Abstract
The potential for siderophore mutants of Pseudomonas aeruginosa to attenuate virulence during infection, and the possibility of exploiting this for clinical ends, have attracted much discussion. This has largely been based on the results of in vitro experiments conducted in iron-limited growth medium, in which siderophore mutants act as social ‘cheats:’ increasing in frequency at the expense of the wild type to result in low-productivity, low-virulence populations dominated by mutants. We show that insights from in vitro experiments cannot necessarily be transferred to infection contexts. First, most published experiments use an undefined siderophore mutant. Whole-genome sequencing of this strain revealed a range of mutations affecting phenotypes other than siderophore production. Second, iron-limited medium provides a very different environment from that encountered in chronic infections. We conducted cheating assays using defined siderophore deletion mutants, in conditions designed to model infected fluids and tissue in cystic fibrosis lung infection and non-healing wounds. Depending on the environment, siderophore loss led to cheating, simple fitness defects, or no fitness effect at all. Our results show that it is crucial to develop defined in vitro models in order to predict whether siderophores are social, cheatable and suitable for clinical exploitation in specific infection contexts.
Similar content being viewed by others
Introduction
Bacteria can be social organisms, displaying coordinated behaviours, including quorum sensing (QS), biofilm formation and the production of diffusible exoproducts, which can be shareable ‘public goods’ (West et al., 2007). Iron-scavenging siderophores, exoproteases, biofilm polymers, toxins and QS signals all have the potential to be public goods because the ability of a cell to benefit from their presence (take up a siderophore or the products of extracellular proteolysis) are not contingent on that cell having secreted the molecule itself. Cells which do not produce molecules that enhance growth or survival grow poorly or not at all in monoculture. However, in co-culture with wild-type cells, they avoid the metabolic costs of exoproduct synthesis and reap the benefits of their neighbours’ investment. Their evolutionary fitness relative to the wild-type increases and these mutants are commonly referred to as ‘cheats’ (Rainey and Rainey, 2003; Griffin et al., 2004; Harrison et al., 2006; Diggle et al., 2007; Jiricny et al., 2010; Darch et al., 2012; Raymond et al., 2012; Mund et al., 2017). Circumstances under which the benefits of cheating are constrained, allowing cooperation to persist in populations, have been predicted by theory (Hamilton, 1964; Székely et al., 2010). Because bacteria are amenable to evolution experiments, they have been widely used to test theoretical predictions in vitro (for example, Griffin et al., 2004; Diggle et al., 2007; Kümmerli et al., 2009a). Siderophore production by fluorescent Pseudomonas is a particularly tractable workhorse for sociomicrobiology and has served as a useful model system to facilitate tests of key evolutionary hypotheses (Table 1, also Morgan et al., 2012; Zhang and Rainey, 2013; Luján et al., 2015).
Many exoproducts that could be considered public goods play important roles in infection. As the field of sociomicrobiology progressed, this led to suggestions that cooperation among pathogens could be usefully manipulated, especially in hard-to-treat chronic infections (for example, Foster, 2005; Harrison et al., 2006; Brown et al., 2009; Rumbaugh et al., 2009). The opportunistic pathogen Pseudomonas aeruginosa causes various chronic infections in immunocompromised hosts, notably lung infections in people with cystic fibrosis (CF) and wound infections in people with burns or diabetic ulcers (Hirsch and Tam, 2010; Friedrich et al., 2015). Siderophore-null mutants of this species can invade in vitro populations and cheat their way to high density, resulting in population reduction (Griffin et al., 2004; Table 1). Siderophore-negative mutants are commonly isolated from chronic P. aeruginosa infections (De Vos et al., 2001; Jiricny et al., 2014; Andersen et al., 2015). Researchers have therefore hypothesised that the presence of these mutants is due to cheating in vivo, and that siderophore mutants could be used as ‘Trojan horses’ to ferry antibiotic-susceptibility alleles into infections and render them more sensitive to treatment (Harrison et al., 2006; Brown et al., 2009; Jiricny et al., 2014; Andersen et al., 2015).
A potential problem with this reasoning is the implicit assumption that gene expression, the accessibility of exoproducts to non-producers and the fitness consequences of mutations are comparable in vitro and in vivo, and in acute and chronic infection contexts. Technical advances have revealed how much bacterial transcriptomes (Croucher and Thomson, 2010; Dötsch et al., 2015) and the fitness of loss-of-function mutants (Van Opijnen et al., 2009) vary across environments. When the environment that bacteria experience in a specific infection context is carefully modelled in the lab, the importance of this in determining the fitness consequences of a particular genotype become clear (Palmer et al., 2007; Harrison et al., 2014; Turner et al., 2015).
Typically, experiments on siderophore cooperation and cheating are conducted in iron-limited minimal medium, creating clear costs and benefits to siderophore production and allowing null mutants to be cheats. While this may recapitulate the iron restriction encountered by pathogens colonising healthy tissues during acute infection, chronic infections present a very different environment. Tissue damage, inflammation and disease-specific changes in-host phenotype (for example, increased mucus volume and adhesiveness in CF lungs, Boucher, 2007) are accompanied by changes in the growth substrates available to bacteria. In CF lungs, bacteria use amino acids released by damaged tissues, or from mucus, as carbon sources (Palmer et al., 2005, 2007; Flynn et al., 2016), and iron is plentiful (Tyrrell and Callaghan, 2016). Consequently, bacterial gene expression, and presumably the roles played by virulence factors, differ in chronic and acute contexts (Goodman et al., 2004). More specifically, in CF, there is evidence that P. aeruginosa switches from siderophore-mediated uptake of iron (III) to utilising available iron (II) and heme (Cornelis and Dingemans, 2013; Marvig et al., 2014). Further, experiments have typically used well-mixed liquid medium where exoproducts and cells move freely. Adding an element of spatial structure to the in vitro environment locally restricts the accessibility of siderophores, rendering them less ‘public’ and reducing the ability of non-producing mutants to cheat (Kümmerli et al., 2009c; Luján et al., 2015; Leinweber et al., 2017). Because chronic infections caused by bacterial biofilm in host tissues are structured (Bjarnsholt, 2013), similar restrictions may apply in vivo. Clearly, there is a pressing need to re-assess the role played by siderophore mutants in environments that better model chronic infection. This is now possible because lab models of the environments encountered in CF lungs and soft-tissue wounds have been developed (Palmer et al., 2007; Werthén et al., 2010; Harrison et al., 2014; Turner et al., 2015; Harrison and Diggle, 2016).
To assess and extend the potential of laboratory experiments to yield clinically useful data on the evolutionary dynamics of P. aeruginosa siderophore mutants, we reviewed published experimental work. Our goal was to characterise any biases in the literature which could restrict its applicability to the various environments in which this flexible opportunist can thrive. We found two such biases: (a) many experiments (by ourselves and others) used an uncharacterised, UV-generated mutant (PAO6609, often referred to as PAO9) as a siderophore cheat; and (b) a lack of studies employing a well-defined model of chronic infection. We defined two empirical aims to address these biases.
First, we conducted whole-genome sequencing of PAO6609 to determine the genetic basis of its siderophore-null phenotype, and reveal any other mutations that could affect the outcome of competition with the wild type. Second, we determined the fitness consequences of siderophore loss-of-function mutations in relatively well-characterised models of CF lung infection and chronic soft-tissue wounds. Because we found many mutations that potentially affect metabolism and growth in PAO6609, we used defined siderophore deletion mutants of the siderophores pyoverdine and pyochelin for these experiments. We explicitly explored differences between liquid chronic infection medium and structured infection models because spatial structure affects the social dynamics of public goods (Kümmerli et al., 2009c; Luján et al., 2015; Leinweber et al., 2017; Mund et al., 2017), and because biofilm formation in structured models of lung (Harrison and Diggle, 2016) and wound (Werthén et al., 2010) infection could alter gene expression and the fitness consequences of mutation (Whiteley et al., 2001; Bjarnsholt et al., 2013). Our results demonstrate that whether siderophore mutants act as cheats is dependent on the environment, and suggest that these mutants may not act as cheats in vivo.
