Abstract
Two new antibacterial phenazines were isolated from the culture broth of Brevibacterium sp. KMD 003 obtained from a marine purple vase sponge of the genus Callyspongia, collected in Kyeongpo, Gangwondo, Korea. The structures of these compounds were determined to be 6-hydroxymethyl-1-phenazine-carboxamide (1) and 1,6-phenazinedimethanol (2) through analyses of HR–EI–MS and NMR data. Compounds 1 and 2 showed antibacterial activities against Enterococcus hirae and Micrococcus luteus with 5 μ M MIC values.
Similar content being viewed by others
Introduction
Marine sponges are animals belonging to the phylum porifera and are known to be an abundant source of bioactive secondary metabolites. They are host organisms for various microorganisms, and a number of cytotoxic, anti-microfouling and antimicrobial metabolites have been reported from bacteria associated with marine sponges.1, 2, 3, 4 Marine sponges of genus Callyspongia have been shown to contain bioactive secondary metabolites such as pyridine alkaloids,5 polyacetylenes,6 peptides,7 sulfated meroterpenoids8 and polyketide.9 However, little is known about the secondary metabolite production of microbes derived from Callyspongia. To date, only two cultured microbes, a carotenoid-producing Streptomyces sp.10 and a macrolide-producing fungus Cladosporium herbarum,11 have been isolated from Callyspongia diffusa and C. aerizusa, respectively.
In an effort to investigate the chemical potential of microbes growing in marine environments, we initiated the investigation of the microbial diversity associated with marine sponges and their secondary metabolites production. During this survey, we isolated a dark green-pigmented bacterium, Brevibacterium sp. KMD 003, from the marine purple vase sponge, Callyspongia sp. From the liquid culture of KMD 003, we isolated two new antimicrobial phenazines, 6-hydroxymethyl-1-phenazine-carboxamide (1) and 1,6-phenazinedimethanol (2). Compounds 1 and 2 displayed potent antimicrobial activity against Enterococcus hirae and Micrococcus luteus.
Phenazine-related compounds have been shown to display a variety of biological activities such as antibiotic, antitumor, antimalaria and antiparasitic activities.12, 13, 14, 15 Phenazines have also attracted great interest due to their physiological roles related to pigment production, quorum sensing and biofilm formation in microbial communities.16 Here, we report the isolation, structure elucidation and antimicrobial activity of two new phenazines (1 and 2) isolated from Callyspongia-derived Brevibacterium sp.
Results and discussion
The sequence analysis of the 16S rRNA gene placed the strain KMD 003 within the genus Brevibacterium on the basis of the 99.9 and 99.7% sequence identity with Brevibacterium sanguinis17 and B. celere,18 respectively. The cultured strain KMD 003 was deposited with Korean Culture Center of Microorganisms (KCCM 90080). B. sanguinis and B. celere, the nearest neighbors of strain KMD 003 in the phylogenetic tree, were isolated from blood cultures of a patient with human immunodeficiency virus and degraded thallus of a brown alga, respectively. A few Brevibacterium species have previously been shown to produce the antibiotic phenazine pigments, 1,6-dihydroxyphenazine-5-oxide,19 1,6-dihydroxyphenazine-5,10-oxide,20 iodinin21 and 1,6-dihydroxyphenazine.21 However, no biologically valuable organic compounds have been reported from the bacterial strains, B. sanguinis and B. celere.
