Abstract
Soil is likely the most complex ecosystem on earth. Despite the global importance and extraordinary diversity of soils, they have been notoriously challenging to study. We show how pioneering microfluidic techniques provide new ways of studying soil microbial ecology by allowing simulation and manipulation of chemical conditions and physical structures at the microscale in soil model habitats.
Similar content being viewed by others
Soil—a challenging habitat to study
Soil presents microbes with physical and chemical environments that vary strongly across three-dimensional (3D) space and time. Even single soil aggregates can consist of contrasting mineral materials in a matrix of air, water and organic matter, exhibiting alternating hotspots and ‘desert areas’ of nutrients and toxins. This complex environment challenges soil organisms that require food and shelter within the labyrinth of pores in the soil. Fragmenting barriers are likely responsible for seemingly paradoxical observations, such as the accumulation of organic material in the direct vicinity of decomposers, and make soils one of the most species-dense ecosystems in the world (Eisenhauer et al., 2017; Rillig et al., 2017).
Earlier approaches to address the recognized spatial and chemical complexity of soil ecosystems, such as glass bead or artificial soil model systems, and in situ analysis of soil with microelectrodes or tomography, have provided important insights into soil function. However, they have not been able to simulate, or manipulate, the spatio-temporal heterogeneity of soils at the microscale, the scale that microorganisms experience. Innovative microtechnologies are now emerging (Text Box 1), allowing researchers to mimic and manipulate spatially and chemically complex growth environments at the scale of soil particles (Figure 1). These approaches are opening new possibilities for addressing unanswered questions in soil microbial ecology (Table 1) by allowing researchers to construct microscopic habitats, and study interactions among individuals, distinct populations, multi-species and organism–environment interactions at the relevant scale, including at the level of single cells and hyphal tips.
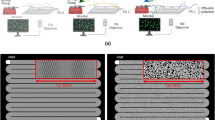
Fabrication of microfluidic devices. A common method to make microfluidic devices is to make a master by photolithography, which is then used to mold PDMS silicone. (a) Deposition of photoresist on a silicon or glass wafer. The thickness of the photoresist is defined by spinning the wafer at a certain rotational speed for a certain time. (b) UV light exposure through mask. UV light illuminates the desired pattern through a photomask and catalyses photoresist crosslinking. (c) Development of the exposed wafer. Non-crosslinked photoresist is removed using a solvent bath. The pattern is now visible on the surface of the master. (d) PDMS molding. PDMS is poured on the developed master and allowed to polymerize in an oven, forming a flexible polymer block. (e) Surface activation. Holes for desired inlets are punched into the PDMS slab, and both PDMS and the glass slide are activated in a plasma chamber. Other materials such as membranes or other PDMS layers can also be used to seal the chip. (f) Sealing the chip by placing the surfaces in contact, which form covalent bonds between the PDMS and the glass surface. (g) Hyphae of Mycetinis scorodonius growing in a pillar system with 100 μm wide pillars. (h) Nematode that migrated into a chip channel from a natural soil inoculum. (i) Microfluidic chip where a dye gradient is generated by sequential mixing and introduced into a culture chamber. (j) Zoom-in on the gradient generator showing and dye diffusion.
In this perspectives paper, we show how microfluidics can be applied in soil microbial ecology, we present inspirational examples of microfluidic approaches in other fields such as biomedical cell research, and critically reflect on potential drawbacks and challenges to studying soil ecology with this technology.
Novel experimental possibilities using microfluidics
Microfluidic platforms have the potential to address four major challenges in studying soil systems, namely (1) their enormous spatio-temporal heterogeneity, (2) the lack of current methods to mimic soil realistically at the appropriate scale, (3) the difficulties in studying interactions among soil microbes and (4) the lack of optical access in real soil systems. We discuss these issues in turn, highlighting important advances driving the transfer of this technology to soil systems, but also identify potential challenges.