Materials and methods
Bacterial strains
P. aeruginosa PAO1 was used as a wild-type siderophore producer and defined ΔpvdD and ΔpvdDΔpchEF mutants in this background (Ghysels et al., 2004) were used as single pyoverdine and double pyoverdine/pyochelin mutants, respectively. Recent literature shows that PAO1 strains fall into two discrete phenotypic groups termed P1 and P2, based on presence or absence of indels in the mexT locus (Luong et al., 2014). We Sanger sequenced the mexT locus of these strains and aligned it against the University of Washington PAO1 genome (NCBI reference sequence NC_002516), which has an 8 bp insertion in mexT producing the P2 phenotype. None of our strains had this insertion, nor did they have a commonly-reported 2 bp deletion present in other P2 strains (Luong et al., 2014). Thus our strains have the P1 phenotype (data not shown). The UV-induced mutant PA6609 was derived from PAO6049 (Hohnadel et al., 1986); PAO6049 is a Tn5-induced methionine auxotroph derived from PAO1 (Rella et al., 1985). See Supplementary Figure S1 for the genealogy of PAO6609.
Collating and analysing experimental work on siderophore cheating
To identify published experiments on siderophore cheating, we used Scopus.com to (i) conduct a literature search and (ii) locate articles citing already-identified experimental and theoretical articles on siderophore social evolution. For each experiment in each article, we recorded which strains/clones and media were used; whether an explicit test for cooperation was conducted (relative fitness of mutant in pure/mixed culture with a producer strain and growth or final density of pure/mixed cultures); the starting frequency of mutants in co-culture experiments; the location of the key data in the article; whether cheating was observed and any other key conclusions. The diverse starting frequencies and mutants used make this data unsuitable for formal meta-analysis, so we simply present the key characteristics and findings of the studies in a format that facilitates qualitative comparison.
Whole-genome sequencing of PAO6609 (PAO9)
PAO6609 was cultured overnight in 10 ml Lysogeny Broth at 37 °C on an orbital shaker. Genomic DNA was extracted using a Sigma-Aldrich (Gillingham, UK) GenElute Bacterial Genomic DNA Kit. Library preparation was performed using the Nextera XT library preparation kit (Illumina Inc., San Diego, CA, USA), and 2 × 300 bp paired-end sequencing performed on the Illumina MiSeq platform using a V3 sequencing cartridge, to ~50 × coverage. Reads were assembled using SPAdes run with the –careful flag, and annotated using Prokka. This produced an assembly of 6 232 039 bp comprising 89 contigs with an N50 of 220 789. The assembly was compared with the reference PAO1 genome (NCBI reference sequence NC_002516) using BLAST and the comparison visualised using ACT to search for gene acquisition or loss events. SNP typing was performed by mapping the raw reads of PAO6609 against the PAO1 reference genome (NC_002516.2) using SMALT and SAMtools. A total of 98.3% of 1.17M reads mapped to the reference, from which high fidelity SNPs were called using a cutoff of minimum allele frequency of 0.8, minimum quality score 30, and minimum depth of 8.
Growth conditions for cheating experiments
We used five different growth environments. In each case, cultures were grown in 24-well microtitre plates and incubated on a rocking platform at 37 °C. (i) 2 ml casamino acids medium (CAA: 5 g casamino acids, 1.18 g K2HPO4·3H2O, 0.25 g MgSO4·7H2O, per litre) supplemented with 20 mm NaHCO3 and 100 μg ml−1 human apo-transferrin (Sigma, Gillingham, UK), cultured for 24 h. (ii) 2 ml artificial sputum medium (ASM) following recipe in Palmer et al. (2007); cultured for 24 or 48 h. (iii) ex vivo pig lung (EVPL) model comprising 5 mm2 pig bronchiole+400 μl ASM following protocol in Harrison and Diggle (2016); cultured for 96 h. (iv) 2 ml synthetic wound fluid (SWF: 50% v/v peptone water/foetal bovine serum following Werthén et al. (2010)), cultured for 24 or 48 h. (v) 400 μl synthetic chronic wound (SCW: SWF solidified with rat tail collagen following protocol in Werthén et al. (2010)), cultured for 48 h. For environments (ii–v), we measured wild-type siderophore production and calculated the fitness of siderophore mutants relative to the wild type in clonal and mixed culture. Four or five replica cultures were inoculated with (a) wild-type, (b) ΔpvdD (c) ΔpvdDΔpchEF, (d) 50% wild-type+50% ΔpvdD or (e) 50% wild-type+50% ΔpvdDΔpchEF bacteria for each environment/culture time combination, and each experiment was repeated twice to yield two experimental blocks. For environment i (CAA), we simply measured siderophore production by four replica populations of the wild type for comparison with environments (ii-v).
The density of the inoculum was kept as consistent as possible across environments and population types, as cell density can affect the outcome of wild-type—siderophore-null co-culture experiments (Ross-Gillespie et al., 2009). For experiments in CAA, ASM and SWF, each culture was inoculated with a total of two colonies of the relevant genotype(s), picked from LB plates using a 200 μl pipette tip. For experiments in EVPL and SCW, each culture was inoculated with two colonies of the relevant genotype(s), picked from LB plates using an insulin syringe fitted with a 30 G needle.
To construct time courses of bacterial growth in ASM, two colonies of (a) wild-type, (b) ΔpvdD or (c) ΔpvdDΔpchEF were inoculated into four 2 ml aliquots of ASM in a 24-well microtitre plate. This was incubated at 37 °C in a Tecan Infinite 200 Pro multimode reader for 18 h; every 20 min, the plate was shaken for 2 s and absorbance of each well read at 400 nm. (Reading at more traditional values of 600–700 nm risks interference from absorbance by pyoverdine in wild-type cultures).
Calculation of relative fitness
The fitness of mutants relative to the wild type in mixed cultures, v, was calculated as x2(1-x1)/x1(1-x2), where x1 is the starting frequency of the mutant and x2 is the end frequency. To calculate the relative fitness of mutants in pure culture, mutant populations were randomly paired with pure wild-type cultures from the same experimental block and the same formula used. It follows from the definition that v<1 reflects a decrease in mutant frequency over the period of competition, that is, the mutant is less fit than the wild type, while v>1 reflects an increase in mutant frequency, that is, the mutant is fitter than the wild type. If mutant and wild type have the same fitness, then v=1.
Statistical analyses
The data were analysed in R 3.3.0 (R Development Core Team, 2016) using general linear models and ANOVA. To meet model assumptions of homoscedasticity and normality of residuals, the fitness data from SWF were log-transformed and fitness data from ASM and EVPL square-root transformed; the fitness data from SCWs did not require transformation. All data on pyoverdine production, pyochelin production and growth for wild-type monocultures in the different growth environments were log transformed to meet model assumptions. Where missing values caused non-orthogonality, the car package (Fox and Weisberg, 2011) was used to perform ANOVA with Type II sums of squares, returning correct F-ratios for main effects in models containing an interaction. Fitted means and confidence intervals were retrieved, and post hoc contrasts tested, using the 'lsmeans' package, with default correction of multiple P-values for false discovery rate (Lenth, 2013). Post hoc multiple comparisons against a control were implemented using the 'glht' command in the 'multcomp' package (Hothorn et al., 2008).
Data availability
The raw sequence data has been deposited in the European Nucleotide Archive (Accession number: ERR1725797). Summary information on mutations in PAO6609 (Figure 1) and the raw data for experiments depicted in Figures 2, 3, 4, 5,Supplementary Figure S2 are available as Supplementary Information at the ISME Journal website.
Location of one deletion and 90 SNPs in the genome of PAO6609 (a.k.a. PAO9), mapped against a PAO1 reference sequence (NC_002516.2). SNPs are colour coded by functional class of the locus affected. 11 SNPs that that result in amino acid substitution and which seem most likely to affect growth and virulence are highlighted and the gene name given.