To investigate secondary metabolite production by KMD 003, the strain was cultured in 10 1-l Erlenmeyer flasks each containing 500 ml of TCG liquid medium (total 5 l) ((3 g tryptone (Difco, Sparks, MD, USA), 5 g casitone (Difco) and 4 g glucose (Difco) in 1 l filtered seawater). Metabolite production in the culture broth was monitored daily by HPLC analysis using an Agilent 1100 LC-MS system (Agilent Technologies Inc., Santa Clara, CA, USA) with a Phenomenex Luna 5 C18(2) analytical column (Phenomenex Inc., Torrance, CA, USA) (4.6 × 150 mm, flow rate 0.7 ml min−1) and a gradient elution of 10–100% acetonitrile in water for 30 min. Two major peaks were observed at retention time 11.3 and 12.2 min on the fourth day of culture. At the end of the culture period (day 7), the cultured broth of the strain KMD 003 was extracted with Amberlite XAD-7 resin (Sigma-Aldrich, St Louis, MO, USA) (20 g l−1) and acetone. The crude extract was subjected to preparative HPLC using C18 column and gradient elution of 10–100% aqueous acetonitrile for the separation of the two major peaks. The subfraction including the major peaks was purified by repeated HPLC using a silica column and an isocratic elution of methylene chloride–methanol (50:1) to afford compounds 1 and 2.
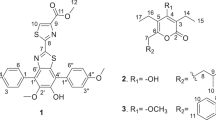
Compound 1 was isolated as a yellow amorphous powder. The molecular formula was assigned to be C14H11N3O2 by HR–EI–MS ((M)+=253.0848) (Table 1 and Supplementary Figure S1). The UV spectrum of 1 showing maximal absorptions at 204, 251 and 367 nm was similar to those of the core phenazine structure, which was confirmed by comparison analysis with our in-house HPLC–UV database (Supplementary Figure S2). The 1H NMR spectrum (Supplementary Figure S3) showed six aromatic protons signals at δH 8.04 ( × 2), 8.06, 8.28, 8.43 and 8.68, and one oxymethylene doublet signal at δH 5.32. Two-dimensional (2D) NMR analysis, using heteronuclear multiple bond correlation (HMBC) and heteronuclear single quantum coherence (HSQC) experiments, showed that these proton signals correlated with 13C NMR signals at δC 132.5, 127.8, 130.6, 127.9, 131.1, 134.5 and 58.8. Other characteristic features of the 1H NMR spectrum of 1 were the presence of a primary alcohol at δH 5.49 (1H, t, J=5.0 Hz) and two protons of a primary amide at δH 8.12 (1H, br s) and 9.77 (1H, br s). As expected, the amide carbon signal was observed at δC 166.1 in the 13C NMR spectrum (Supplementary Figure S4). In addition, the 13C NMR spectrum of 1 showed additional signals attributed to six aromatic quaternary carbons at δC 133.7, 140.5, 140.9, 141.5, 141.7 and 142.2. Comprehensive collation of 2D NMR data (Supplementary Figures S5–S7) from 1H-1H COSY, HSQC and HMBC experiments led to the construction of a phenazine structure with a primary amide and a hydroxymethyl functionality. The HMBC correlations between H-2 (δH 8.68) and C-1′ (δC 166.1), and between 1′-NH (δH 8.12) and C-1 (δC 131.1) allowed the amide group to be positioned at C-1. The position of the hydroxymethlene group was also determined to be C-6 by the HMBC correlation of a methylene proton signal at δH 5.32 with C-6 (δC 141.7) and C-7 (δC 127.8). In addition, the NOE correlation between 1′-NH at δH 9.77 and H-9 (δH 8.28), in 2D NOESY experiment with 700 ms mixing time (Supplementary Figure S8), further supported that the functionalities were not at C-1 and C-9, but compound 1 was 1,6-disubstituted phenazines. These data allowed us to assign the structure of compound 1 as 6-hydroxymethyl-1-phenazine-carboxamide (Figure 1).