Simulating structural and chemical habitat heterogeneity
A major disadvantage of classical solid nutrient medium model systems is the absence of habitat heterogeneity. When physical barriers are absent, organisms and compounds have no constraints to dispersal. Microfluidic technology can be used to increase environmental realism by incorporating structures at the micro- to nanometer scale that confine microorganisms and prevent unintended cellular dispersal and diffusion of substrates or signaling molecules. Complex topologies can be created using two-dimensional silicone molding systems (Figures 1a–d), silicon and glass etching or 3D printing. These micro-structured chip systems provide a new platform for studying microbial interactions with their physical environment, for example, the algorithms used by fungi to explore space in search of nutrients (Held et al., 2010). Conglomerations of soil particles of different sizes can be mimicked by imprinting arrays of cylinders or walls with varying diameters and shapes (Figures 1g and h), simulating soil porosity, aggregation and surface roughness (Figure 2a) (Deng et al., 2015). By simulating different degrees of soil aggregation, researchers could mirror the effects of various soil cultivation practices. This new approach would allow for the analysis of soil structure effects and compaction on microbial performance, and the effect of microbes and their exudates on soil characteristics, such as moisture retention (Deng et al., 2015). These new microfluidic techniques provide a great compliment to previous studies simulating soil structure through controlled assembly of different soil materials (Pronk et al., 2017), different types of transparent materials (for example, glass beads, ‘aquabeads’ or other synthetic polymers, Downie et al., 2012), and recent developments of 3D-printed soil structure proxies, designed to, for example, test how fungi explore pore spaces (Otten et al., 2012).
Five aspects of how microfluidics can be used to mimic the soil-environment and study microbial behavior in a small-structured environment. (a) Simulating physical heterogeneity. Pillars and walls of different sizes and shapes can be used to simulate differences in soil structure and porosity to study how variation in physical heterogeneity affects, for example, microbial establishment, behavior (Held et al., 2010; Deng et al., 2015), and feedback interactions with their environment. (b) Creating chemical gradients and patches. Chemical gradients or plume-like injections can be created inside the chips to mimic spatial heterogeneity of nutrients or other soluble compounds and study, for example, chemotaxis (Stocker et al., 2008). (c) Manipulating microbial interactions. Arenas for the study of microbial physiology, behavior and interactions can be fabricated, allowing minute control over when and where microbes enter the system, with the possibility to restrict encounters to few individual cells or hyphae (Stanley et al., 2014; Hol et al., 2016). (d) Culturing the unculturable. With the development of the Ichip (Nichols et al., 2010), new possibilities have opened up for culturing soil bacteria that have not previously been possible in solid medium cultures. The main design factor thought to facilitate this is the micro-confinement of individual cells in diffusion chambers sealed off with membranes, still allowing for metabolic transfer to and from the surrounding environment. This strongly expands the species pool for laboratory studies, and facilitates identification of their special requirements for pure culture isolation. (e) Studying rhizosphere interactions. Plant roots can be grown from seeds, for example, through pipette tips, into channels of microfluidic devices (Grossmann et al., 2011), permitting close monitoring of root morphology, and giving us the ability to control nutrient supply as well as microbial exposure within the root system. This will open up possibilities to study, for example, the colonization success and succession of root symbiosis such as those involving rhizobia and mycorrhizas, or monitoring of pathogens under differentiated nutrient conditions.
Chemical heterogeneity, a hallmark of soil aggregates, has previously been simulated at the macroscale as patches using cut agar blocks (Boswell, 2003), in the form of gradients using tilted Petri dishes, or at the microscale by diffusion through porous materials such as ceramic plates (Wolfaardt et al., 2008). While all these techniques still can be useful for simulating soil structure in experiments, they do not provide the combination of real-time optical access and precise control of physical heterogeneity, chemical conditions and liquids that microfluidics offer. A microfluidic approach to simulate chemical heterogeneity is to include a gradient generator design in the chip with stepwise mixing of fluids (Figures 1i and j), or to use diffusion gradients in hydrogels. With controlled in- and outflow of liquids, the heterogeneity is spatially stable and manipulated over time to study foraging, chemotaxis, co-existence or niche separation.
