoniae.
Similar content being viewed by others
Introduction
The predominant mechanism for resistance to β-lactam antibiotics in Gram-negative bacteria is the synthesis of β-lactamases. Many types of β-lactamases have been classified into class A, B, C and D according to amino-acid homology.1 AmpC β-lactamases belongs to the molecular class C and functional classification group 1, which are characterized by their hydrolysis of cephalosporins and resistance to clavulanic acid.2 AmpC β-lactamases appeared initially in organisms such as Enterobacter cloacae, Citrobacter freundii, Serratia marcescens and Pseudomonas aeruginosa that could, by mutation, overproduce their chromosomal AmpC β-lactamases.3 Later, resistance appeared in bacterial species that lack an inducible AmpC enzyme, such as Klebsiella pneumoniae, Escherichia coli, Salmonella spp. and Proteus mirabilis.4
MIR-1 mobilized from an Enterobacter isolate was the first genetically characterized plasmid-mediated AmpC.5 Since then, plasmid-mediated AmpC β-lactamases have been detected in many regions of the world. The overproduction of AmpC β-lactamases by Gram-negative organisms results in resistance to most β-lactam antibiotics including the carbapenems.6 They are often encoded by transposons or integrons, which could be easily transferred from one replicon to another, and this phenomenon could contribute to the spread of multidrug-resistant E. coli, K. pneumoniae and other Enterobacteriaceae.7 Thus, we further reported the phenotypic and molecular characterization of the novel AmpC enzyme ACT-6 involved in this study.
Materials and methods
Bacterial strains
K. pneumoniae E701 was isolated from the sputum specimen of a 70-year-old male patient who suffered from lung cancer in September 2007 at Yijishan Hospital of Wannan Medical College. It is a tertiary-care general hospital with 1200 beds in Wuhu, Anhui, China. K. pneumoniae E701 was isolated from the sputum specimen before the treatment. It was identified using the Microscan Walkaway-96 System (Siemens, Munich, Germany). Next, PCR amplification of the complete 16S rRNA gene was performed with genomic DNA in accordance with a previously published protocol.8 Amplification product was subjected to direct sequencing, and a 100% match to GenBank sequence accession number X87276 of K. pneumoniae was noted. E. coli J53, E. coli JM109, E. coli V517, E. coli ATCC 25922 and E. coli ATCC 35218 were stored at the Anhui Center for Surveillance of Bacterial Resistance.
Susceptibility testing
MICs of antimicrobial agents were determined by means of the agar dilution method according to a standard procedure described by the Clinical and Laboratory Standards Institute in 2008.9 Quality control strains were E. coli ATCC 25922 and E. coli ATCC 35218 with every batch of clinical strains to ensure accurate and comparable performance of assays. The plates were incubated in ambient air at 35 °C for 18 h. The inoculating concentration of bacteria was 1–1.5 × 108 colony-forming units per ml, equivalent to a 0.5 McFarland standard. An inoculum of 104 colony-forming units per spot was delivered with a multipoint inoculator (AQS, West Sussex, UK).
PCR amplification and DNA sequencing
PCR analysis were performed with sets of primers specific for AmpC enzyme and the conserved segment of class 1 integron from both the parental and exconjugant strains.10, 11 Purified PCR products were ligated with the pMD18-T easy vector (TaKaRa, Dalian, China), then expressed in E. coli JM109 on a Luria-Bertani agar plate with blue-white selection. Nucleotide sequences were determined by bidirectional sequencing of PCR production with a 3730 automatic DNA sequencer three times.
Conjugation experiment
E. coli J53, Azir, resistant to sodium azide, was used as the recipient strain. Equal volumes of the donor and the recipient strain were mixed and cultured in Luria-Bertani broth. The mixtures were then incubated at 37 °C for 16 h. The transconjugant was selected on tryptone soya agar supplemented with sodium azide (100 μg ml−1, to inhibit growth of the donor strain) and cefoxitin (2 μg ml−1, to inhibit growth of the recipient strain).