Compound 2 was obtained as a yellow amorphous powder that was determined to have the molecular formula C14H12N2O2 by interpretation of HR–EI–MS ((M)+=240.0900) (Supplementary Figure S9) and NMR data (Table 1). The UV and IR spectra (Supplementary Figures S10 and S11) of 2 displayed similar absorption bands with those of compound 1, indicating that 2 contains a phenazine component. Proton NMR spectral data analysis (Supplementary Figures S12 and S13) indicated signals attributable to a 1,2,3-trisubstitued phenyl group (δH 7.95 (1H, dd, J=8.5, 7.0 Hz), 7.98 (1H, br dd, J=7.0, 2.0 Hz) and 8.12 (1H br dd, J=8.5, 2.0 Hz)) and a hydroxymethyl group (δH 5.31 (2H, d, J=5.0 Hz) and 5.43 (1H, t, J=5.0 Hz, OH)). The HSQC spectrum (Supplementary Figure S14) showed that these three aromatic protons signals at δH 7.95, 7.98 and 8.12 correlated with carbon signals at δC 131.2, 127.1 and 128.1, respectively The HSQC spectrum also showed the correlation between the hydroxylmethyl proton signal at δH 5.31 and an oxymethylene carbon signal at δC 59.5. The 13C NMR spectrum (Supplementary Figure S15) of 2 showed additional signals attributed to three aromatic quaternary carbons at δC 140.9, 141.4 and 142.0. Key HMBC correlations allowed the hydroxymethyl group to be positioned at C-1 (δC 141.4) (Supplementary Figure S16). Overall analysis of the NMR data for compound 1 indicated a molecular formula of C7H6NO, exactly one-half of the molecular formula, C14H12N2O2, determined by HRMS. Thus, it became clear that compound 1 is a symmetrical phenazine composed of two identical 1-hydroxymethyl phenyl groups. The substituted position of two hydroxymehtyl group, 1,6- or 1,9-substitution, was determined to be C-1 and C-6 by NOE correlation between hydroxymethyl (δH 5.31) and H-9 signal (δH 8.12) in 2D NOESY experiment with 700 ms mixing time (Supplementary Figure S17). These data allowed us to assign the structure of compound 2 as 1,6-phenazinedimethanol (Figure 1).
Although compounds 1 and 2 seem quite simple, small modifications of the core phenazine structure give rise to a dynamic change of color, redox potential and solubility.16 Moreover, phenazine-1-carboxamide (PCN) and phenazine-1-carboxylic acid (PCA), the structural neighbors of 6-hydroxymethyl-1-phenazinecarboxylic acid (1), have important physiological roles, including Fe acquisition,22 and contribute to pathogen inhibition23 and microbial biofilm formation.24 Compound 1 is composed of two functionalities, a hydroxymethyl and a carboxamide, whereas compound 2 includes two hydroxymethyl functionalities, the first examples of these structural compositions. Mono-functionalized phenazine structures including a hydroxymethyl or a carboxamide can be found in 1-phenazinemethanol or PCN. This class of compounds has a variety of ecological functions and therapeutic potential. 1-Phenazinemethanol possesses antifungal activity against Gaeumannomyces graminis var. tritici and Pythium spp.,25 and PCN displayed antimicrobial activity especially against plant pathogenic Gram-positive bacteria and fungi.23 PCN is synthesized in Pseudomonas aeruginosa by the conversion of PCA in the pyocyanin biosynthetic pathway, which consists of two core loci responsible for the synthesis of PCA and three additional genes encoding unique enzymes involved in the conversion of PCA to pyocyanin, 1-hydroxyphenazine and PCN.26 In addition, PCN has important physiological roles, including Fe acquisition, due to its redox-activity like other phenazines such as pyocyanin, PCA and 1-hydroxyphenazine inter alia.22 PCA, along with its hydroxylated analog, 2-hydroxy-PCA, contributes to pathogen inhibition and microbial biofilm formation.24 PCA also increases oxidant formation and alters the expression of IL-8 (interleukin-8) and ICAM-1 (intercellular adhesion molecule-1) in human airway epithelial cells by oxidant-dependent mechanisms.27
Compounds 1 and 2 showed antimicrobial activity toward three human pathogenic microorganisms (MIC50=20 μM against B. subtilis, 5 μM against E. hirae and 5 μM against M. luteus). In previously reported literature,23 the structural neighbors of compound 1, PCA and PCN, efficiently inhibited the growth of Bacillus cereus (MIC <2 μM), whereas showed only modest antimicrobial activity against M. luteus (MIC 20 μM). Compounds 1 and 2 were only weakly cytotoxic against eukaryotic cell lines with an IC50 value 205.8 and 174.3 μM on HL-60 human leukemia cell lines, respectively, whereas showed no cytotoxicity against five human solid tumor cell lines.
