Abstract
Four new terpenoglycoside antibiotics, phenalinolactones A–D were isolated from Streptomyces sp. Tü 6071. The structures were elucidated on the basis of detailed NMR and MS analyses. Phenalinolactones combine a diterpenoid tricycle, a 2,3,6-trideoxysugar, a pyrrole-carboxylic acid and an uncommonly oxidized unsaturated γ-lactone in a unique manner. Phenalinolactones show an inhibitory activity against Gram-positive bacteria.
Similar content being viewed by others
Introduction
In our search for new secondary metabolites by HPLC-diode array analysis of actinomycete extracts, strain Streptomyces sp. Tü 6071 was subjected to a closer scrutiny because of its production of chemically diverse metabolites. On the basis of their UV–visible spectra and retention times, the already known macrolactam antibiotics maltophilin2 and dihydromaltophilin,3 as well as the structurally related alteramide4 were identified in the mycelium extract by our in-house developed HPLC–UV–vis database.5 Furthermore, four new cyclic decapeptides, streptocidins A–D were isolated from the mycelium of strain Tü 6071.6 Besides these compounds, a third unknown metabolite family was detected in the culture filtrate extract having characteristic UV–vis spectra, which were different from those of all reference compounds of the database. This report describes the fermentation, isolation and structure elucidation of four novel diterpenoid compounds, named phenalinolactones A–D (1–4). Their structures are shown in Figure 1. Only a few diterpenoid compounds of actinomycetes origin are known containing a tricyclic aglycon similar to that of phenalinolactones. One example is brasilicardin A, which is produced by Nocardia brasiliensis that exhibits a potent immunosuppressive and cytotoxic activity.7, 8, 9
The genetic organization of the phenalinolactone gene cluster in Streptomyces sp. Tü 6071 was published 2006 by the group of Andreas Bechthold.10
Materials and methods
General experimental procedures
All homo- and heteronuclear 1D- and 2D NMR experiments were carried out on an Inova 500 spectrometer (Varian, Palo Alto, CA, USA). Chemical shifts are expressed in δ values with solvents as internal standards. ESI-MS and HR-ESI-MS data were collected on a Finnigan LC-Q mass spectrometer (Thermo Fisher Scientific, Waltham, MA, USA) and an Apex-Q III mass spectrometer (Bruker-Daltonics, Bremen, Germany), respectively. IR spectra were recorded on a FT IR-1600 instrument (Perkin-Elmer, Waltham, MA, USA) as KBr pellets, and the UV spectra on a Varian Cary 3E spectrophotometer (Varian). Optical rotation values were measured with a Perkin-Elmer 343 polarimeter and the CD spectra with a J 500 spectrometer (Jasco, Easton, MA, USA).
Producing strain
Strain Tü 6071 was isolated from a soil sample collected at Cape Coast, Ghana and was determined by its morphological and chemotaxonomic features, as well as sequencing of the almost complete 16S rDNA gene as a new member of the genus Streptomyces.6 The strain has been deposited in our strain collection at the Mikrobiologisches Institut, University of Tübingen, Germany.
HPLC-DAD screening
The chromatographic system consisted of a HP 1090M liquid chromatograph equipped with a diode array detector and a HP Kayak XM 600 ChemStation and HPLC software revision A.08.03 (Agilent Technologies, Waldbronn, Germany). Multiple wavelength monitoring was performed at 210, 230, 260, 280, 310, 360, 435 and 500 nm, and UV–vis spectra were measured from 200 to 600 nm.
A 10-ml aliquot of the fermentation broth was centrifuged. The supernatant was adjusted to pH 5 and extracted with the same volume of EtOAc. After centrifugation, the organic layer was concentrated to dryness in vacuo and resuspended in 1 ml MeOH. A 10-μl aliquot of the samples was injected onto an HPLC column (125 × 4.6 mm i.d.), fitted with a guard-column (20 × 4.6 mm i.d.), which was packed with 5-μm Nucleosil-100 C-18 (Maisch, Ammerbuch, Germany). Samples were analyzed by linear gradient elution using 0.1% ortho-phosphoric acid as solvent A and CH3CN as solvent B at a flow rate of 2 ml min−1. The gradient was from 0 to 100% solvent B in 15 min with a 2-min hold at 100% of solvent B. The evaluation of the chromatograms was done by means of our HPLC-UV-vis database that contained about 930 entries, mostly antibiotics.