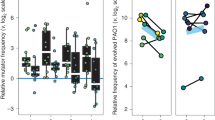
(a, b) Relative fitness of ΔpvdD and ΔpvdDΔpchEF mutants in pure culture (grey bars) and in mixed culture with an isogenic wild type (white bars) in ASM after (a) 24 and (b) 48 h of growth. After eliminating variation due to experimental block, we found significant effects on 24-h fitness of mutant genotype (ANOVA: F1,33=6.96, P=0.012), presence/absence of the wild type (F1,33=26.8, P<0.001) and their interaction (F1,33=4.49, P<0.042). Neither mutant had a relative fitness significantly different from 1 when grown in pure culture (post hoc t-tests, P⩾0.17). ΔpvdD mutant fitness was unaffected by the wild type (relative fitness in mixed culture not significantly different from 1; post hoc t-test, P=0.074), but the ΔpvdDΔpchEF mutant was outcompeted in mixed culture (post hoc t-test, P<0.001). The only significant determinant of 48-h fitnes was genotype (ANOVA eliminating experimental block: genotype F1,35=8.34, P=0.007; presence/absence of the wild type: F1,33=0.277, P=0.602; interaction term: F1,33=3.62, P=0.065). Post hoc t-tests showed that the ΔpvdD mutant was less fit than the wild type when grown in pure culture (P=0.018) but as fit as the wild type in mixed culture (P=0.279). The ΔpvdDΔpchEF mutant had a relative fitness <1 in both pure and mixed culture (P⩽0.001). (c) Cheating by ΔpvdD over 48 h of co-culture results in mixed wild-type+mutant cultures showing the same reduction in total population density as pure mutant cultures (ANOVA eliminating experimental block, F2,26=12.6, P<0.001; Dunnett’s test for mutant/mixed populations versus wild type: P=0.009 and <0.001, respectively; post hoc t-test for mutant versus mixed populations, P=0.159). In all panels, bars show means of 9–10 replicates split across two replica experiments, with associated 95% confidence interval. In both panels, results of post hoc tests for relative fitness=1 are shown with *=P<0.02; n.s.=not significant.
Relative fitness of ΔpvdD and ΔpvdDΔpchEF mutants in pure culture (grey bars) and in mixed culture with an isogenic wild type (white bars) in ex vivo pig lung+ASM after 96 h of growth. Bars show means of eight replicates spread across two replica experiments, with associated 95% confidence interval. (ANOVA eliminating experimental block: genotype F1,27=2.24, P=0.146; presence/absence of the wild type: F1,27=10.7, P=0.003; interaction term: F1,27=0.137, P=0.714). Neither strain had a relative fitness signficantly different from 1 in pure culture (post hoc t-tests, P=0.074 for ΔpvdD, P=0.472 for ΔpvdDΔpchEF). In co-culture, relative fitness was not significantly different from 1 for ΔpvdD (P=0.074), but <1 for ΔpvdDΔpchEF (P=0.007). In both panels, results of post hoc tests for relative fitness=1 are shown with *=P=0.007; n.s.=not significant.
Relative fitness of ΔpvdD and ΔpvdDΔpchEF mutants in pure culture (grey bars) and in mixed culture with an isogenic wild type (white bars) in SWF after (a) 24 and (b) 48 h of growth. Bars show means of 10 replicates split across two replica experiments, with associated 95% confidence interval. Twenty-four hour fitnss was detemrined only by the presence/absnece of the wild type (ANOVA eliminating experimental block: genotype F1,35=0.621, P=0.436; presence/absence of wild type: F1,35=35.2, P<0.001; interaction term: F1,35=2.82, P=0.102); The ΔpvdD mutant’s relative fitness was not significantly different from 1 in pure culture (post hoc t-test, P=0.051), and the ΔpvdDΔpchEF mutant slightly and significantly >1 (P=0.006), but both were less fit than the wild type in mixed culture (P=0.032 and <0.001, respectively). Forty-eight-hour fitness was not affected by genotype or co-culture (ANOVA eliminating experimental block: genotype: F1,35=0.045, P=0.833; presence/absence of wild type: F1,35=1.34, P=0.255; interaction term: F1,35=0.078, P=0.781): both mutants had a relative fitness of 1 regardless of wild type presence (post hoc t-tests, P⩾0.104). In both panels, results of post hoc tests for relative fitness=1 are shown with *=P<0.02; n.s.=not significant. Confidence intervals and P-values in (b) are taken from a model that excluded two outliers where relative fitness was zero (indicated by open diamonds): including these outliers in the analysis did not change any of the conclusions given in the text.
Relative fitness of ΔpvdD and ΔpvdDΔpchEF mutants in pure culture (grey bars) and in mixed culture with an isogenic wild type (white bars) in synthetic wounds after 48 h of growth. Relative fitness was unaffected by genotype or culture condition (ANOVA eliminating experimental block: genotype F1,35=0.055, P=0.817; presence/absence of wild type: F1,35=0.113, P=0.739; interaction term: F1,35=2.32, P=0.137; post hoc t-tests for relative fitness=1, P⩾0.270). Bars show means of 10 replicates split across two replica experiments, with associated 95% confidence interval and result of post hoc tests (all n.s.).
Results
Defining key research priorities: analysis of published experiments.
A review of the published experiments on P. aeruginosa siderophore mutants and cheating identified 36 experiments in 23 published articles: these are summarised in Table 1. It should be noted that the motivations behind these experiments were diverse, and generally sought to understand the evolutionary dynamics of siderophores in iron-limited environments without any direct reference to potential clinical relevance. Nevertheless, we can use these experiments to gain a picture of the state of the art in siderophore research and assess how far this could inform understanding of chronic infection. Two-thirds of the experiments used the well-characterised lab strain PAO1 as the siderophore-producing wild type, and six experiments used clinical or environmental isolates. Of the 35 experiments that did not focus on natural isolates, 15 used defined siderophore mutants. These included deletions of the pvdD/pvdF and/or pchEF loci, which are involved in the biosynthesis of the siderophores pyoverdine and pyochelin, respectively. Of the remainder, 10 used the UV-induced mutant PAO6609 (Supplementary Figure S1) and five used spontaneous or evolved mutants.
We recorded whether each experiment included a specific test for cheating. We determined a ‘cheating’ mutant to be a non-producer which (1) had lower fitness (grew less well) than the producer genotype in monoculture, and (2) gained a growth advantage in mixed culture, such that it grew as well or better than the wild type, that is, the mutant was maintained in the population and may have increased in frequency. Thus, we defined a specific test for cheating as a quantitative comparison of the growth or fitness of producers and non-producers in monoculture and co-culture, and recorded the location of the data pertaining to this test in each publication.
Out of 36, 34 experiments in Table 1 were conducted in CAA medium or minimal salts supplemented with CAA as a carbon source; in all but two cases, the media was made more iron-limited by the addition of the human iron chelator transferrin: one study did not use apotransferrin at all and one explicitly compared CAA with and without apotransferrin. This medium was not designed to reflect any specific natural environment: it was optimised to ensure that siderophores were necessary for growth, and acted as a public good, when bacteria were cultured in it (Griffin et al., 2004). This makes it ideal for experiments to test evolutionary theory about why and how different social strategies can be selected for. Experiments in iron-limited CAA have, for instance, revealed that the potential for siderophore mutants to cheat can be reduced or curtailed by the scale of competition in a metapopulation (Griffin et al., 2004) or by growth in a structured environment, where spatial segregation prevents mutants from accessing siderophores produced by wild types (Kümmerli et al, 2009c; Leinweber et al., 2017). As an opportunist with a large, flexible metabolome and secretome, P. aeruginosa can persist in a range of environments. Some of these may be approximated by iron-limited minimal media. For instance, experiments conducted in CAA may therefore be useful for understanding the dynamics of siderophore genotypes/phenotypes at the onset of acute infection in healthy host tissues, where iron is sequestered by high-affinity host chelators (including transferrin), or in nutrient-poor abiotic environments.