methods
General experimental procedures
UV spectra were obtained on an Agilent 8453 UV-Spectrophotometer. FT-IR spectra were measured on a Bruker Tensor27 Spectrophotometer (Bruker, Ettlingen, Germany) at Kangnung-Wonju National University. 1H-, 13C NMR and 2D NMR spectra were obtained in DMSO-d6 (δH 2.55 and δC 39.5) on a Varian Unity Plus 500 MHz NMR System (Varian, Palo Alto, CA, USA). Low-resolution ESI–MS was measured on an Agilent Technologies VS/Agilent 1100 system at Kangnung-Wonju National University. HR–EI–MS was measured on a Hewlett-Packard 5890A (Hewlett-Packard, Palo Alto, CA, USA) at the National Center for Inter-University Facilities of Seoul National University. Lichroprep RP-18 (Merck, Darmstadt, Germany, 40–63 μm) was used for column chromatography. A Gilson 321 HPLC system (Gilson, Middleton, WI, USA), with a Delta pak C18 (300 × 30.00 mm, 15 μm), a Luna C18(2) column (250 × 10.00 mm, 10 μm) and UV detector (254 nm) was used for preparative HPLC. A Waters 1525 system (Waters, Milford, MA, USA) with PDA detector and a Luna C18(2) column (150 × 4.6 mm, 5 μm) were used for HPLC analysis.
Isolation of KMD 003 strain, cultivation, extraction
The bacterial strain, Brevibacterium sp. KMD 003, was isolated from the tissue of the marine purple vase sponge, Callyspongia sp., collected at a depth of 12 m near Kyung-Po beach in Korea (June 2007). The sponge was washed with autoclaved seawater and the tissues ground and diluted with autoclaved seawater (1:10). The diluted suspension (100 μl) was spread on a TCG agar plate. The TCG agar medium contained 3 g tryptone (Difco), 5 g casitone (Difco), 4 g glucose (Difco) and 18 g agar (Difco) in 1 l filtered seawater (pH 7.0). The plate was incubated for 2 weeks at 25 °C under aerobic conditions. KMD 003 was isolated as a dark green-pigmented colony and it was on a TCG agar plate before secondary culturing in TCG liquid medium (25 ml) while shaking at 200 r.p.m. for 7 days at 25 °C. Stocks of the isolated bacterial strain were generated and stored at −80 °C in liquid culture medium containing 15% (v/v) glycerol.
The strain was further cultured in 10 1-l Erlenmeyer flasks each containing 500 ml of TCG liquid medium (total 5 l). The culture flasks were incubated at 25 °C for 7 days with shaking at 200 r.p.m. At the end of the culture period (day 7), Amberlite XAD-7 resin (20 g l−1) was added to each flask, followed by shaking for one additional hour. The resin was collected by filtration through cheesecloth, washed with deionized water and eluted twice with acetone. The acetone solution was then concentrated under reduced pressure to yield 150 mg of crude extract.