Stocker et al. (2008) proposed a chip design that allows for a plume-like injection of soluble compounds into the chip systems, simulating ephemeral microscale nutrient patches in marine systems. This technique could be adapted to simulate the patchy distribution of nutrients in soils at the microscale. Further, multilayer chips can combine chemical gradients with structural heterogeneity, as gradients are placed over, for example, a maze, resulting in highly controllable, spatially resolved chemostats over a heterogeneous micro-landscape.
How do microbes forage across time and space? In contrast to our understanding of macrofaunal foraging strategies, we have been constrained in identifying consistent search patterns of microbes. Pioneering work tracking Escherichia coli in a microfluidic maze demonstrated how bacteria self-organize into waves and how pre-stages of biofilms search for microcavities with nutrients (Park et al., 2003). Similarly, using microfluidic techniques, the colonization of distinct patches and sub-micron cavities by E. coli and Bacillus subtilis have been successfully described by systematically decreasing corridor size between a densely populated patch and a nutrient-rich empty patch (Männik et al., 2009). These types of experiments provide quantitative means for studying microbial dispersal across simulated soil aggregates, and could help answer fundamental questions in ecology such as the factors driving the maintenance of metapopulations by habitat heterogeneity (Keymer et al., 2006).
Soil realism at the appropriate scale
The relevant spatial heterogeneity of the environment experienced by soil organisms is likely in the size range of their own cells (that is, micrometers). An environment at the microscale exhibits properties that may appear non-intuitive for us who live in a macroscale world. The surface-to-volume ratio increases with decreasing dimensions, increasing the importance of surface interactions compared to volume effects. For example, surface effects, such as surface tension, capillary forces, adhesion and viscous drag, become very important at the microscale. The dominance of viscosity over inertia causes liquids to move with laminar instead of turbulent flow so that mixing mainly occurs via diffusion (Figure 1j). This means that microchannels in microfluidic systems better reflect soil conditions, where soil pore water often forms a thin film on mineral particles over which substrates can diffuse. Microfluidic systems can be inoculated with microbial isolates using 3D bacterial community printing techniques that allow precise control of the density, shape and size of the microbial communities (Connell et al., 2013). Devices can further be scaled depending on the organism of interest, from bacteria (Park et al., 2003) to roots (Grossmann et al., 2011), to determine how structure affects microbial foraging decisions.
Facilitating microbial interactions
A major aim of soil ecology is to understand species interactions, and how these drive nutrient cycling. Microfluidic techniques offer methods for confining and directing microbes such that community composition can be controlled and specific cell-to-cell interactions can be monitored. They enable the study of exchange of metabolic compounds, signaling molecules, antibiotic resistance and interactions across kingdoms. Recent work using a device to track interactions between bacteria and fungi demonstrated how bacteria can induce phenotypic responses in fungal hyphae (Stanley et al., 2014; Figure 2c). With this type of chip, the physical aspects of species interactions can be resolved such that we understand the mechanisms by which swarming soil bacteria can drive the dispersal of non-mobile fungi, and reciprocally how fungi provide physical bridges for the bacteria to cross air gaps in the soil (Ingham et al., 2011). The manipulability of microfluidics allows researchers to ask questions about partner interactions and control, for example, eavesdropping at the soil–root interface to understand symbiotic communication (Figure 2e), or the ways in which predator–prey dynamics oscillate across fragmented landscapes (Hol et al., 2016; Figure 2c).
The development of the iChip exemplifies a successful use of microchips to improve culturing techniques for soil microbes, and facilitates culturing of previously unculturable species. (Nichols et al., 2010; Figure 2d). Physical microscale structure is the key factor to balance co-existence of the numerous bacterial species, as it prevents dominant species from overgrowing slower growing species. Combined with controlled diffusion of metabolites between cavities and the surrounding environment, this structuring can provide essential food sources to neighboring species. These techniques could inspire a strategy to increase the pool of currently cultivable fungi, by allowing rarer and more nutritionally demanding species to be isolated and screened for their metabolic profiles, such as antibiotics production.