Four studies explicitly tested the effect of environmental variables on siderophore production in monoculture. Leinweber et al. (2017) showed that the ability of a double pyoverdine/pyochelin mutant to cheat in CAA is curtailed when the medium is not made strictly iron limited with apotransferrin. Brockhurst et al. (2008) showed that increasing resource supply (carbohydrates and amino acids) reduced the cost of siderophore production and so reduced the cheating benefits of siderophore loss. Dumas et al. (2013) dissected the relative roles of pyoverdine and pyochelin, suggesting that while pyoverdine is the primary siderophore expressed under severe iron limitation, under moderate iron limitation pyochelin production dominates. The authors used parameters estimated from their experiments to simulate wild-type—siderophore mutant competition in a range of environmental scenarios and found that the fitness consequences of siderophore-null mutations varied. On the basis of these results, the authors suggested that pyoverdine-deficient mutants may be favoured in chronic infection due to better growth in high-iron, low-pH environments, rather than because they are cheats. The importance of environmental iron regime and pyoverdine/pyochelin switching in determining the fitness consequences of siderophore mutants has also been explored in a suite of experiments using the closely-related bacterium P. fluorescens: pyoverdine mutants of this species were shown to cheat under severe, but not moderate, iron limitation (Zhang and Rainey, 2013; Kümmerli and Ross-Gillespie, 2014). Finally, Vasse et al. (2017) showed that the presence of an antibiotic could enhance the fitness benefits accruing to a pyoverdine mutant in co-culture with the wild type.
Further, the strength of siderophore cheating may be constrained. Cheating could become self-limiting as increases in mutant frequency alter the cost:benefit ratio of losing siderophore production, or the cost:benefit ratio may be different in actively-growing versus established populations. In many studies, in Table 1 that explored a range of starting frequencies, mutants act as cheats only when they are initially inoculated at frequencies⩽0.1, which raises questions about the ability of these mutants to cheat at and persist at higher frequencies (although a few studies did report cheating from very high starting frequencies—for example, Kümmerli et al., 2009b; Leinweber et al., 2017). In some cases, even though co-culture increased mutant relative fitness to>1, there was no detrimental effect on total population density. These observations call into question the ability of siderophore mutants to act as ‘Trojan horses’ to treat infection, as does the observation in one study (Ghoul et al., 2016) that a siderophore mutant only cheats if added to wild-type cultures before stationary phase.
Therefore, whether siderophore-null mutants act as cheats in chronic infection—and why apparently comparable experiments can give different results—remains an open question. We identified two key areas that require close attention in order to gain a more realistic picture of the consequences of siderophore null mutations in chronic infections.
First, the repeated use of PAO6609 is potentially problematic. This is a UV-induced mutant derived from PAO6049 (Hohnadel et al., 1986), which is a methionine auxotroph derived from PAO1 (Rella et al., 1985) following several deliberate and spontaneous mutational events. We have reconstructed the complex genealogy of PAO6609 and provide this information as a flow chart in Supplementary Figure S1. The exact nature of the mutation leading to the siderophore-negative phenotype in PAO6609 is not known. Further, as UV mutagenesis is non-specific, it is likely that this strain carries additional mutations that influence other important aspects of its phenotype, or moderate the fitness consequences of siderophore loss. Without knowing the exact genotype of PAO6609, we cannot be sure that empirical results obtained using it are the result of its siderophore phenotype alone.
Second, while some of the studies in Table 1 explicitly addressed how environmental variables affect siderophore dynamics, none of them set out to model conditions relevant to chronic infection. It is possible that increased availability of iron in chronically-damaged tissues means that the benefits of siderophore production are reduced and that production is downregulated, removing any advantage to siderophore-null genotypes. Alternatively, siderophores may be beneficial but not exploitable by potential cheats: this could result from spatial structuring, persistence in stationary phase, and/or a reliance on siderophore recycling rather than continued production (Kümmerli and Brown, 2010).
Whole-genome sequence of a commonly used siderophore cheat, PAO6609
We performed whole-genome sequencing of this strain and mapped the raw sequence data against a PAO1 reference genome (NCBI Reference Sequence: NC_002516.2, Stover et al., 2000). The results are summarised in Figure 1. PAO6609 harbours a single ~6.6 kb deletion, removing most of the non-ribosomal peptide synthetase locus pvdJ, which is involved in synthesising the pyoverdine side chain. This deletion also removes part of a second pyoverdine biosynthetic locus, pvdI. We also found 90 high-resolution SNPs, including expected mutations in amiE and metZ (Supplementary Figure S1). The details of the deletion and SNPS are provided as Supplementary Information. Briefly, none of the SNPs were in loci associated with pyoverdine or pyochelin biosynthesis. However, we found SNPs resulting in missense mutations in ten genes. These were cyoE, which is involved in heme metabolism and upregulated when iron is low (Arai, 2011); two loci, which ancide an assimilatory nitrate reductase involved in anaerobic ATP generation (napA, napF); two loci involved with assimilation of nitrogen for biosynthesis (cysG, PA1779); two loci involved in quorum sensing (lasI, qteE); two loci involved in motility (fliO, fimV); and the cpo gene, which is involved in the biosynthesis of many organochlorine compounds. All of these mutations could potentially affect growth and/or virulence. Finally, we found a missense mutation in the rho transcription termination factor. Further work would be needed to deduce the effects of this mutation, but the possibility of generalised defects in the control of gene expression cannot be ruled out. In summary, PAO6609 harbours numerous mutations in addition to the pvdJ partial deletion that likely make it an unreliable strain to use for empirical studies of the fitness and virulence consequences of siderophore loss.
Production of siderophores by P. aeruginosa in models of chronic lung and wound infections
Because of the problems identified with PAO6609, we used defined siderophore mutants to test the potential for social cheating in laboratory media that have been developed to represent specific chronic infection contexts. As discussed above, while most research has focused on pyoverdine, under weaker iron limitation, the metabolically cheaper siderophore pyochelin may take on a primary role in iron chelation (Dumas et al., 2013; Kümmerli and Ross-Gillespie, 2014). We therefore used a standard wild-type lab strain (PAO1) and two isogenic deletion mutants: a pyoverdine knockout (ΔpvdD) and a double pyoverdine/pyochelin knockout (ΔpvdDΔpchEF) constructed using allelic exchange (Ghysels et al., 2004). Both mutants have been reported to act as social cheats in iron-limited CAA, when inoculated in co-culture with the wild type at starting frequencies of up to 50% (Table 1,Kümmerli et al., 2009b, 2015; Kümmerli and Brown, 2010).
We first verified that PAO1 produces siderophores in our chronic infection models, and compared production levels with those in iron-limited CAA. Detectable levels of pyoverdine and pyochelin were produced in all environments (Supplementary Figure S2a). Cell densities also differed between environments (Supplementary Figure S2b). Therefore, we calculated relative production of siderophores per CFU and used this measure of production to compare investment between the different growth environments. Per-cell levels of pyoverdine and pyochelin were positively correlated and, consistent with the suggestion of Dumas et al. (2013); the pyochelin:pyoverdine ratio was significantly higher in chronic infection models and media than in CAA (Supplementary Figure S2a). (Note that replica cultures in ex vivo lung and synthetic wounds were more variable than replica cultures in liquid media).
Social dynamics of siderophore mutants in artificial CF sputum are influenced by genotype and culture time
Mutant and wild-type bacteria were grown in pure culture and each mutant was grown in mixed culture with the wild type, with a starting frequency of 50%, in ASM. After 24 and 48 h of growth, total population densities were determined and the relative fitness of each mutant in pure and mixed culture calculated. As shown in Figure 2a, neither mutant had a relative fitness significantly different from 1 when grown in pure culture. ΔpvdD mutant fitness was unaffected by the wild type, but the ΔpvdDΔpchEF mutant was outcompeted in mixed culture. These results are not consistent with either mutant acting as a cheat: they do not show a disadvantage when cultured alone, and do not benefit from growth with the wild type.
After 48 h (Figure 2b), the results were different. The ΔpvdD mutant now showed cheating dynamics: it was less fit than the wild type when grown in pure culture but as fit as the wild type in mixed culture. The ΔpvdDΔpchEF mutant had a relative fitness <1 in both pure and mixed culture. As predicted for a textbook ‘cheat,’ the presence of the ΔpvdD mutant reduced total population density: mixed cultures reached similar densities to pure ΔpvdD cultures (Figure 2c).