Preparation of probes and Southern hybridization
Plasmid DNAs were extracted from the clinical strain and transconjugant by the rapid alkaline lysis protocol.12 After agarose gel electrophoresis, DNA fragments in the gel were transferred onto Hybond-N+ nylon membranes (Amersham Biosciences, London, UK) according to the manufacturer's instructions for the following hybridization experiment. Purified PCR-generated probes whose sequences were specific for a complete codon region of the class 1 integron and the novel ACT-like gene, respectively, were labeled with the Digoxin Random Labeling and Detection Kit (Boster, Wuhan, China).
Cloning and expression of the ampC gene
The different PCR primers were designed in order to introduce the EcoRI restriction (TaKaRa) up and the PstI restriction (TaKaRa) downstream of the genes, respectively. Then the whole open reading frame containing the ampC genes was amplified by PCR and cloned in the PMD-18T easy vector (TaKaRa). The plasmid of the clones was digested by EcoRI and PstI restriction enzymes. The 1.2-kb fragment containing the ampC genes was gel purified and ligated into the pHSG398 vector (2227 bp, including a chloramphenicol resistance gene), which was digested by the same restriction enzyme mentioned above. Then the recombinant plasmid was introduced into E. coli JM109 made competent by the calcium chloride method. After transformation, several strains were cultured on Luria-Bertani agar plates supplemented with cefoxitin (2 μg ml−1) and chloromycetin (50 μg ml−1).
AmpC β-lactamase purification and isoelectric focusing
The crude enzyme extracted from the transformant was carried out by AKTA Purifier 100 (Amersham Pharmacia, Stockholm, Sweden) with the eluant A (20 mmol l−1, Tris-HCl, 2 mmol l−1 dithiothreitol, 10 mmol l−1 EDTA, 10 mmol l−1 NaCl, pH 7.5). Purified enzymes were used for subsequent β-lactamase assays, checked by SDS–polyacrylamide gel electrophoresis, and stained with Coomassie brilliant blue G-250 (Sigma, Santa Clara, CA, USA). Analytical isoelectric focusing of crude cell extracts, detection of β-lactamase bands by nitrocefin, and detection of the activities of the β-lactamase bands separated by analytical isoelectric focusing against β-lactams by a substrate-overlaying procedure were carried out as described previously.13 Reference strains producing TEM-1, TEM-2, TEM-12, SHV-1, SHV-5 and MIR-1 were used as controls, as described previously.13
Enzyme assay
ACT-6 was assayed against various β-lactam substrates at 30 °C in 50 mM phosphate-buffered saline (pH 7.0) using an autospectrophotometer type 752 (Spectrum Instruments, Shanghai, China). The wavelength and changes in extinction coefficients of β-lactams were previously described.14 For all assays, controls were performed by omitting the specific substrate from the reaction mixture. For the good substrates, the kinetic parameters were determined from the initial rates of reactions, using the Hanes–Woolf linearization of the Henri–Michaelis–Menten equation.15 Inhibition studies were as described previously.16 The IC50 was determined as the β-lactamase inhibitor (clavulanate, sulbactam, tazobactam) that reduced the hydrolysis rate of 100 μM nitrocefin by 50% when the enzyme was preincubated with various concentrations of the inhibitor for 30 min at 25 °C before the addition of the substrate.17
Nucleotide sequence accession number
The nucleotide sequence of the blaACT-6 gene was submitted to the GenBank database and assigned an accession number FJ237366.
Results
Antimicrobial susceptibility
The MICs of representative antimicrobial agents for parental strain K. pneumoniae E701, as well as the transconjugant and transformant are listed in Table 1. K. pneumoniae E701 and its transconjugant had almost the same resistance spectrum, which exhibited the high resistance rate to seven β-lactam antimicrobial agents, one β-lactam in combinations with β-lactamase inhibitor, three fluoroquinolones and two aminoglycosides. In comparison with K. pneumoniae E701, the transformant had the same high resistance rate, but was susceptible to three fluoroquinolones and two aminoglycosides (MICs, ⩽1 μg ml−1). Additionally, the MIC of the transformant to piperacillin in combination with the β-lactamase inhibitor tazobactam showed no significant synergy, which is one of the characteristics of the AmpC enzyme. All strains were susceptible to cefepime (MIC⩽2 μg ml−1), imipenem and meropenem (MIC⩽1 μg ml−1).