Fermentation and isolation
Batch fermentations of strain Tü 6071 were carried out in a 10-lt stirred tank fermenter (Biostat S, B. Braun, Melsungen, Germany) in a medium consisting of 2% mannitol and 2% soybean meal in tap water; pH was adjusted to 7.5 before sterilization. The fermenter was inoculated with 5 vol% of shake cultures grown in 500-ml Erlenmeyer flasks, with one baffle for 72 h on a rotary shaker at 120 r.p.m. and 27 °C in the same medium. The fermentation was carried out for 8 days at 27 °C with an aeration rate of 0.5 volume air per volume per min and an agitation at 250 r.p.m.
Hyphlo Super Cel (3%) was added to the fermentation broth, which was separated by multiple sheet filtration into culture filtrate and mycelium. The culture filtrate (7 l) containing 85 mg l−1 of the main component 1 was adjusted to pH 5 (1 M HCl). Non-polar impurities were removed by petroleum benzene extraction and were discarded. The residue was extracted twice with EtOAc, the organic layer was concentrated in vacuo to dryness, dissolved in CH2Cl2 and subjected to a LiChroprep-Diol column (40 × 2.6 cm i.d.; E Merck, Darmstadt, Germany). Separation of components 1 to 4 was accomplished by a linear gradient using CH2Cl2–MeOH, starting at CH2Cl2 up to CH2Cl2–MeOH (75:25) within 3 h at a flow rate of 5 ml min−1. Discrete phenalinolactone-containing fractions were concentrated to dryness, dissolved in a small volume of MeOH and purified by Sephadex LH-20 chromatography (90 × 2.5 cm i.d.; Amersham, Freiburg, Germany) using MeOH as eluent. Pure phenalinolactone compounds were obtained by preparative RP-HPLC on 10-μm Nucleosil-100 C-18 (25 × 1.6 cm i.d.; Maisch) and linear gradient elution with 0.5% HCOOH–MeOH, starting from 60 to 100% MeOH within 20 min at a flow rate of 24 ml min−1. Phenalinolactones were obtained as white powders after lyophilization in tert-BuOH.
Feeding of 13C-labelled precursors
Strain Tü 6071 was grown in 2-lt Erlenmeyer flasks, with one baffle containing 500 ml medium (same as mentioned above) on a rotary shaker at 120 r.p.m. and 27 °C. A measure of 500 mg of either 1-13C-labelled acetate or glucose was dissolved in 50 ml of sterile water. Using a pulse-feeding method, the solution was added to the growing cultures at 96, 98, 100, 102 and 104 h after inoculation. The cultures were collected after 118 h and processed like described before.