Time course experiments (Supplementary Figure S3) suggested that the ΔpvdD mutant has a disadvantage in pure culture in ASM because it ceases logarithmic growth earlier than the wild type, reaching a lower yield in stationary phase. The ΔpvdDΔpchEF mutant has the double disadvantage of a longer lag phase and an earlier cessation of logarithmic growth. This is consistent with the differing fitness of the mutants at 24 vs 48 h: continued growth and siderophore production by the wild type from 24–48 h presumably provide an opportunity for the ΔpvdD mutant to cheat, as levels of available iron start to fall.
Siderophore mutants are outcompeted by the wild type in a CF bronchiolar biofilm model
ASM models the chemistry of CF mucus (Palmer et al., 2007; Turner et al., 2015), but liquid culture lacks realistic spatial structure. To allow bacteria to form biofilms associated with bronchiolar surfaces (Bjarnsholt et al., 2013), we repeated the monoculture and 50% co-culture experiments in an ex vivo model comprising a section of pig bronchiole cultured in ASM for 4 days. P. aeruginosa forms a loose sleeve of mucoid biofilm around the tissue (Harrison and Diggle, 2016). As shown in Figure 3, the only significant predictor of relative fitness was presence/absence of the wild type. Both strains were as fit as the wild type in pure culture but showed a trend towards being outcompeted in mixed culture. This trend was significant for the ΔpvdDΔpchEF mutant but not for the ΔpvdD mutant.
Siderophore mutants do not suffer a long-term fitness disadvantage in synthetic wound fluid
We next repeated the pure/mixed culture experiments in synthetic wound fluid (SWF: Werthén et al., 2010). The only significant predictor of relative fitness at 24 h was presence/absence of the wild type (Figure 4a). The ΔpvdD mutant was as fit as the wild type in pure culture, and the ΔpvdDΔpchEF mutant slightly and significantly fitter, but both mutants were outcompeted by the wild type in mixed culture. After 48 h (Figure 4b), there was no difference between genotypes or culture conditions: both mutants had a relative fitness of 1 regardless of wild-type presence. Dropping two outliers from the 48-h data set resulted in tighter confidence intervals but did not change the results.
Siderophore mutants do not have any fitness disadvantage in synthetic chronic wounds
As with the CF lung model, we wished to add spatial structure to SWF to better model a soft tissue infection. SWF was solidified with collagen, allowing bacteria to form 3D biofilm, (Werthén et al., 2010) and experiments repeated in the resulting solid plugs. After 48 h in synthetic wounds, (Figure 5), both mutants had a relative fitness of 1 regardless of culture condition.
Discussion
Many experiments have been conducted to explore the social evolution of siderophore production by P. aeruginosa. These have been used to suggest explanations for the appearance of siderophore mutants in chronic infections; and how this behaviour could be exploited for clinical ends. Our targeted literature review revealed potential restrictions on the generality of predictions made from these experiments about the dynamics of siderophore mutants in vivo. First, many experiments used an undefined mutant (PA6609) as a siderophore cheat. We sequenced this strain and found that while it has a deletion of one of the pyoverdine biosynthetic loci, it also carries numerous mutations in genes likely to affect growth and metabolism in unpredictable ways. It is therefore difficult to disentangle the effects of siderophore phenotype vs other phenotypes on its evolutionary dynamics. Second, most published experiments were carried out in iron-limited minimal broth. Inferences made from these experiments may provide information on the fitness consequences of siderophore loss in acute infections (Granato et al., 2016). However, a growing body of evidence shows a significant mismatch between contrived lab media and clinically-relevant growth conditions. In chronic infections, bacteria experience specialised and idiosyncratic environments that affect the cost:benefit ratio of siderophore production, the ability of cells to access each other’s siderophores and the growth rate of bacterial populations. Experiments that dissect the fitness consequences of siderophore loss in environmentally-explicit models of infection are essential if we are to understand the natural history of siderophore production in chronic infections. Defined chronic infection models are also vital if we want to draw clinically-relevant conclusions about the likely success of therapeutic interventions based on disrupting bacterial social interactions or using ‘public good’ mutants as Trojan horses.
We tested whether pyoverdine or pyoverdine/pyochelin deletion mutants acted as cheats in four laboratory models designed to mimic chronic infections: structured and unstructured models of CF lung infections and non-healing soft-tissue wounds. Wild-type P. aeruginosa produced siderophores in all tested environments. However, there was environment-dependent variation in siderophore production, and the ratio of pyochelin:pyoverdine was higher in chronic infection models than in CAA (Supplementary Figure S1). This is consistent with a suggestion by other authors that pyochelin production may be favoured when iron is more easily available (Dumas et al., 2013; Hunter et al., 2013; McCallin et al., 2015). The pyoverdine mutant was less fit than the wild type in monoculture in artificial CF sputum after 48 h growth, demonstrating a benefit to pyoverdine production in this environment, and was able to cheat on the wild type. In all other environments tested, there was no effect of losing pyoverdine production on fitness. Production of pyoverdine in these environments may therefore be a maladaptive response, or may have benefits other than simple growth rate enhancements (for example, copper detoxification: Koh and Henderson, 2015). There was a non-significant trend towards this mutant being outcompeted by the wild type in ex vivo bronchiolar biofilm. This may be due to poor biofilm production, even when iron is plentiful (Banin et al., 2005; Harrison and Buckling, 2009). The pyoverdine/pyochelin mutant was less fit than the wild type in artificial CF sputum and model bronchiolar biofilms, but had no disadvantage in synthetic wound fluid or synthetic wound biofilm. This is most likely due to ready availability of iron as heme groups in serum. Our results cast significant doubt on the ability of siderophore mutants to cheat in chronic infections, and therefore on their utility as Trojan horses.
The models we use are not perfect, but they have been carefully constructed to reflect key aspects of CF lung or wound physiology and chemistry, and validated by comparing bacterial growth, gene expression, metabolism and/or biofilm morphology with qualitative or quantitative data from clinical fluids or tissues (Palmer et al., 2007; Werthén et al., 2010; Harrison and Diggle, 2016; Price et al., 2016). Alternative optimised media are available (for example, other versions of synthetic CF sputum: Sriramulu et al., 2005; Turner et al., 2015 and Kirchner et al., 2012), and P. aeruginosa can be grown in chronic infection-like media and models for long time periods, with different studies using various growth periods. We decided to focus on the four media presented, and conduct our experiments over the most commonly used time periods (based on our sampling of the literature) for each growth medium or model. Clearly, there are many variables to be considered with regard to modelling chronic infection environments, and we stress that the present article presents the data from a representative sample of potential models. We would not venture to argue that the conditions we use are the ‘correct’ ones: rather, we present our results as examples of how improved lab models of specific infection contexts can be developed and used, and to demonstrate how different the fitness effects of a bacterial phenotype can be in different environments.
When increased levels of public goods production increase the population growth rate or carrying capacity, cheating mutants are predicted to be under negative frequency-dependent selection (Ross-Gillespie et al., 2007). It is therefore usual to conduct cheating assays using a range of starting frequencies, including <50%. We chose to initially conduct cheating assays using only a 50% starting frequency, as both mutants we used have been reported to act as cheats under this condition in iron-limited CAA (Kümmerli et al., 2009b, 2015; Kümmerli and Brown, 2010). Further, if mutants that cheat from low starting frequencies cannot maintain this advantage as they become more common, then the likely clinical significance of cheating as a determinant of virulence—and the potential power of a ‘Trojan horse’ approach to managing infection—is called into question. Further investigation of different starting frequencies was not necessary: in the case of ASM, the single mutant showed cheating dynamics even at this high starting frequency, and in all other media, both mutants had equal fitness to the wild type in pure culture so cannot be called cheats regardless of the outcome of competition at any starting frequency.