Taxonomy
The strain KMD 003 was identified based on 16S rRNA gene sequencing analysis. The chromosomal DNA of strain KMD 003 was extracted using the G-spin Genomic DNA Extraction Kit (iNtRON Biotechnology, Daejoen, Korea). The 16S rRNA gene of strain KMD 003 was amplified by PCR using universal primers 27f and 1492r corresponding to positions 27 in the forward direction and 1492 in the reverse direction of the Eschrichia coli 16S rRNA gene.28 The DNA sequencing reaction was carried out by using an ABI Prism Bigdye terminator cycle sequencing ready reaction kit V.3.1 (Applied Biosystems, Foster City, CA, USA). The PCR cycle-sequencing product was purified by using Montage dye remove kit (Millipore, Bedford, TX, USA) according to the manufacturer’s protocol. 16S rRNA gene sequence was determined on a Perkin-Elmer model ABI 3730XL capillary DNA sequencer (Applied Biosystems). The 16S rRNA gene sequence of strain KMD 003 was compared with primary sequence information within the GenBank/EMBL/DDBJ nucleotide sequence database using the BLAST algorithm.29
Isolation and purification of phenazines 1 and 2
The crude extract was subjected to preparative HPLC using gradient elution of 10–100% aqueous acetonitrile for 1 h (flow rate 12 ml min−1, Waters Delta pak C18 column 15 μm 300 × 30 mm) to yield eight subfractions (fractions I–VIII). Fraction VI (33.6 mg) was fractionated by reversed-phase HPLC using a gradient elution of 30–100% aqueous methanol for 1 h (flow rate 4 ml/min, column: Phenomenex Luna C18(2) 10 μm 250 × 10 mm) to give 15 subfractions (fraction VI-1–VI-15). Compound 1 (5.4 mg) was purified from subfraction VI-9 (9.6 mg) by repeated HPLC using a Phenomenex Luna Silica (2) column (250 × 10 mm, 10 μm) and an isocratic elution of methylene chloride–methanol (50:1). Compound 2 (5.0 mg) purified from subfraction VI-10 (7.8 mg) was purified by repeated HPLC in the same manner as the purification of compound 1.
6-Hydroxymethyl-1-phenazine-carboxamide (1)
1H-NMRa (500 MHz, DMSO-d6) δ 5.32. (2H, d, J=5.0 Hz, H2-1″), 5.49 (1H, t, J=5.0 Hz, 1″-OH), 8.04 (2H, mb, H-8 and H-7), 8.06 (1H, dd, J=8.5, 7.0 Hz, H-3), 8.12 (1H, br s, amide NH2), 8.28 (1H, mb, H-9), 8.43 (1H, dd, J=8.5, 1.5 Hz, H-4), 8.68 (1H, dd, J=7.0, 1.5 Hz, H-2), 9.77 (1H, br s, amide NH2). aThe reference for chemical shifts: the signal of DMSO-d6 (δH 2.55). bResonance multiplicity was not able to be assigned due to peak overlapping and second-order effects; 13C NMRa (125 MHz, DMSO-d6) δ 58.8 (C-1″), 127.8 (C-7), 127.9 (C-9), 130.6 (C-3), 131.1(C-1), 132.5 (C-8), 133.7 (C-4), 134.5 (C-2), 140.5 (C-10a), 140.9 (C-5a), 141.5 (C-9a), 141.7 (C-6), 142.2 (C-4a), 166.1 (C-1′). aThe reference for chemical shifts: the signal of DMSO-d6 (δC 39.5).
1,6-Phenazinedimethanol (2)
1H-NMRa (500 MHz, DMSO-d6) δ 5.31. (4H, d, J=5.0 Hz, H2-1′ and H2-1″), 5.43 (2H, t, J=5.0 Hz, 1′-OH and 1″-OH), 7.95 (2H, dd, J=8.5, 7.0 Hz, H-3 and H-8), 7.98 (2H, br dd, J=7.0, 2.0 Hz, H-2 and H-7), 8.12 (2H, br dd, J=8.5, 2.0 Hz, H-4 and H-9); 13C NMRa (125 MHz, DMSO-d6) δ 59.5 (C-1′ and C-1″), 127.1 (C-2 and C-7), 128.1 (C-4 and C-9), 131.2 (C-3 and C-8), 140.9 (C-5a and C-10a), 141.4 (C-1 and C-6), 142.0 (C-4a and C-9a). aThe reference for chemical shifts: the signals of DMSO-d6 (δH 2.55 and δC 39.5).