Structure determination
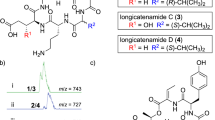
Phenalinolactone A (1): White solid: Rf 0.23 (CHCl3–MeOH, 9:1); [α]20D=−27.1° (c 1.0, CH3OH); UV (MeOH) λmax (log ɛ) 275 (4.44), 216 (3.76) nm; UV (MeOH+HCl) λmax (log ɛ) 276 (4.39), 243 (4.06) nm; UV (MeOH+NaOH) λmax (log ɛ) 322 (2.89), 278 (4.56), 224 (3.92) nm; CD (MeOH) λmax ([Θ]) 281 (14382), 256 (−22441) nm; IR (KBr) νmax 3431, 2955, 1756, 1689, 1622, 1489, 1461, 1433, 1383, 1322, 1261, 1222, 1150, 1122, 1089, 1063, 1017 cm−1; 1H NMR (500 MHz, CDCl3) δ 9.24 (1H, bs, NH), 7.17 (1H, d, J=5.5 Hz, 16-OH), 6.84 (1H, dd, J=3.0, 3.0 Hz, H-3′), 6.27 (1H, bs, OH), 5.98 (1H, dd, J=3.0, 3.0 Hz, H-4′), 5.87 (1H, d, J=5.5 Hz, H-16), 5.57 (1H, bs, H-12), 5.04 (1H, dd, J=12.0, 4.0 Hz, H-3), 4.74 (1H, d, J=12.0 Hz, H-20β), 4.63 (1H, d, J=2.5 Hz, H-1′′), 4.38 (1H, d, J=9.5 Hz, H-21β), 3.95 (1H, d, J=12.0 Hz, H-20α), 3.89 (1H, dq, J=9.0, 6.0, 6.0, 6.0 Hz, H-5′′), 3.58 (1H, m, H-1), 3.52 (3H, s, H3-7′′), 3.40 (1H, d, J=9.5 Hz, H-21α), 3.39 (1H, bs, H-14), 3.34 (1H, dm, J=19.0 Hz, H-11β), 3.03 (1H, ddd, J=10.0, 9.0, 4.0 Hz, H-4′′), 2.35 (1H, m, H-11α), 2.32 (3H, s, H3-6′), 2.15 (1H, m, H-3′′β), 2.06 (3H, s, H3-26), 1.90 (3H, m, H-2β, H-6β, H-2′′β), 1.83 (1H, dm, J=13.5 Hz, H-6α), 1.71 (2H, m, H-2α, H-2′′α), 1.65 (1H, m, H-7β), 1.62 (3H, bs, H3-23), 1.55 (1H, m, H-3′′α), 1.30 (1H, d, J=7.5 Hz, H-9), 1.27 (3H, d, J=6.0 Hz, H3-6′′), 1.27 (3H, s, H3-22), 1.22 (1H, dm, J=11.5 Hz, H-5), 1.09 (1H, m, H-7α), 1.01 (3H, s, H3-24) p.p.m.; 13C NMR (125.7 MHz, CDCl3) δ 170.3 (s, C-25), 169.2 (s, C-18), 160.5 (s, C-1′), 139.2 (s, C-19), 134.4 (s, C-5′), 131.4 (s, C-13), 130.0 (s, C-15), 122.9 (d, C-12), 120.7 (s, C-2′), 116.7 (d, C-3′), 109.1 (d, C-4′), 98.4 (d, C-16), 98.2 (d, C-1′′), 81.6 (d, C-4′′), 78.4 (d, C-1), 70.2 (d, C-3), 68.8 (d, C-5′′), 68.2 (t, C-21), 62.4, (t, C-20), 56.7 (q, C-7′′), 55.5 (d, C-9), 47.0 (d, C-5), 44.9 (s, C-4), 43.9 (s, C-10), 40.6 (d, C-14), 40.5 (t, C-7), 39.7 (s, C-8), 34.0 (t, C-2), 29.2 (t, C-2′′), 27.7 (q, C-24), 24.3 (t, C-11), 23.8 (t, C-3′′), 22.0 (q, C-23), 21.2 (q, C-26), 20.2 (t, C-6), 17.7 (q, C-6′′), 13.2 (q, C-6′), 10.3 (q, C-22), p.p.m.; HRESI-MS m/z 738.3465230 [M+Na]+, calculated and found for C38H53NO12Na.