Our results underline the importance of the in-host environment in determining bacterial genotype-fitness maps, and hence the necessity of carefully-designed lab models (Roberts et al., 2015). We previously reported that P. aeruginosa QS mutants, which can cheat in acute infections (Rumbaugh et al., 2009), are not cheats in an ex vivo model of chronic infection (Harrison et al., 2014). This is consistent with previously published in vitro experiments showing that the fitness effects of QS mutations depend on the specific carbon sources present (D’Argenio et al., 2007; Ghoul et al., 2014). Together with the present results, this demonstrates the unreliability of extrapolating predictions about bacterial social evolution from one infection context to another, or from simple lab media to specialised in vivo environments. In chronic lung and wound biofilm infections, availability of free iron, weakly-chelated iron or heme most likely reduces the benefit of producing siderophores and so production levels (Supplementary Figure S2). If siderophores are less important for growth, and/or reduced production and strong spatial structure in tissue- or mucus-embedded biofilms restrict siderophore availability, then clinical strategies to reduce bacterial load based on manipulating siderophore social interactions will not work.
So why do siderophore-null mutants arise during chronic infection? Are they cheats, or simply better adapted to local growth conditions? Are they not selected at all, but present transiently and/or at low frequencies? We cannot answer these questions with the current data.
Careful choice and optimisation of in vitro models that allow for long-term evolution experiments in realistic environments will be invaluable in providing answers. Alongside such models, there is also a need for quantitative, rather than qualitative, data from patients on the prevalence of siderophore mutants (see Mowat et al., 2011 for an example of a quantitative approach to studying haplotype turnover in CF). There is also a need for more considered choice of ‘wild type’ and mutant genotypes, paying particular attention to genotypes that commonly arise during chronic infection. The fitness consequences of siderophore loss could depend on the nature of the mutations involved, and on background genotype. In future, it would be useful to conduct experiments with a range of clinical isolates that are most typical of those seen in patients, and/or with constructed mutants that recapitulate these. Further, future experiments using chronic infection models should also consider the impact of inter-species interactions in polymicrobial chronic infections communities: P. aeruginosa is part of a diverse microbial flora in CF lung and chronic wound biofilms, and interactions with co-infecting microbes have the potential to affect the costs and benefits of siderophore production (for example, Weaver and Kolter, 2004; Harrison et al., 2008).
For instance, we and other authors have focussed on experiments with mutants with impaired siderophore biosynthesis. However, a recent study of siderophore mutants isolated from CF patients found that the majority carried mutations in the regulatory gene pvdS (Andersen et al., 2015). Sequencing clones from an in vitro evolution experiment also revealed pyoverdine-negative phenotypes that stemmed from pvdS mutation (Kümmerli et al., 2015). PvdS is a sigma factor that positively regulates the expression of pyoverdine biosynthesis under iron starvation (Miyazaki et al., 1995), but which also positively regulates the expression of other virulence-related exoproducts (Ochsner et al., 1996; Wilson et al., 2001; Hunt et al., 2002; Gaines et al., 2007).
Longitudinal sputum sampling from CF patients (Andersen et al., 2015) showed that clones with mutations that reduce or ablate pyoverdine production (mainly in pvdS) initially retain receptors for iron-loaded pyoverdine, but gain mutations in the receptor once pyoverdine-producing clones disappear from the sampled populations. This was interpreted by the authors as evidence for cheating driving the loss of siderophores. But in the absence of explicit tests for cheating by these isolates in co-culture with co-isolated wild types, using growth media that mimics lung biofilm, the longitudinal pattern presented does not unequivocally prove this conclusion. Other possible explanations include early selection for loss of pvdS due to other downstream phenotypes and/or reduced transcriptional costs; a relaxation of purifying selection for functional siderophore receptors in the late stages of infection when iron is plentiful; a bias in the data set towards early isolates, which increases the conditional probability that later isolates are also siderophore-null; or a combination of these factors.
Our results demonstrate the necessity of evaluating and modelling in-host environments as carefully as possible if we aim to understand in-host microbiology. Lab experiments in simple in vitro conditions have limited ecological and clinical validity, and so limited predictive power. A plethora of data on the chemical and microbial ecology of chronic infection is now available, providing abundant material for researchers wishing to study the natural history of pathogens in the lab (for example, for CF: Yang et al., 2011; Folkesson et al., 2012; Quinn et al., 2014; Cowley et al., 2015; Kyle et al., 2015; Roberts et al., 2015; Turner et al., 2015; Flynn et al., 2016). This information is ripe for consideration by microbiologists who justifiably see great potential for manipulating the in-host ecology of pathogens in order to halt the progression of debilitating chronic infection.
References
Andersen SB, Marvig RL, Molin S, Krogh Johansen H, Griffin AS . (2015). Long-term social dynamics drive loss of function in pathogenic bacteria. Proc Natl Acad Sci 112: 10756–10761.
Arai H . (2011). Regulation and function of versatile aerobic and anaerobic respiratory metabolism in Pseudomonas aeruginosa. Front Microbiol 2: 103.
Banin E, Vasil ML, Greenberg EP . (2005). Iron and Pseudomonas aeruginosa biofilm formation. Proc the Natl Acad Sci USA 102: 11076–11081.
Bjarnsholt T . (2013). The role of bacterial biofilms in chronic infections. APMIS 121: 1–58.
Bjarnsholt T, Alhede M, Alhede M, Eickhardt-Sørensen SR, Moser C, Kühl M et al. (2013). The in vivo biofilm. Trends Microbiol 21: 466–474.
Boucher RC . (2007). Airway surface dehydration in cystic fibrosis: pathogenesis and therapy. Annu Rev Med 58: 157–170.
Brockhurst MA, Buckling A, Racey DA, Gardner A . (2008). Resource supply and the evolution of public-goods cooperation in bacteria. BMC Biol 6: 20.
Brown SP, West SA, Diggle SP, Griffin AS . (2009). Social evolution in micro-organisms and a Trojan horse approach to medical intervention strategies. PhilosTrans R Soc B Biol Sci 364: 3157–3168.
Cornelis P, Dingemans J . (2013). Pseudomonas aeruginosa adapts its iron uptake strategies in function of the type of infections. Front Cell Infect Microbiol 14: 75.
Marvig RL, Damkiær S, Khademi SMH, Markussen TM, Molin S, Jelsbak L . (2014). Within-host evolution of Pseudomonas aeruginosa reveals adaptation toward iron acquisition from hemoglobin. mBio 5: e00966–14.
Cowley ES, Kopf SH, LaRiviere A, Ziebis W, Newman DK . (2015). Pediatric cystic fibrosis sputum can be chemically dynamic, anoxic, and extremely reduced due to hydrogen sulfide formation. mBio 6: e00767–15.
Croucher NJ, Thomson NR . (2010). Studying bacterial transcriptomes using RNA-seq. Curr Opin Microbiol 13: 619–624.
D’Argenio DA, Wu M, Hoffman LR, Kulasekara HD, Déziel E, Smith EE et al. (2007). Growth phenotypes of Pseudomonas aeruginosa lasR mutants adapted to the airways of cystic fibrosis patients. Mol Microbiol 64: 512–533.
Darch SE, West SA, Winzer K, Diggle SP . (2012). Density-dependent fitness benefits in quorum-sensing bacterial populations. Proc Natl Acad Sci USA 109: 8259–8263.
De Vos D, De Chial M, Cochez C, Jansen S, Tummler B, Meyer JM, Cornelis P . (2001). Study of pyoverdine type and production by Pseudomonas aeruginosa isolated from cystic fibrosis patients: prevalence of type II pyoverdine isolates and accumulation of pyoverdine-negative mutations. Arch Microbiol 175: 384–388.
Diggle SP, Griffin AS, Campbell GS, West SA . (2007). Cooperation and conflict in quorum-sensing bacterial populations. Nature 450: 411–414.
Dötsch A, Schniederjans M, Khaledi A, Hornischer K, Schulz S, Bielecka A et al. (2015). The Pseudomonas aeruginosa transcriptional landscape is shaped by environmental heterogeneity and genetic variation. mBio 6: e00749–00715.
Dumas Z, Kümmerli R . (2012). Cost of cooperation rules selection for cheats in bacterial metapopulations. J Evol Biol 25: 473–484.
Dumas Z, Ross-Gillespie A, Kümmerli R . (2013). Switching between apparently redundant iron-uptake mechanisms benefits bacteria in changeable environments. Proc R Soc B Biol Sci 280: 20131055.
Flynn JM, Niccum D, Dunitz JM, Hunter RC . (2016). Evidence and role for bacterial mucin degradation in cystic fibrosis airway disease. PLoS Pathog 12: e1005846.