Sequence analysis
The complete nucleotide sequence of the bla gene encoding the cephamycin-resistant β-lactamase was obtained with PCR products. PCR analyses revealed the presence of the blaACT-1-like β-lactamase gene in the parental strain K. Pneumoniae E701. One large open reading frame, which corresponded to a putative protein of 381 amino acids was found. The bla gene consisted of 1146 nucleotides. DNA sequencing results indicated that the nucleotide sequence of the novel blaampC gene differed from that ACT-1 by 43 point mutations, with 33 silent mutations. There are 10 positive mutations in the amino-acid substitutions compared with ACT-1 (GenBank accession number U58495). The mutations are located in positions 21 (Thr → Ala), 55 (Glu → Gln), 146 (Lys → Thr), 248 (Gln → Lys), 252 (Ser → Asn), 266 (Asn → Ser), 269 (Arg → Lys), 270 (Lys → Glu), 272 (Leu → Ile) and 378 (Ser → Asp). In addition, the class 1 integron of the ACT-6 consisted of 1849 nucleotides. PCR analysis revealed that the composition of it was dfr A12, hypothetical protein gene and aadA2.
Plasmid DNA and location of the β-lactamase gene and class 1 integron
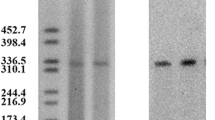
The Southern hybridization analysis was performed with DNA obtained from plasmid preparations. Two plasmids (54.0 and 6.0 kb) were detected in K. pneumoniae E701 and the transconjugant (Figure 1, lane B, lane C). For K. pneumoniae E701 and the transconjugant, only one positive hybridization signal was detectable in the 54.0 kb plasmid band that was hybridized with the blaampC and class 1 integron-specific probes.
Plasmid DNAs analysis of K.pneumoniae E701. Plasmid DNAs were extracted from K.pneumoniae E701 (lane B) and its transconjugant (lane C), run on a 1.0% agarose gel, and compared with standard plasmid molecular sizes of E. coli V517 (lane A). The hybridization signal was in the plasmid (54 kb). Lanes D and E correspond to lanes B and C, respectively.
Purification of β-lactamase and isoelectric focusing
Purities of ACT-6 β-lactamase from the transformant were estimated to be 90% and had a single apparent molecular size of ∼40 kDa on SDS–polyacrylamide gel electrophoresis. One β-lactamase of the AmpC-containing isolate had an apparent isoelectric point (pI) estimated to be 8.4, and the enzyme was not inhibited by clavulanic acid. The other enzyme detected in the isolate had a pI of 8.0, was inhibited by clavulanic acid, and was tentatively classified as extended-spectrum β-lactamase. The pIs of the transconjugant were consistent with that of the clinical strain, and one enzyme pI of 8.4 was also observed in the transformant.
Kinetic parameters
Kinetic parameters (Km and kcat) and hydrolytic efficiency (kcat/Km) of ACT-6 β-lactamase were determined for a representative set of β-lactam antimicrobial agents (Table 2). Measurable hydrolysis rates were observed for penicillin G and cephalosporins. The novel enzyme had lower Km values against penicillin G than for cefoxitin, cefotaxime, cefuroxime and ceftazidime, which were determined to be good substrates. Furthermore, the affinity of ACT-6 β-lactamase for ceftazidime resulted in the lowest hydrolytic efficiency. The inhibition profile showed that the clavulanic acid and sulbactam were not effective inhibitors for ACT-6. However, the tazobactam IC50 values for the ACT-6 lower than that for other AmpC type enzymes, which can be considered to be an effective inhibitor for the ACT-6.18
Discussion
Plasmid-mediated cephalosporinases conferring resistance to oxyimino-cephalosporins, monobactam and cephamycins as well as to β-lactamase resistance inhibitors have been described from 1988 to the present.2 ACT-1 is the first plasmid-mediated AmpC-type β-lactamase derived from Enterobacter, which has been completely sequenced and reported in 1997. The fact that the ACT-1 β-lactamase gene is found on the chromosomes of some strains but on a plasmid in other strains suggests that the blaACT-1 gene is located on a transposon, which is easily mobilized from plasmid to chromosome and vice versa.19 In this study, ACT-6 which shares 96% amino acid identity with ACT-1 is a novel plasmid-mediated AmpC β-lactamase from K. pneumoniae E701 and the phenotypic and molecular characterization of it has rarely been described in other areas.