Phenalinolactone B (2): White solid: Rf 0.23 (CHCl3–MeOH, 9:1); IR (KBr) νmax 3431, 2955, 1756, 1689, 1622, 1489, 1461, 1433, 1383, 1322, 1261, 1222, 1150, 1122, 1089, 1063, 1017 cm−1; 1H NMR (500 MHz, CDCl3) δ 9.51 (1H, bs, NH), 7.13 (1H, d, J=5.5 Hz, 16-OH), 6.96 (1H, ddd, J=2.5, 2.5, 1.5 Hz, H-5′), 6.90 (1H, ddd, J=3.5, 2.5, 1.5 Hz, 3′-H), 6.35 (1H, bs, OH), 6.24 (1H, ddd, J=3.0, 2.5, 2.5 Hz, H-4′), 5.83 (1H, d, J=5.5 Hz, H-16), 5.51 (1H, bs, H-12), 4.99 (1H, dd, J=12.5, 4.5 Hz, -3H), 4.71 (1H, d, J=12.0 Hz, H-20β), 4.59 (1H, d, J=2.5 Hz, H-1′′), 4.34 (1H, d, J=9.5 Hz, H-21β), 3.93 (1H, d, J=12.0 Hz, H-20α), 3.84 (1H, dq, J=9.0, 6.0, 6.0, 6.0 Hz, H-5′′), 3.53 (1H, m, H-1), 3.47 (3H, s, H3-7′′), 3.36 (1H, d, J=9.5 Hz, H-21α), 3.35 (1H, bs, H-14), 3.29 (1H, dm, J=20.0 Hz, H-11β), 2.98 (1H, ddd, J=10.5, 10.5, 4.5 Hz, H-4′′), 2.30 (1H, m, H-11α), 2.11 (1H, m, H-3′′β), 2.01 (3H, s, H3-26), 1.86 (2H, m, H-2β, H-2′′β), 1.79 (2H, m, H2-6), 1.69 (2H, m, H-2α, H-2′′α), 1.62 (1H, dm, J=14.0 Hz, H-7β), 1.58 (3H, bs, H3-23), 1.52 (1H, m, H-3′′α), 1.26 (1H, d, J=7.5 Hz, H-9),1.22 (3H, d, J=6.0 Hz, H3-6′′), 1.21 (3H, s, H3-22), 1.18 (1H, dm, J=9.0 Hz, H-5), 1.04 (1H, m, H-7α), 0.97 (3H, s, H3-24) p.p.m.; 13C NMR (125.7 MHz, CDCl3) δ 170.4 (s, C-25), 169.3 (s, C-18), 160.6 (s, C-1′), 139.3 (s, C-19), 131.4 (s, C-13), 130.0 (s, C-15), 123.5 (d, C-5′), 122.8 (d, C-12), 122.2 (s, C-2′), 115.8 (d, C-3′), 110.5 (d, C-4′), 98.4 (d, C-16), 98.1 (d, C-1′′), 81.5 (d, C-4′′), 78.4 (d, C-1), 70.2 (d, C-3), 68.8 (d, C-5′′), 68.1 (t, C-21), 62.7, (t, C-20), 56.7 (q, C-7′′), 55.5 (d, C-9), 47.0 (d, C-5), 44.9 (s, C-4), 43.9 (s, C-10), 40.5 (d, C-14), 40.4 (t, C-7), 39.6 (s, C-8), 34.0 (t, C-2), 29.2 (t, C-2′′), 27.7 (q, C-24), 24.3 (t, C-11), 23.8 (t, C-3′′), 22.0 (q, C-23), 21.2 (q, C-26), 20.2 (t, C-6), 17.7 (q, C-6′′), 10.3 (q, C-22) p.p.m.; ESI-MS m/z 724.6 [M+Na]+, 1447.2 [2M-H+2Na]+, 700.7 [M-H]−; C37H51NO12.