Folkesson A, Jelsbak L, Yang L, Johansen HK, Ciofu O, Hoiby N et al. (2012). Adaptation of Pseudomonas aeruginosa to the cystic fibrosis airway: An evolutionary perspective. Nat Rev Microbiol 10: 841–851.
Foster KR . (2005). Hamiltonian medicine: Why the social lives of pathogens matter. Science 308: 1269–1270.
Fox J, Weisberg S . (2011)An R Companion to Applied Regression. SAGE Publications: Thousand Oaks, CA, USA.
Friedrich M, Lessnau K-D, Cunha BA . (2015), Pseudomonas aeruginosa Infections. In: Talavera F, Brusch JL (eds). Medscape Drugs and Diseases. Available at: http://emedicine.medscape.com/article/226748.
Gaines JM, Carty NL, Tiburzi F, Davinic M, Visca P, Colmer-Hamood JA et al. (2007). Regulation of the Pseudomonas aeruginosa toxA regA and ptxR genes by the iron-starvation sigma factor PvdS under reduced levels of oxygen. Microbiology 153: 4219–4233.
Ghoul M, West SA, Diggle SP, Griffin AS . (2014). An experimental test of whether cheating is context dependent. J Evol Biol 27: 551–556.
Ghoul M, West SA, McCorkell FA, Lee Z-B, Bruce JB, Griffin AS . (2016). Pyoverdin cheats fail to invade bacterial populations in stationary phase. J Evol Biol 29: 1728–1736.
Ghysels B, Dieu BTM, Beatson SA, Pirnay JP, Ochsner UA, Vasil ML et al. (2004). FpvB, an alternative type I ferripyoverdine receptor of Pseudomonas aeruginosa. Microbiology 150: 1671–1680.
Goodman AL, Kulasekara B, Rietsch A, Boyd D, Smith RS, Lory S . (2004). A signaling network reciprocally regulates genes associated with acute infection and chronic persistence in Pseudomonas aeruginosa. Dev Cell 7: 745–754.
Granato ET, Harrison F, Kümmerli R, Ross-Gillespie A . (2016). When is a bacterial ‘virulence factor’ really virulent? bioRxiv doi:10.1101/061317.
Griffin AS, West SA, Buckling A . (2004). Cooperation and competition in pathogenic bacteria. Nature 430: 1024–1027.
Hamilton WD . (1964). The genetical evolution of social behaviour I & II. J Theor Biol 7: 1–52.
Harrison F, Browning LE, Vos M, Buckling A . (2006). Cooperation and virulence in acute Pseudomonas aeruginosa infections. BMC Biol 4: 21.
Harrison F, Paul J, Massey RC, Buckling A . (2008). Interspecific competition and siderophore-mediated cooperation in Pseudomonas aeruginosa. ISME J 2: 49–55.
Harrison F, Buckling A . (2009). Siderophore production and biofilm formation as linked social traits. ISME J 3: 632–634.
Harrison F, Buckling A . (2011). Wider access to genotypic space facilitates loss of cooperation in a bacterial mutator. PLoS One 6: e17254.
Harrison F, Muruli A, Higgins S, Diggle SP . (2014). Development of an ex vivo porcine lung model for studying growth Virulence, And signaling of Pseudomonas aeruginosa. Infect Immun 82: 3312–3323.
Harrison F, Diggle S . (2016). An ex vivo lung model to study bronchioles infected with Pseudomonas aeruginosa biofilms. Microbiology. 162: 1755–1760.
Hirsch EB, Tam VH . (2010). Impact of multidrug-resistant Pseudomonas aeruginosa infection on patient outcomes. Expert Rev Pharmacoecon Outcomes Res 10: 441–451.
Hohnadel D, Haas D, Meyer JM . (1986). Mapping of mutations affecting pyoverdine production in Pseudomonas aeruginosa. FEMS Microbiol Lett 36: 195–199.
Hothorn T, Bretz F, Westfall P . (2008). Simultaneous inference in general parametric models. Biom J 50: 346–363.
Hunt TA, Peng W-T, Loubens I, Storey DG . (2002). The Pseudomonas aeruginosa alternative sigma factor PvdS controls exotoxin A expression and is expressed in lung infections associated with cystic fibrosis. Microbiology 148: 3183–3193.
Hunter RC, Asfour F, Dingemans J, Osuna BL, Samad T, Malfroot A, Cornelis P, Newman DK . (2013). Ferrous iron is a significant component of bioavailable iron in cystic fibrosis airways. mBio 4: e00557–13.
Jiricny N, Diggle SP, West SA, Evans BA, Ballantyne G, Ross-Gillespie A et al. (2010). Fitness correlates with the extent of cheating in a bacterium. J Evol Biol 23: 738–747.
Jiricny N, Molin S, Foster K, Diggle SP, Scanlan PD, Ghoul M et al. (2014). Loss of social behaviours in populations of Pseudomonas aeruginosa infecting lungs of patients with cystic fibrosis. PLoS One 9: e83124.
Koh E-I, Henderson JP . (2015). Microbial Copper-binding siderophores at the host-pathogen interface. J Biol Chem 290: 18967–18974.
Kirchner S, Fothergill JL, Wright EA, James CE, Mowat E, Winstanley C . (2012). Use of artificial sputum medium to test antibiotic efficacy against Pseudomonas aeruginosa in conditions more relevant to the cystic fibrosis lung. J Vis Exp 64: 3857.
Kümmerli R, Gardner A, West SA, Griffin AS . (2009a). Limited dispersal, budding dispersal, and cooperation: an experimental study. Evolution 63: 939–949.
Kümmerli R, Jiricny N, Clarke LS, West SA, Griffin AS . (2009b). Phenotypic plasticity of a cooperative behaviour in bacteria. J Evol Biol 22: 589–598.
Kümmerli R, Griffin AS, West SA, Buckling A, Harrison F . (2009c). Viscous medium promotes cooperation in the pathogenic bacterium Pseudomonas aeruginosa. Proc R Soc B Biol Sci 276: 3531–3538.
Kümmerli R, Brown SP . (2010). Molecular and regulatory properties of a public good shape the evolution of cooperation. Proc Natl Acad Sci USA 107: 18921–18926.
Kümmerli R, Van Den Berg P, Griffin AS, West SA, Gardner A . (2010). Repression of competition favours cooperation: experimental evidence from bacteria. J Evol Biol 23: 699–706.
Kümmerli R, Ross-Gillespie A . (2014). Explaining the sociobiology of pyoverdin producing Pseudomonas: a comment on Zhang and Rainey (2013). Evolution 68: 3337–3343.
Kümmerli R, Santorelli LA, Granato ET, Dumas Z, Dobay A, Griffin AS et al. (2015). Co-evolutionary dynamics between public good producers and cheats in the bacterium Pseudomonas aeruginosa. J Evol Biol 28: 2264–2274.
Leinweber A, Fredrik Inglis R, Kummerli R . (2017). Cheating fosters species co-existence in well-mixed bacterial communities. ISME J 11: 1179–1188.
Lenth RV . (2013), lsmeans: R Package Version 1.06-05.
Luján AM, Gómez P, Buckling A . (2015). Siderophore cooperation of the bacterium Pseudomonas fluorescens in soil. Biol Lett 11: 20140934.
Luong PM, Shogan BD, Zaborin A, Belogortseva N, Shrout JD, Zaborina O et al. (2014). Emergence of the P2 phenotype in Pseudomonas aeruginosa PAO1 strains involves various mutations in MexT or MexF. J Bacteriol 196: 504–513.
McCallin K, Cowley E, Reyes MC, Van Sambeek L, Hunter R, Asfour F et al. (2015), Sputum iron levels during cystic fibrosis pulmonary exacerbation: a longitudinal study. American Thoracic Society International Conference Abstracts B52: Pediatric Cystic Fibrosis pp A3343-A3343.
Miyazaki H, Kato H, Nakazawa T, Tsuda M . (1995). A positive regulatory gene, pvdS, for expression of pyoverdin biosynthetic genes in Pseudomonas aeruginosa PAO. Mol Gen Genet MGG 248: 17–24.