No soil-borne symbiosis has been documented in microfluidic chips yet. However, chip designs for studying initiation or symbiotic nutrient trading dynamics in rhizobia or mycorrhizal systems, or for increasing knowledge about other endophytes, may be inspired by hydroponic chip-plant cultures (Grossmann et al., 2011; Figure 2e). Chips can also be developed to help us better understand lichen formation and lesser-known symbiotic associations in soils, such as diverse uncultured methane-oxidizing consortia (Hays et al., 2015).
Optical transparency
Soils are naturally opaque. This means that imaging, even with modern microscopy, including micro-spectroscopy with synchrotron light, requires extensive sample preparation in the form of embedding and thin cutting of soil particles or the application of vacuum conditions. These procedures are known to produce artefacts such as changes in drying pore structures or chemical alterations. Recently, the development of ‘transparent soils’ helped to study rhizosphere activity in real time (Downie et al., 2012). Microfluidic devices allow for this type of analysis at an even finer scale (Grossmann et al., 2011), and are commonly produced in transparent materials such as glass, or transparent polymers. This facilitates both direct microscopy and extensive in situ analyses of microscopic samples (Nge et al., 2013).
Unfortunately, the most commonly used chip material polydimethylsiloxane (PDMS) adsorbs analytic radiation such as infrared light and does not allow for example, IR-, RAMAN- or scanning transmission X-ray microscopy. Nevertheless, chemical changes of substrates and metabolic activity can be made visible via fluorophores, and tracked both inside or around cells for advanced image and video analysis of soil organisms, cell growth, movement and substrate usage.
Inspiration from neighboring fields
The use of microfluidics is rapidly expanding in different fields of research. During the past decades it has been especially successful in biomedicine, where among other things it is used to mimic in vivo human cell culturing conditions (so called organs-on-a-chip) to study cell differentiation and tissue organization (Bhatia and Ingber, 2014). These advances can serve as important inspiration for developing microfluidic chips in soil ecology.
In neurophysiology, microfluidic systems facilitate the growth of neuronal axons under controlled conditions: chemical gradients, microgrooves and funnel-shaped micro-channels direct axonal growth to study the formation of neural networks and signaling (Millet and Gillette, 2012). Since neurons share many characteristics with other tip-growing cells, this research can serve as inspiration for the development of tools to study behavior and network formation of roots, hyphae, streptomycetes or other tip-growing organisms. The chip designs used in neuroscience are adaptable for studying similar functions within fungal networks, such as the processes of self-organization. Because of the 3D complexity of nervous tissue, multilayer microfluidic devices incorporating hydrogels have been developed. 3D micropatterning techniques have been used to control the area of cell adhesion and neurite projection by etching out collagen gel using an infrared laser beam (Odawara et al., 2013). Following these examples, systems could be constructed to study soil microbial ecology in 3D space. These designs rely on multi-step chip fabrication and combinations of various techniques and materials that are technically challenging, but will be highly rewarding as pioneering techniques to follow 3D processes across heterogeneous landscapes.
Further, there is a large body of research focused on biomedically relevant cell–cell interactions, with the aim of co-culturing human cells (for example, gut or tumor cells) and bacteria (Li et al., 2016). Eukaryotic cells can be placed into desired patterns with microscale resolution onto chip matrices by micropatterning of adhesive and non-adhesive agents (Millet and Gillette, 2012). Important technical problems have been solved, such as introducing cell types with contrasting requirements and growth behavior, or facilitating chemical interactions while constraining direct cell contact via membranes or nanochannels. Gut-on-a-chip designs, which aim to study human microbiomes (Li et al., 2016) provide a blueprint for studies of the rhizobiome and hyphobiome studies in which thin samples of root or fungal cells can interact with bacterial colonizers. Following emerging work on plant leaf imprints (Zhang et al., 2014), imprints of surface structures, such as roots and soil aggregates, can become key components in the study of microbial habitat niche separation.