Phenalinolactone C (3): White solid: Rf 0.23 (CHCl3–MeOH, 9:1); IR (KBr) νmax 3431, 2955, 1756, 1689, 1622, 1489, 1461, 1433, 1383, 1322, 1261, 1222, 1150, 1122, 1089, 1063, 1017 cm−1; 1H NMR (500 MHz, CDCl3) δ 9.24 (1H, bs, NH), 7.17 (1H, d, J=5.5 Hz, 16-OH), 6.84 (1H, dd, J=3.0, 3.0 Hz, 3′-H), 6.27 (1H, bs, OH), 5.98 (1H, dd, J=3.0, 3.0 Hz, H-4′), 5.87 (1H, d, J=5.5 Hz, H-16), 5.57 (1H, bs, H-12), 5.04 (1H, dd, J=12.0, 4.0 Hz, H-3), 4.74 (1H, d, J=12.0 Hz, H-20β), 4.63 (1H, d, J=2.5 Hz, H-1′′), 4.45 (2H, s, H-6′), 4.38 (1H, d, J=9.5 Hz, H-21β), 3.95 (1H, d, J=12.0 Hz, H-20α), 3.89 (1H, dq, J=9.0, 6.0, 6.0, 6.0 Hz, H-5′′), 3.58 (1H, m, H-1), 3.52 (3H, s, H3-7′′), 3.40 (1H, d, J=9.5 Hz, H-21α), 3.39 (1H, bs, H-14), 3.35 (3H, s, H3-7′), 3.34 (1H, dm, J=19.0 Hz, H-11β), 3.03 (1H, ddd, J=10.0, 9.0, 4.0 Hz, H-4′′), 2.35 (1H, m, H-11α), 2.15 (1H, m, H-3′′β), 2.06 (3H, s, H3-26), 1.90 (3H, m, H-2β, H-6β, H-2′′β), 1.83 (1H, dm, J=13.5 Hz, H-6α), 1.71 (2H, m, H-2α, H-2′′α), 1.65 (1H, m, H-7β), 1.62 (3H, bs, H3-23), 1.55 (1H, m, H-3′′α), 1.30 (1H, d, J=7.5 Hz, H-9), 1.27 (3H, d, J=6.0 Hz, H3-6′′), 1.27 (3H, s, H3-22), 1.22 (1H, dm, J=11.5 Hz, H-5), 1.09 (1H, m, H-7α), 1.01 (3H, s, H3-24) p.p.m.; 13C NMR (125.7 MHz, CDCl3) δ 170.3 (s, C-25), 169.2 (s, C-18), 160.5 (s, C-1′), 139.2 (s, C-19), 134.4 (s, C-5′), 131.4 (s, C-13), 130.0 (s, C-15), 122.9 (d, C-12), 120.7 (s, C-2′), 116.7 (d, C-3′), 109.7 (d, C-4′), 98.4 (d, C-16), 98.2 (d, C-1′′), 81.6 (d, C-4′′), 78.4 (d, C-1), 70.2 (d, C-3), 68.8 (d, C-5′′), 68.2 (t, C-21), 67.0 (t, C-6′), 62.4, (t, C-20), 58.1 (q, C-7′), 56.7 (q, C-7′′), 55.5 (d, C-9), 47.0 (d, C-5), 44.9 (s, C-4), 43.9 (s, C-10), 40.6 (d, C-14), 40.5 (t, C-7), 39.7 (s, C-8), 34.0 (t, C-2), 29.2 (t, C-2′′), 27.7 (q, C-24), 24.3 (t, C-11), 23.8 (t, C-3′′), 22.0 (q, C-23), 21.2 (q, C-26), 20.2 (t, C-6), 17.7 (q, C-6′′), 10.3 (q, C-22) p.p.m.; ESI-MS m/z 768.6 [M+Na]+, 790.6 [M-H+2Na]+, 745.5 [M-H]−; C39H55NO13.