Antibacterial activity
The antibacterial activity of compounds 1 and 2 were tested in a range of 1.25–10 μg ml−1 against six pathogenic microorganisms, Escherichia coli (KCTC 2593), Bacillus subtilis (KCTC 1021), Staphylococcus aureus (KCTC 1916), Methicillin resistance S. aureus MRSA 2659, M. luteus (KCCM 11548) and E. hirae (KCCM 11768). All strains except for B. subtilis were grown at 37 °C, and B. subtilis was grown at 30 °C in nutrient agar (Difco, USA). Antibacterial activity was determined when the density of the growth control reached an absorbance of 0.150–0.200 at 600 nm. Each pathogenic microorganism was seeded in 96-well plates at 100 μl per well, and incubated for 24 h. Compounds 1 and 2 (1.25, 2.5, 5 and 10 μg ml−1) were then inoculated and incubated in 96-well plates at 30 °C for B. subtilis and 37 °C for other pathogens. Growth density was checked every 6 h (0–42 h) at 600 nm.
Cell lines and cytotoxicity
Cytotoxicity was measured by the MTT colorimetric method against HCT116, A549, AGS, MCF-7, HepG2 and HL-60 tumor cell lines. Each cancer cell was seeded in 96-well plates at a density of 104 cells per well, and incubated in 5% CO2 for 24 h at 37 °C. The cells were then treated with varying concentrations (1.56, 3.13, 6.25, 12.5, 25 and 50 μg ml−1) of compounds and incubated in 5% CO2 for 24 h at 37 °C. After 24 h, cells were incubated in 5% CO2 for 1 h at 37 °C with 10 μl of WST-8 (2-(2-methoxy-4-nitrophenyl)-3-(4-nitrophenyl)-3-(4-nitrophenyl)-5-(2,4-disulfophenyl)-2H-tetrazolium, monosodium salt) solution to each well of the plate. Absorbance was then measured at 450 nm using a microplate reader, and experiments were performed in triplicate for each concentration of compounds.
References
Tanaka, J. et al. Diverse metabolites of coral reef organisms. Pure Appl. Chem. 77, 83–89 (2005).
Wang, G. Y. S. et al. Three novel anti-microfouling nitroalkyl pyridine alkaloids from the okunawan marine sponge Callyspongia sp. Tetrahedron Lett. 37, 1813–1816 (1996).
Umeyama, A., Nagano, C. & Arihara, S. Three novel C21 polyacetylenes from the marine sponge Callyspongia sp. J. Nat. Prod. 60, 131–133 (1997).
Gray, C. A. et al. Sulfated meroterpenoids from the Brazilian sponge Callyspongia sp. are inhibitors of the antileishmaniasis target adenosine phosphoribosyl transferase. J. Org. Chem. 71, 8685–8690 (2006).
(a) Buchanan, M. S. et al. Niphatoxin C, a cytotoxic tripyridine alkaloid from Callyspongia sp. J. Nat. Prod. 70, 2040–2041 (2007). (b) Davies-Coleman, M.T. et al. A new EGF-active polymeric pyridinium alkaloid from the sponge Callyspongia fibrosa. J. Org. Chem. 58, 5925–5930 (1993). (c) Wang, G.-Y.-S., Kuramoto, M. & Uemura, D. Three novel anti-microfouling nitroalkyl pyridine alkaloids from the Okinawan marine sponge Callyspongia sp. Tetrahedron Lett. 37, 1813–1816 (1996).
(a) Youssef, D. T. A. Van Soest, R. W. M. & Fusetani, N. Callyspongamide A, a new cytotoxic polyacetylenic amide from the Red Sea sponge Callyspongia fistularis. J. Nat. Prod. 66, 861–862 (2003). (b) Nakao, Y., Uehara, T., Matunaga, S., Fusetani, N. & Van Soest, R.W.M. Callyspongynic acid, a polyacetylenic acid which inhibits α glucosidase, from the marine sponge Callyspongia truncate. J. Nat. Prod. 65, 922–924 (2002). (c) Uno, M., Ohta, S., Ohta, E. & Ikegami, S. Callyspongins A and B: novel polyacetylene sulfates from the marine sponge Callyspongia truncata that inhibit fertilization of starfish gametes. J. Nat. Prod. 59, 1146–1148 (1996).