Research opportunities awaiting exploration
While pioneering work has successfully exploited microfluidic techniques to study biological processes beyond medical applications, there are a number of research questions in soil ecology and general microbiology that would benefit from further development of microengineered systems. Fungi are especially understudied. Despite the importance of fungi for ecosystem services and industrial applications, we lack even the most basic understanding of their behavior at the small scale. Microsystems can help reveal how hyphal tips interact with their surroundings, identify environment-dependent foraging strategies, and study nutrient redistribution within mycelial networks. The latter may even be relevant for our understanding of self-organizing systems, as basic rules likely govern survival strategies of fungal networks.
The technical possibilities afforded by lab-on-a-chip approaches have the potential to fuel ecologists’ creativity. Electrochemical sensors could facilitate the study of microbial redox processes or geoelectrical responses (Nge et al., 2013), and it is also possible to separate cells of interest from complex samples based on size or mechanical properties using various microfluidic methods utilizing magnetic, electrical, optical, acoustic, mechanical, hydrodynamic or inertial forces (Lenshof and Laurell, 2010). Analytical techniques such as flow cytometry and mass spectrometry can be used to collect data from microfluidic devices to further help researchers understand the metabolic capabilities of microbial consortia, and allow us to ask larger evolutionary questions about the division of labor that emerges across communities (Hays et al., 2015). Likewise, spatially resolved chemostats inside chips allow for extraction of single cells or hyphal tips from defined surroundings for genomic or transcriptomic analyses.
Challenges and limitations in future microfluidic developments
While the list of advantages and new possibilities presented by application of microfluidics to soil microbial ecology is inspiring, there are limitations and challenges that should be taken into consideration when working with microfluidic systems. Soil ecologists need to be aware that even if microfluidics enables the simulation of more true-to-life soil properties compared to solid nutrient media, the systems are still highly unnatural. Microfluidic chips are often fabricated from the silicone rubber PDMS, which is naturally hydrophobic. It can be turned hydrophilic via plasma treatment, but mimicking natural soils via controlled alterations of hydrophilic and hydrophobic surfaces is still problematic. Surface chemistry of soil minerals can be mimicked by surface coating, but high spatial resolution and precision are challenging. Real soils, with all their physio-chemical interactions, are difficult to simulate, and while microtechnologies conveniently allow for single or a few chosen experimental factors to be individually manipulated, this reductionist approach may neglect some of the interactive effects between soil properties.
The largest current obstacle for soil ecologists to start using microfluidics may be the accessibility of the technology: While it is possible to buy commercial, ready-made chips, mainly adapted for biomedical applications (Volpatti and Yetisen, 2014), many cutting-edge questions in soil science (Antwis et al., 2017; Table 1), still require the development of custom designs and technical add-ons requiring interdisciplinary collaborations with engineering laboratories. However, once a specific chip design is finalized, experiments in chips can be precisely reproduced with ease, since numerous replicas of the chips can be made, and each chip can contain hundreds of internal replications of the same design features. As an alternative option, the movement of do-it-yourself biology is promoting low-tech production solutions, such as laser cutting chips in plastic materials (Walsh et al., 2017), that could serve research questions at the larger microscale (for example, rhizosphere studies).
While there are potential drawbacks to technology-driven science, microbial ecology is ready for the precision afforded by microtechnologies, which will complement other emerging approaches, such as tomography, NanoSIMS, STXM and 3D-printing soil structures (Otten et al., 2012). Microfluidics offers the ability to tackle emerging questions in soil ecology such as the hypothesis of soil aggregates as evolutionary incubators for microbes (Rillig et al., 2017), or phenomena such as the farming of bacteria (that is, cultivating and harvesting) by fungi when local resources become depleted in the soil (Pion et al., 2013) at relevant spatial scales. This has the potential to propel the field in new exciting directions (Table 1). While our ability to predict interactions in complex soils will require a range of approaches and tools, precisely manipulating conditions and inducing topological constraints on structure, nutrients and interactions will allow us to make strides in quantitatively probing soil systems.