Phenalinolactone D (4): White solid: IR (KBr) 3431, 2955, 1756, 1689, 1622, 1489, 1461, 1433, 1383, 1322, 1261, 1222, 1150, 1122, 1089, 1063, 1017 cm−1; 1H NMR (500 MHz, CDCl3) δ 9.15 (1H, bs, NH), 7.15 (1H, d, J=4.5 Hz, 16-OH), 6.83 (1H, dd, J=2.5, 2.5 Hz, 3′-H), 5.94 (1H, dd, J=2.5, 2.5 Hz, H-4′), 5.83 (1H, d, J=4.5 Hz, H-16), 5.50 (1H, bs, H-12), 5.01 (1H, dd, J=12.0, 4.5 Hz, H-3), 4.80 (1H, d, J=11.5 Hz, H-20β), 4.60 (1H, d, J=2.0 Hz, H-1′′), 4.39 (1H, d, J=9.5 Hz, H-21β), 3.85 (1H, m, H-5′′), 3.82 (1H, d, J=11.5 Hz, H-20α), 3.48 (3H, s, H3-7′′), 3.38 (1H, d, J=9.5 Hz, H-21α), 3.37 (1H, bs, H-14), 3.03 (1H, ddd, J=11.0, 10.0, 4.5 Hz, H-4′′), 2.30 (3H, s, H3-6′), 2.29 (1H, m, H-11β), 2.11 (1H, m, H-3′′β), 2.05 (1H, m, H-11α), 2.02 (3H, s, H3-26), 1.85 (3H, m, H-1β, H-6β, H-2′′β), 1.70 (3H, m, H-2β, H-6α, H-2′′α), 1.64 (2H, m, H-2α, H-7β), 1.60 (3H, bs, H3-23), 1.55 (1H, m, H-3′′α), 1.27 (1H, dm, J=11.5 Hz, H-5), 1.21 (3H, d, J=6.0 Hz, H3-6′′), 1.14 (3H, s, H3-22), 1.09 (2H, m, H-1α, H-7α), 1.07 (1H, d, J=7.5 Hz, H-9), 0.96 (3H, s, H3-24) p.p.m.; 13C NMR (125.7 MHz, CDCl3) δ 170.5 (s, C-25), 169.2 (s, C-18), 160.4 (s, C-1′), 139.0 (s, C-19), 134.1 (s, C-5′), 132.5 (s, C-13), 130.0 (s, C-15), 122.1 (d, C-12), 120.9 (s, C-2′),116.7 (d, C-3′), 109.1 (d, C-4′), 98.5 (d, C-16), 98.1 (d, C-1′′), 81.6 (d, C-4′′), 73.9 (d, C-3), 68.8 (t, C-21), 68.6 (d, C-5′′), 62.6, (t, C-20), 56.7 (q, C-7′′), 54.7 (d, C-9), 49.1 (d, C-5), 45.1 (s, C-4), 40.5 (t, C-7), 40.3 (d, C-14), 39.3 (s, C-8), 38,4 (s, C-10), 38.2 (t, C-1), 29.3 (t, C-2′′), 27.6 (q, C-24), 23.8 (t, C-3′′), 23.4 (t, C-2), 23.1 (t, C-11), 22.1 (q, C-23), 21.3 (q, C-26), 20.7 (t, C-6), 17.7 (q, C-6′′), 14.3 (q, C-22), 13.2 (q, C-6′) p.p.m.; ESI-MS m/z 722.6 [M+Na]+, 698.3 [M-H]−; C38H53NO11.
Results and discussion
Taxonomy of the producing strain
As described in a previous communication, strain Tü 6071 was assigned as a member of the genus Streptomyces, because of its morphological and chemotaxonomical characteristics, and the similarity of the almost complete 16S rDNA sequence with type strains belonging to the family Streptomycetaceae.6
Screening, fermentation and isolation
Freshly isolated actinomycetes strains from a soil collected in the tropical rain forest at Cape Coast, Ghana were grown in submerged culture in different complex media in the 100 ml-shake flask scale. Extracts were prepared from the mycelia and culture filtrates at various cultivation times, and were analyzed by HPLC-diode array monitoring to determine their chemical diversity with the aim to detect novel secondary metabolites. A family of four unknown metabolites having characteristic and nearly identical UV–vis spectra was identified in the culture filtrate extract from strain Tü 6071.
Batch fermentations of strain Tü 6071 were carried out in a 10-l stirred tank fermenter, reaching a maximal production of phenalinolactones after a cultivation time of 8 days. The main compound, phenalinolactone A (1), was produced in an amount of 60 mg l−1. Metabolites 1–4 were isolated from the culture filtrate by ethyl acetate extraction and subsequent chromatographic separation steps on diol-modified silica gel, Sephadex LH-20 and preparative RP-HPLC on Nucleosil-100 C-18 material.
Structure determination
The 13C NMR spectra of 1 showed 38 signals, of which 19 had positive values in an APT-spectrum (CH3- and CH-groups, respectively). The rest exhibited negative values (CH2 groups and quaternary carbon atoms). These results, together with the chemical shifts, indicated the presence of 3 ester/amide groups, 8 quaternary carbon atoms, 12 CH-, 8 CH2- and 6 CH3 groups as well as 1 methoxy group. Eight of the mentioned atoms were located in a range characteristic for aromatics and olefins. The 1H NMR spectra displayed 52 protons and 3 of them were exchangeable with D2O. Taking this into account, the molecular formula was demonstrated to be C38H53NO12, using HRESI-MS.