(a) Capon, R. J. et al. Phoriospongin A and B: two new nematocidal depsipeptides from the Australian marine sponges Phoriospongia sp. and Callyspongia bilamellata. J. Nat. Prod. 65, 358–363 (2002). (b) Berer, N., Rudi, A., Goldberg, I., Benayahu, Y. & Kashman, Y. Callynormine A, a new marine cyclic peptide of a novel class. Org. Lett. 6, 2543–2545 (2004).
Gray, C. A. et al. Sulfated meroterpenoids from the Brazilian sponge Callyspongia sp. are inhibitors of the antileishmaniasis target adenosine phosphoribosyl transferase. J. Org. Chem. 71, 8685–8690 (2006).
Kobayashi, M., Higuchi, K., Murakami, N., Tajima, H. & Aoki, S. Callystatin A, a potent cytotoxic polyketide from the marine sponge, Callyspongia truncata. Tetrahedron Lett. 38, 2859–2862 (1997).
Dharmaraj, S., Ashokkumar, B. & Dhevendaran, K. Food-grade pigments from Streptomyces sp. isolated from the marine sponge Callyspongia diffusa. Food Res. Int. 42, 487–492 (2009).
Jadulco, R. et al. New macrolides and furan carboxylic acid derivative from the sponge-derived fungus Cladosporium herbarum. J. Nat. Prod. 64, 527–530 (2001).
Singh, M. P. et al. Biological and mechanistic activities of phenazine antibiotics produced by culture LL-14I352. J. Antibiot. 50, 785–787 (1997).
Giddens, S. R. & Bean, D. C. Investigations intothe in vitro antimicrobial activity and mode of action of the phenazine antibiotic D-alanylgriseoluteic acid. Int. J. Antimicrob. Agents 29, 93–97 (2007).
Laursen, J. B. & Nielsen, J. Phenazine natural products: biosynthesis, synthetic analogues, and biological activity. Chem. Rev. 104, 1663–1685 (2004).
de Andrade-Neto, V. F. et al. Antimalarial activity of phenazines from lapachol β-lapachone and its derivatives against Plasmodium falciparum in vitro and Plasmodium berghei in vivo. Bioorg. Med. Chem. Lett. 14, 1145–1149 (2004).
Price-Whelan, A., Dietrich, L. E. P. & Newman, D. K. Rethinking ‘secondary’ metabolism: physiological roles for phenazine antibiotics. Nat. Chem. Biol. 2, 71–78 (2006).
Wauters, G. et al. Identification of a novel Brevibacterium species isolated from humans and description of Brevibacterium sanguinis sp. nov. J. Clin. Microbiol. 42, 2829–2832 (2004).
Ivanova, E. P. et al. Brevibacterium celere sp. nov., isolated from degraded thallus of a brown alga. Int. J. Syst. Evol. Microbiol. 54, 2107–2111 (2004).
Gerber, N. N. & Lechevalier, M. P. 1,6-phenazinediol-5-oxide from microorganisms. Biochemistry 4, 176–180 (1965).
Clemo, G. R. & Daglish, A. F. Structure of the pigment of Chromobacterium iodinum. Nature 162, 776 (1948).
Irie, T., Kurosawa, E. & Nagaoka, J. The constitution of the pigments of Brevibacterium crystalloiodinum, Sasaki, Yoshida et Sasaki. Bull. Chem. Soc. Jpn. 33, 1057–1059 (1960).