References
Antwis RE, Griffiths SM, Harrison XA, Aranega-Bou P, Arce A, Bettridge AS et al. (2017). Fifty important research questions in microbial ecology. FEMS Microbiol Ecol 93: 1–10.
Bhatia SN, Ingber DE . (2014). Microfluidic organs-on-chips. Nat Biotechnol 32: 760–772.
Boswell G . (2003). Growth and function of fungal mycelia in heterogeneous environments. Bull Math Biol 65: 447–477.
Connell JL, Ritschdorff ET, Whiteley M, Shear JB . (2013). 3D printing of microscopic bacterial communities. Proc Natl Acad Sci USA 110: 18380–18385.
Deng J, Orner EP, Chau JF, Anderson EM, Kadilak AL, Rubinstein RL et al. (2015). Synergistic effects of soil microstructure and bacterial EPS on drying rate in emulated soil micromodels. Soil Biol Biochem 83: 116–124.
Downie H, Holden N, Otten W, Spiers AJ, Valentine TA, Dupuy LX . (2012). Transparent soil for imaging the rhizosphere. PLoS ONE 7: 1–6.
Eisenhauer N, Antunes PM, Bennett AE, Birkhofer K, Bissett A, Bowker MA et al. (2017). Priorities for research in soil ecology. Pedobiologia 63: 1–7.
Grossmann G, Guo W-J, Ehrhardt DW, Frommer WB, Sit RV, Quake SR et al. (2011). The RootChip: an integrated microfluidic chip for plant science. Plant Cell 23: 4234–4240.
Hays SG, Patrick WG, Ziesack M, Oxman N, Silver PA . (2015). Better together: engineering and application of microbial symbioses. Curr Opin Biotechnol 36: 40–49.
Held M, Lee AP, Edwards C, Nicolau DV . (2010). Microfluidics structures for probing the dynamic behaviour of filamentous fungi. Microelectron Eng 87: 786–789.
Hol FJH, Rotem O, Jurkevitch E, Dekker C, Koster DA, Dethlefsen L et al. (2016). Bacterial predator-prey dynamics in microscale patchy landscapes. Proc Biol Sci 283: 517–523.
Ingham CJ, Kalisman O, Finkelshtein A, Ben-Jacob E . (2011). Mutually facilitated dispersal between the nonmotile fungus Aspergillus fumigatus and the swarming bacterium Paenibacillus vortex. Proc Natl Acad Sci USA 108: 19731–19736.
Keymer JE, Galajda P, Muldoon C, Park S, Austin RH . (2006). Bacterial metapopulations in nanofabricated landscapes. Proc Natl Acad Sci USA 103: 17290–17295.
Lenshof A, Laurell T . (2010). Continuous separation of cells and particles in microfluidic systems. Chem Soc Rev 39: 1203.
Li R, Lv X, Zhang X, Saeed O, Deng Y . (2016). Microfluidics for cell-cell interactions: a review. Front Chem Sci Eng 10: 90–98.
Männik J, Driessen R, Galajda P, Keymer JE, Dekker C . (2009). Bacterial growth and motility in sub-micron constrictions. Proc Natl Acad Sci USA 106: 14861–14866.
Millet LJ, Gillette MU . (2012). New perspectives on neuronal development via microfluidic environments. Trends Neurosci 35: 752–761.
Nge PN, Rogers CI, Woolley AT . (2013). Advances in microfluidic materials, functions, integration, and applications. Chem Rev 113: 2550–2583.
Nichols D, Cahoon N, Trakhtenberg EM, Pham L, Mehta A, Belanger A et al. (2010). Use of Ichip for high-throughput in situ cultivation of ‘uncultivable’ microbial species. Appl Environ Microbiol 76: 2445–2450.