On the basis of 2D NMR correlations (1H-COSY, HMBC, TOCSY and solvent shifts: CDCl3 vs C6D6) as well as biosynthetic considerations, the structure of phenalinolactone A (1) was assigned (Figure 2). The 2,3,6-trideoxysugar moiety bound to the tricyclic diterpenoid aglycon exhibited a 3JHH-coupling of 9.0 Hz between H′′-4 and H′′-5, thus arguing for a diaxial position of the respective protons. This led to the conclusion that in the phenalinolactones, an amicetose rather than the epimeric form rhodinose is incorporated. Following Klyne's rule, the sugar should be either an α-L- or a β-D-amicetose. The question, which of the two isomeric forms is present, was solved by the 1H NMR signal of the anomeric proton, which showed as a broadened doublet. With a value of 6.0 Hz, its width at half maximum militates for an equatorial position of H′′-1, for a diaxial coupling of more than 9.0 Hz to the adjacent protons can be excluded. Thus, one dihedral angle has to be close to 90°. This finding was affirmed by the 1JCH-coupling constant of H′′-1, which is 168 Hz: α-glycosides usually exhibit values of 168–171 Hz, whereas β-glycosides showed smaller constants of 158–162 Hz.
The IR (KBr) absorption band observed at νmax 1758 cm−1 is characteristic for unsaturated furanones, and thus confirmed the existence of a lactone ring. The double bond 13C NMR shifts were remarkably upfield because of the attached substituents. The chemical shifts of the similarly substituted 4,5-dimethyl-3-hydroxy-2,5-dihydrofuran-2-one (δC 137.5 and 133.5) showed a good agreement.11 C-16 (δC=98.4) is attached to two oxygen atoms, one of which is OH (δH=7.17) with a doublet coupling (J=5.5 Hz) to H-16 (δH=5.87). Owing to the fact that 1 shows only one set of NMR signals, the hemiacetal is stereochemically homogenous.
The relative stereochemistry of phenalinolactone A (1) was determined by NOESY spectra (Figure 3). Presumably because of a relatively free rotatability of the lactone moiety with respect to the tricyclic scaffold, no conclusion could be drawn regarding the stereochemistry at C-16. It has to be mentioned though, that the stereochemistry is only relative. Ent-1 is equally possible.
The structures of the derivatives 2, 3 and 4 were established on the basis of MS and NMR data. It is assumed that their relative configuration is the same as in 1 because of very similar chemical shifts and coupling constants of the NMR signals of the terpenoid moiety. Only compound 4 showed signal differences for C-1 and attached atoms.
With the assumption that the diterpenoid part of the aglycon is composed of four isoprene units and C-15 of the lactone derives from one of those units, an interesting biosynthesis must have taken place. If like in analogy to the assembly of brasilicardin A, a C3 unit, possibly serine, was attached to the diterpene, a rearrangement must have occurred. Feeding of 13C-labelled acetate and glucose to evaluate the building blocks of the lactone ring as well as the pathway to the isoprene units did not result in sufficient amounts of any of the phenalinolactones.
Biological activity
Antimicrobial spectra of phenalinolactones were determined in an agar plate diffusion assay and minimal inhibition concentrations by a broth dilution method. The main compounds 1 and 2 exhibited a moderate growth inhibition towards Bacillus subtilis DSM 10, with minimal inhibition concentration values of 10 and 3 μg ml−1, respectively, against Rhodococcus erythropolis DSM 1069, with minimal inhibition concentration values of 3 and 10 μg ml−1, respectively, and against Streptomyces viridochromogenes Tü 57, with a minimal inhibition concentration of 30 μg ml−1. Gram-negative bacteria, yeasts and filamentous fungi were not sensitive to phenalinolactones. The minor congeners 3 and 4 were not tested.