(a) Wang, Y. & Newman, D. K. Redox reactions of phenazine antibiotics with ferric (Hydr)oxides and molecular oxygen. Environ. Sci. Technol. 42, 2380–2386 (2008). (b) Hernandez, M.E., Kappler, A. & Newman, D.K. Phenazines and other redox-active antibiotics promote microbial mineral reduction. Appl. Environ. Microbiol. 70, 921–928 (2004).
Jayatilake, G. S., Thornton, M. P., Leonard, A. C., Grimwade, J. E. & Baker, B. J. Metabolites from an Antarctic sponge-associated bacterium, Pseudomonas aeruginosa. J. Nat. Prod. 59, 293–296 (2006).
Maddula, V. S. R. K., Pierson, E. A. & Pierson, L. S. Altering the ratio of phenazines in Pseudomonas chlororaphis (aureofaciens) Strain 30–84: effects on biofilm formation and pathogen inhibition. J. Bacteriol. 190, 2759–2766 (2008).
Gurusiddaiah, S., Weller, D. M., Sarkar, A. & Cook, R. J. Characterization of an antibiotic produced by a strain of Pseudomonas fluorescens inhibitory to Gaeumannomyces graminis var. tritici and Pythium spp. Antimicrob. Agents Chemother. 29, 488–495 (1986).
Mavrodi, D. V. et al. Functional analysis of genes for biosynthesis of pyocyanin and phenazine-1-carboxamide from Pseudomonas aeruginosa PAO1. J. Bacteriol. 183, 6454–6465 (2001).
Look, D. C. et al. Pyocyanin and its precursor phenazine-1-carboxylic acid increase IL-8 and intercellular adhesion molecule-1 expression in human airway epithelial cells by oxidant-dependent mechanisms. J. Immunol. 175, 4017–4023 (2005).
Brosius, J., Palmer, M. L., Kennedy, P. J. & Noller, H. F. Complete nucleotide sequence of a 16S ribosomal RNA gene from Escherichia coli. Proc. Natl. Acad. Sci. USA 75, 4801–4805 (1978).
Altschul, S. F., Gish, W., Miller, W., Myers, E. W. & Lipman, D. J. Basic local alignment search tool. J. Mol. Biol. 215, 403–410 (1990).
Acknowledgements
This research is a result of financial support from the Korea Institute of Science and Technology Institute under grant 2Z03270, and from the Marine Biotechnology Program funded by the Ministry of Land, Transport and Maritime Affairs, Republic of Korea. We thank Dr W. Strangman at the University of British Columbia, for scientific advice and English revision.
Author information
Authors and Affiliations
Corresponding author
Additional information
Supplementary Information accompanies the paper on The Journal of Antibiotics website (http://www.nature.com/ja)
Supplementary information
Rights and permissions
About this article
Cite this article
Choi, E., Kwon, H., Ham, J. et al. 6-Hydroxymethyl-1-phenazine-carboxamide and 1,6-phenazinedimethanol from a marine bacterium, Brevibacterium sp. KMD 003, associated with marine purple vase sponge. J Antibiot 62, 621–624 (2009). https://doi.org/10.1038/ja.2009.92
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ja.2009.92
Keywords
This article is cited by
-
Exploring the biosynthetic gene clusters in Brevibacterium: a comparative genomic analysis of diversity and distribution
BMC Genomics (2023)
-
Disruption of Firmicutes and Actinobacteria abundance in tomato rhizosphere causes the incidence of bacterial wilt disease
The ISME Journal (2021)
-
Marine Brevibacterium aureum Extract and Its Constituent’s Saphenic Acid a Derivative of 1-phenazinecarboxylic Acid (Tubermycin B), Initiate Apoptosis via Inhibition of NF-κB and MAPK Expression
Toxicology and Environmental Health Sciences (2018)
-
Antibacterial activity of extracellular compounds produced by a Pseudomonas strain against methicillin-resistant Staphylococcus aureus (MRSA) strains
Annals of Clinical Microbiology and Antimicrobials (2013)