The nucleotide sequence of blaACT-6 showed a high degree of similarity to the sequence of ACT-1. GenBank database searches revealed that blaACT-6 had great homology with the genes of ACT-1, ACT-2 and ACT-3 β-lactamases (96.3, 96.7 and 96.3%, respectively). Analysis of the internal region of the ACT-6 amino-acid sequence suggested the presence of 10 positive mutations compared with ACT-1, nine positive mutations compared with ACT-2 and 10 positive mutations compared with ACT-3 (see ref. 20). The amino acid sequence of the enzymes revealed an active site serine in the motif Ser-X-X-Lys (where X is any amino acid) at residues 64 to 67 of the mature protein. A Lys-Ser/Thr-Gly motif has been found at residues 315 to 317 and plays an essential role in forming the tertiary structure of the active site. A tyrosine residue at position 150 forms part of the class C-typical motif Tyr-X-Asn and is also important (but not essential) for catalysis of β-lactam hydrolysis.21 This domain had 100% identity to the corresponding domains in the AmpC protein sequence of ACT-1, ACT-2, ACT-3 β-lactamase. In our study, the 10 substitution sites did not belong to the active sites, nor were they adjacent to the indirect active sites. We therefore proposed that the substitutions could not result in the resistance pattern changes, in other words, it meant neutral mutations. Compared with ACT-1, the number of acidic amino acids has increased, which probably played a major role in the non-uniformity with regard to the pIs of ACT-1 (9.0) and ACT-6 (8.4) β-lactamases.
The K. pneumoniae isolate carrying the blaACT-6 gene was resistant to many antimicrobial agents besides cefepime and the carbapenems in our study, and its transconjugant had the same resistance spectrum. This phenomenon indicated that plasmids encoding AmpC enzymes often carry multiple other resistances, including resistance to aminoglycosides, chloramphenicol, sulfonamide, tetracycline, trimethoprim or mercuric ion.5, 19, 22, 23, 24 Many resistance genes, including those for Ambler class A, B and D β-lactamases, are located in gene cassettes with a downstream 59-base element that acts as a specific recombination site for incorporation into integrons.25 Analysis of published sequences indicates that AmpC genes found on plasmids are not linked to 59-base elements. Verdet et al.26 identified a 50-bp sequence near blaDHA-1 that is present also in In6 and In7. A virtually identical sequence occurs near genes for FOX-4, CMY-1, CMY-8, and MOX-1. Such a sequence could represent a recombination site different from the 59-base element. In our study we amplify the conserved segment of class 1 integron for the K. pneumoniae E701, the composition of it explained for the aminoglycoside resistance, and the resistance was also found in the transconjugant. The location of blaACT-6 and class 1 integron were the same size plasmid. Therefore, it can be speculated that K. pneumoniae E701 have accumulated an array of antibiotic resistance genes through transposition of the plasmid, which carries integrons with gene cassettes conferring resistance to aminoglycosides into the vicinity of the blaACT-6. In this work, the plasmid extracted from transconjugant was not entirely sequenced. So we have not the direct evidence to proved that the speculation above.
There are more than 30 kinds of plasmid-mediated AmpC enzyme reported in the world, and the majority of the plasmid-borne ampC genes have been described in K. pneumoniae.5, 19, 27, 28 Most strains producing plasmid-determined AmpC enzymes had been isolated from patients after several days in the hospital or empirically treated with β-lactam antimicrobial agents.7, 19, 29 AmpC-producing nosocomial isolates can be responsible for outbreaks, for example, MIR-1 (11 patients),5 a BIL-1(CMY-2)-like enzyme (five patients),30 ACC-1 (13 patients)31 and ACT-1 (17 patients).19 Integrons are genetic elements that can integrate, by site-specific recombination, gene cassettes, which usually confer antibiotic resistance.32 Antibiotic exposure is probably the major determinant of increased integron prevalence.11 Therefore, it is necessary to avoid the irrationally empirical use of antimicrobial agents, strengthen the surveillance of antimicrobial resistance in local areas, and exchange data between areas. Rational use of antimicrobial agents could improve the situation. This work is of extremely important epidemiology significance to prevent the dissemination of resistant genes.