The influence of the phenalinolactones on the growth of tumor cells was tested according to NCI guidelines (National Cancer Institute, Bethesda, MD, USA).12 No growth inhibitory activities towards gastric adenocarcinoma (HM02), breast carcinoma (MCF2) and hepatocellular carcinoma (HepG2) were observed. It would be worthwhile screening the novel terpenoglycoside metabolites in a multitude of biological assays for prospective activities.
References
Nachtigall, J. et al. Two new aurachins from Rhodococcus sp. Acta 2259. J. Antibiot. 63, 567–569 (2010).
Jakobi, M. et al. Maltophilin: a new antifungal compound produced by Stenotrophomonas maltophila R3089. J. Antibiot. 49, 1101–1104 (1996).
Graupner, P. R. et al. Dihydromaltophilin; a novel fungicidal tetramic acid containing metabolite from Streptomyces sp. J. Antibiot. 50, 1014–1019 (1997).
Shigemori, H., Bae, M.- A., Yazawa, K., Sasaki, T. & Kobayashi, J. Alteramide A, a new tetracyclic alkaloid from a bacterium Alteromonas sp. associated with the marine sponge Halichondria okadai. J. Org. Chem. 57, 4317–4320 (1992).
Fiedler, H.- P. Biosynthetic capacities of actinomycetes. 1. Screening for secondary metabolites by HPLC and UV-visible absorbance spectral libraries. Nat. Prod. Lett. 2, 119–128 (1993).
Gebhardt, K., Pukall, R. & Fiedler, H.- P. Streptocidins A∼D, novel cyclic decapeptide antibiotics produced by Streptomyces sp. Tü 6071. I. Taxonomy, fermentation, isolation and biological activities. J. Antibiot. 54, 428–433 (2001).
Shigemori, H. et al. Brasilicardin A, a novel tricyclic metabolite with potent immunosuppressive activity from actinomycete Nocardia brasiliensis. J. Org. Chem. 63, 6900–6904 (1988).
Komaki, H. et al. Brasilicardin A, a new terpenoid antibiotic from pathogenic Nocardia brasiliensis: fermentation, isolation and biological activity. J. Antibiot. 52, 13–19 (1999).
Komaki, H. et al. Antitumor activity of brasilicardin A, a novel terpenoid antibiotic from Nocardia brasiliensis. J. Antibiot. 53, 75–77 (2000).
Dürr, C. et al. Biosynthesis of the terpene phenalinolactone in Streptomyces sp. Tü 6071: analysis of the gene cluster and generation of derivatives. Chem. Biol. 13, 365–377 (2006).
Pouchert, C. J. & Behnke, J. J. The Aldrich Library of 13C and 1H FTNMR Spectra 1st edn (Aldrich Chemical Company Inc., Milwaukee, WI, USA, 1993).
Grever, M. R., Shepartz, S. A. & Chabner, B. A. The National Cancer Institute: cancer drug discovery and development program. Semin. Oncol. 19, 622–638 (1992).
Acknowledgements
We are grateful to Mr G Grewe, University of Tübingen for assistance in fermentations, Mrs J Gerber-Nolte, University of Göttingen for technical assistance, Dr J Fuchser, Bruker-Daltonik, Bremen, Germany for measuring HRESI-MS and Prof Dr W Beil, Medizinische Hochschule Hannover, Germany for performing cytotoxicity assays.
Author information
Authors and Affiliations
Corresponding author
Additional information
Art.no. 57 in ‘Biosynthetic capacities of actinomycetes’. Art.no. 56 (see ref. 1).
Rights and permissions
About this article
Cite this article
Gebhardt, K., Meyer, S., Schinko, J. et al. Phenalinolactones A–D, terpenoglycoside antibiotics from Streptomyces sp. Tü 6071. J Antibiot 64, 229–232 (2011). https://doi.org/10.1038/ja.2010.165
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ja.2010.165
Keywords
This article is cited by
-
Phenelfamycins G and H, new elfamycin-type antibiotics produced by Streptomyces albospinus Acta 3619
The Journal of Antibiotics (2011)