Odawara A, Gotoh M, Suzuki I . (2013). Control of neural network patterning using collagen gel photothermal etching. Lab Chip 13: 1981–2168.
Otten W, Pajor R, Schmidt S, Baveye PC, Hague R, Falconer RE . (2012). Combining X-ray CT and 3D printing technology to produce microcosms with replicable, complex pore geometries. Soil Biol Biochem 51: 53–55.
Park S, Wolanin PM, Yuzbashyan EA, Lin H, Darnton NC, Stock JB et al. (2003). Influence of topology on bacterial social interaction. Proc Natl Acad Sci USA 100: 13910–13915.
Pion M, Spangenberg JE, Simon A, Bindschedler S, Flury C, Chatelain A et al. (2013). Bacterial farming by the fungus Morchella crassipes. Proc Biol Sci 280: 20132242.
Pronk GJ, Heister K, Vogel C, Babin D, Bachmann J, Ding GC et al. (2017). Interaction of minerals, organic matter, and microorganisms during biogeochemical interface formation as shown by a series of artificial soil experiments. Biol Fertil Soils 53: 9–22.
Rillig MC, Muller LA, Lehmann A . (2017). Soil aggregates as massively concurrent evolutionary incubators. ISME J 11: 1943–1948.
Stanley CE, Stöckli M, van Swaay D, Sabotič J, Kallio PT, Künzler M et al. (2014). Probing bacterial-fungal interactions at the single cell level. Integr Biol 6: 935–945.
Stocker R, Seymour JR, Samadani A, Hunt DE, Polz MF . (2008). Rapid chemotactic response enables marine bacteria to exploit ephemeral microscale nutrient patches. Proc Natl Acad Sci USA 105: 4209–4214.
Volpatti LR, Yetisen AK . (2014). Commercialization of microfluidic devices. Trends Biotechnol 32: 347–350.
Walsh DI, Kong DS, Murthy SK, Carr PA . (2017). Enabling microfluidics: from clean rooms to makerspaces. Trends Biotechnol 35: 383–392.
Wolfaardt GM, Hendry MJ, Birkham T, Bressel A, Gardner MN, Sousa AJ et al. (2008). Microbial response to environmental gradients in a ceramic-based diffusion system. Biotechnol Bioeng 100: 141–149.
Zhang B, Luo Y, Pearlstein AJ, Aplin J, Liu Y, Bauchan GR et al. (2014). Fabrication of biomimetically patterned surfaces and their application to probing plant-bacteria interactions. ACS Appl Mater Interfaces 6: 12467–12478.
Acknowledgements
We thank the anonymous reviewers and the editor for valuable comments. EH, KA and PO acknowledge the Swedish Research Council (VR-621-2014-5912), the Crafoord Foundation, the Wallenberg Foundation and BECC; ETK acknowledges NWO Vidi, the European Research Council (Grant 335542); TSS acknowledges NWO/FOM.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Rights and permissions
This work is licensed under a Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-nd/4.0/
About this article
Cite this article
Aleklett, K., Kiers, E., Ohlsson, P. et al. Build your own soil: exploring microfluidics to create microbial habitat structures. ISME J 12, 312–319 (2018). https://doi.org/10.1038/ismej.2017.184
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ismej.2017.184
This article is cited by
-
Plants and endophytes interaction: a “secret wedlock” for sustainable biosynthesis of pharmaceutically important secondary metabolites
Microbial Cell Factories (2023)
-
Microfluidic hotspots in bacteria research: A review of soil and related advances
Soil Ecology Letters (2023)
-
Unboxing the black box—one step forward to understand the soil microbiome: A systematic review
Microbial Ecology (2023)
-
Hyphosphere microbiome of arbuscular mycorrhizal fungi: a realm of unknowns
Biology and Fertility of Soils (2023)
-
Microbiome engineering for bioremediation of emerging pollutants
Bioprocess and Biosystems Engineering (2023)