References
Ambler, R. P. The structure of β-lactamases. Philos. Trans. R. Soc. Lond. B Biol. Sci. 289, 321–331 (1980).
Bush, K. & Jacoby, G. A. Updated functional classification of β-lactamases. Antimicrob. Agents Chemother. 54, 969–976 (2010).
Sanders, C. C. Chromosomal cephalosporinases responsible for multiple resistances to newer β-lactam antibiotics. Annu. Rev. Microbiol. 41, 573–593 (1987).
Jacoby, G. A. Genetics of extended-spectrum beta-lactamases. Eur. J. Clin. Microbiol. Infect. Dis. 13, 2–11 (1994).
Papanicolaou, G. A., Medeiros, A. A. & Jacoby, G. A. Novel plasmid-mediate β-lactamase (MIR-1) conferring resistance to oxyimino- and α-methoxy β-lactams in clinical isolates of Klebsiella pneumoniae. Antimicrob. Agents Chemother. 34, 2200–2209 (1990).
Jacoby, G. A., Mills, D. M. & Chow, N. Role of β-lactamases and porins in resistance to Ertapenem and other β-lactams in Klebsiella pneumoniae. Antimicrob. Agents Chemother. 48, 3203–3206 (2004).
Philippon, A., Arlet, G. & Jacoby, G. A. Plasmid-determined AmpC-type β-lactamases. Antimicrob. Agents Chemother. 46, 1–11 (2002).
Baker, G. C., Smith, J. J. & Cowan, D. A. Review and re-analysis of domain-specific 16S primers. J. Microbiol Methods. 55, 541–555 (2003).
Clinical and Laboratory Standards Institute. NCCLS performance standards for antimicrobial susceptibility testing; Eighteenth informational supplement. CLSI/NCCLS document M100-S18 28, 1–188 (2008).
Alvarez, M., Tran, J. H., Chow, N. & Jacoby, G. A. Epidemiology of conjugative plasmid-mediated AmpC β-lactamases in the United States. Antimicrob. Agents Chemother. 48, 533–537 (2004).
Skurnik, D. et al. Integron-associated antibiotic resistance and phylogenetic grouping of Escherichia coli isolates from healthy subjects free of recent antibiotic exposure. Antimicrob. Agents Chemother. 49, 3062–3065 (2005).
Kado, C. I. & Liu, S. T. Rapid procedure for detection and isolation of large and small plasmids. J. Bacteriol. 145, 1365–1373 (1981).
Pagani, L. et al. Emerging extended-spectrum β-lactamases in Proteus mirabilis. J. Clin. Microbiol. 40, 1549–1552 (2002).
Zhu, W. M. & Qian, Y. S. Study of kinetics of TEM type β-lactamases. Chin. J. Antibiot. 26, 460–463 (2001).
Bauvois, C. et al. Kinetic properties of four plasmid-mediated AmpC β-lactamases. Antimicrob. Agents Chemother. 49, 4240–4246 (2005).
Rasmussen, B. A. et al. Genetically diverse ceftazidime-resistant isolates from a single center: biochemical and genetic characterization of TEM-10 β-lactamases encoded by different nucleotide sequences. Antimicrob. Agents Chemother. 37, 1989–1992 (1993).
Meziane-Cherif, D., Lambert, T., Dupechez, M., Courvalin, P. & Galimand, M. Genetic and biochemical characterization of OXA-63, a new class D β-lactamase from Brachyspira pilosicoli BM4442. Antimicrob. Agents Chemother. 52, 1264–1268 (2008).
Doi, Y. et al. Inhibitor-sensitive AmpC β-lactamase variant produced by an Escherichia coli clinical isolate resistant to oxyiminocephalosporins and cephamycins. Antimicrob. Agents Chemother. 48, 2652–2658 (2004).
Bradford, P. A. et al. Imipenem resistance in Klebsiella pneumoniae is associated with the combination of ACT-1, a plasmid-mediated AmpC β-lactamase, and the loss of an outer membrane protein. Antimicrob. Agents Chemother. 41, 563–569 (1997).
Chen, Y. et al. ACT-3, a novel plasmid-encoded class C β-lactamase in a Klebsiella pneumoniae isolate from China. J. Antimicrob. Agents. 33, 95–96 (2009).
Dubus, A., Normarck, S., Kania, M. & Page, M. G. P. The role of tyrosine 150 in catalysis of β-lactam hydrolysis by AmpC β-lactamase from Escherichia coli investigated by site-directed mutagenesis. Biochemistry 33, 8577–8586 (1994).
Bauernfeind, A., Stemplinger, I., Jungwirth, R., Wilhelm, R. & Chong, Y. Comparative characterization of the Cephamycinase blaCMY-1 gene and its relationship with other β-lactamase genes. Antimicrob. Agents Chemother. 40, 1926–1930 (1996).
Bauernfeind, A., Wagner, S., Jungwirth, R., Schneider, I. & Meyer, D. A novel class C β-lactamase (FOX-2) in Escherichia coli conferring resistance to cephamycins. Antimicrob. Agents Chemother. 41, 2041–2046 (1997).
Stapleton, P. D., Shannon, K. P. & French, G. L. Carbapenem resistance in Escherichia coli associated with plasmid-determined CMY-4 β-lactamase production and loss of an outer membrane protein. Antimicrob. Agents Chemother. 43, 1206–1210 (1999).
Hall, R. M. & Collis, C. M. Antibiotic resistance in gram-negative bacteria: the role of gene cassettes and integrons. Drug Resist. Updat. 1, 109–119 (1998).
Verdet, C., Arlet, G., Barnaud, G., Lagrange, P. H. & Philippon, A. A novel integron in Salmonella enterica Serovar Enteritidis, carrying the blaDHA-1 gene and its regulator gene ampR, originated from Morganella morganii. Antimicrob. Agents Chemother. 44, 222–225 (2000).
Wachino, J. I. et al. Horizontal transfer of blaCMY-bearing plasmids among clinical Escherichia coli and Klebsiella pneumoniae isolates and emergence of cefepime-hydrolyzing CMY-19. Antimicrob. Agents Chemother. 50, 534–541 (2006).
Bauernfeind, A., Schneider, I., Jungwirth, R., Sahly, H. & Ullmann, U. A novel type of AmpC β-lactamase, ACC-1, produced by a Klebsiella pneumoniae strain causing nosocomial pneumonia. Antimicrob. Agents Chemother. 43, 1924–1931 (1999).
Jenks, P. J., Hu, Y. M., Danel, F., Mehtar, S. & Livermore, D. M. Plasmid-mediated production of class 1(AmpC) β-lactamase by two Klebsiella pneumoniae isolates from the UK. J. Antimicrob. Chemother. 35, 235–236 (1995).
M’Zali, F. H. et al. Transcontinental importation into the UK of Escherichia coli expressing a plasmid-mediated AmpC-type β-lactamase exposed during an outbreak of SHV-5 extended-spectrum β-lactamase in a Leeds hospital. J. Antimicrob Chemother. 40, 823–831 (1997).
Nadjar, D. et al. Outbreak of Klebsiella pneumoniae producing transferable AmpC-type β-lactamase (ACC-1) originating from Hafnia alvei. FEMS Microbiol Lett. 187, 35–40 (2000).
Ploy, M. C. Integron-associated antibiotic resistance in Salmonella enterica Serovar Typhi from Asia. Antimicrob. Agents Chemother. 47, 1427–1429 (2003).
Acknowledgements
This work was supported by the National Natural Science Foundation of China (no 30772286) and Anhui Natural Science Foundation (no 070413110).
Author information
Authors and Affiliations
Corresponding authors
Rights and permissions
About this article
Cite this article
Zhu, Yl., Zhang, Xn., Gao, F. et al. ACT-6, a novel plasmid-encoded class C β-lactamase in a Klebsiella pneumoniae isolate from China. J Antibiot 64, 317–320 (2011). https://doi.org/10.1038/ja.2011.1
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ja.2011.1