Morgan AD, Quigley BJZ, Brown SP, Buckling A . (2012). Selection on non-social traits limits the invasion of social cheats. Ecol Lett 15: 841–846 xw.
Mowat E, Paterson S, Fothergill JL, Wright EA, Ledson MJ, Walshaw MJ, Brockhurst MA, Winstanley C . (2011). Pseudomonas aeruginosa population diversity and turnover in cystic fibrosis chronic infections. Am J Respir Crit Care Med 183: 1674–1679.
Mund A, Diggle SP, Harrison F . (2017). The fitness of Pseudomonas aeruginosa quorum sensing signal cheats is influenced by the diffusivity of the environment. mBio 8: e00353–17.
Ochsner UA, Johnson Z, Lamont IL, Cunliffe HE, Vasil ML . (1996). Exotoxin A production in Pseudomonas aeruginosa requires the iron-regulated pvdS gene encoding an alternative sigma factor. Mol Microbiol 21: 1019–1028.
Palmer KL, Mashburn LM, Singh PK, Whiteley M . (2005). Cystic fibrosis sputum supports growth and cues key aspects of Pseudomonas aeruginosa physiology. J Bacteriol 187: 5267–5277.
Palmer KL, Aye LM, Whiteley M . (2007). Nutritional cues control Pseudomonas aeruginosa multicellular behavior in cystic fibrosis sputum. J Bacteriol 189: 8079–8087.
Price BL, Lovering AM, Bowling FL, Dobson CB . (2016). Development of a novel collagen wound model to simulate the activity and distribution of antimicrobials in soft tissue during diabetic foot infection. Antimicrob Agents Chemother 60: 6880–6889.
Quinn RA, Lim YW, Maughan H, Conrad D, Rohwer F, Whiteson KL . (2014). Biogeochemical forces shape the composition and physiology of polymicrobial communities in the cystic fibrosis lung. mBio 5: e00956–13.
R Development Core Team (2016)R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing: Vienna, Austria. Available at: http://www.R-project.org.
Rainey PB, Rainey K . (2003). Evolution of cooperation and conflict in experimental bacterial populations. Nature 425: 72–74.
Raymond B, West SA, Griffin AS, Bonsall MB . (2012). The dynamics of cooperative bacterial virulence in the field. Science 336: 85–88.
Rella M, Mercenier A, Haas D . (1985). Transposon insertion mutagenesis of Pseudomonas aeruginosa with a Tn5 derivative: application to physical mapping of the arc gene cluster. Gene 33: 293–303.
Roberts AEL, Kragh KN, Bjarnsholt T, Diggle SP . (2015). The limitations of in vitro experimentation in understanding biofilms and chronic infection. J Mol Biol 427: 3646–3661.
Ross-Gillespie A, Gardner A, West SA, Griffin AS . (2007). Frequency dependence and cooperation: Theory and a test with bacteria. Am Nat 170: 331–342.
Ross-Gillespie A, Gardner A, Buckling A, West SA, Griffin AS . (2009). Density dependence and cooperation: theory and a test with bacteria. Evolution 63: 2315–2325.
Ross-Gillespie A, Dumas Z, Kümmerli R . (2015). Evolutionary dynamics of interlinked public goods traits: an experimental study of siderophore production in Pseudomonas aeruginosa. J Evol Biol 28: 29–39.
Rumbaugh KP, Diggle SP, Watters CM, Ross-Gillespie A, Griffin AS, West SA . (2009). Quorum sensing and the social evolution of bacterial virulence. Curr Biol 19: 341–345.
Sriramulu DD, Lünsdorf H, Lam JS, Römling U . (2005). Microcolony formation: a novel biofilm model of Pseudomonas aeruginosa for the cystic fibrosis lung. J Med Microbiol 54: 667–676.
Stover CK, Pham XQ, Erwin AL, Mizoguchi SD, Warrener P, Hickey MJ et al. (2000). Complete genome sequence of Pseudomonas aeruginosa PAO1, an opportunistic pathogen. Nature 406: 959–964.
Székely T, Moore A, Komdeur J . (2010)Social Behaviour: Genes, Ecology and Evolution. Cambridge University Press: Cambridge, UK.
Turner KH, Wessel AK, Palmer GC, Murray JL, Whiteley M . (2015). Essential genome of Pseudomonas aeruginosa in cystic fibrosis sputum. Proc Natl Acad Sci USA 112: 4110–4115.
Tyrrell J, Callaghan M . (2016). Iron acquisition in the cystic fibrosis lung and potential for novel therapeutic strategies. Microbiology 162: 191–205.
Van Opijnen T, Bodi KL, Camilli A . (2009). Tn-seq: high-throughput parallel sequencing for fitness and genetic interaction studies in microorganisms. Nat Methods 6: 767–772.
Vasse M, Noble RJ, Akhmetzhanov AR, Torres-Barceló C, Gurney J, Benateau S et al. (2017). Antibiotic stress selects against cooperation in the pathogenic bacterium Pseudomonas aeruginosa. Proc Natl Acad Sci 114: 546–551.
Weaver VB, Kolter R . (2004). Burkholderia spp. alter Pseudomonas aeruginosa physiology through iron sequestration. J Bacteriol 186: 2376–2384.
Werthén M, Henriksson L, Jensen PØ, Sternberg C, Givskov M, Bjarnsholt T . (2010). An in vitro model of bacterial infections in wounds and other soft tissues. APMIS 118: 156–164.
West SA, Diggle SP, Buckling A, Gardner A, Griffin AS . (2007). The social lives of microbes. Annu Rev Ecol Evol Syst pp 53–77.
Whiteley M, Bangera MG, Bumgarner RE, Parsek MR, Teitzel GM, Lory S et al. (2001). Gene expression in Pseudomonas aeruginosa biofilms. Nature 413: 860–864.
Wilson MJ, McMorran BJ, Lamont IL . (2001). Analysis of promoters recognized by PvdS, an extracytoplasmic-function sigma factor protein from Pseudomonas aeruginosa. J Bacteriol 183: 2151–2155.
Yang L, Jelsbak L, Marvig RL, Damkiær S, Workman CT, Rau MH et al. (2011). Evolutionary dynamics of bacteria in a human host environment. Proc Natl Acad Sci USA 108: 7481–7486.
Zhang X-X, Rainey PB . (2013). Explaining the sociobiology of pyoverdin producing Pseudomonas. Evolution 67: 3161–3174.
Acknowledgements
This work was supported by the Universities of Nottingham and Warwick (FH), a Human Frontier Science Program Young Investigators grant to SPD (RGY0081/2012) and a NERC grant to SPD (NE/J007064/1). Funding for open access charges was provided by RCUK. We thank Pierre Cornelis for the P. aeruginosa wild type and siderophore knockout strains; Gabriella Pessi and Dieter Haas for providing documentation on historical strains; Angus Buckling for PAO6609 and Rolf Kümmerli, Stuart West, Danna Gifford, Steven Higgins and three reviewers for helpful comments on the manuscript.
Author contributions
FH and SPD conceived the study and designed experiments. FH, AM and ACS carried out experimental work. FH and AM analysed the data. SH reconstructed the genealogy of PAO6609. FH drafted the manuscript. All authors contributed to the manuscript development.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies this paper on The ISME Journal website
Rights and permissions
This work is licensed under a Creative Commons Attribution 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder o reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/
About this article
Cite this article
Harrison, F., McNally, A., da Silva, A. et al. Optimised chronic infection models demonstrate that siderophore ‘cheating’ in Pseudomonas aeruginosa is context specific. ISME J 11, 2492–2509 (2017). https://doi.org/10.1038/ismej.2017.103
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ismej.2017.103
This article is cited by
-
Copper selects for siderophore-mediated virulence in Pseudomonas aeruginosa
BMC Microbiology (2022)
-
Iron limitation by transferrin promotes simultaneous cheating of pyoverdine and exoprotease in Pseudomonas aeruginosa
The ISME Journal (2021)
-
In-vivo microscopy reveals the impact of Pseudomonas aeruginosa social interactions on host colonization
The ISME Journal (2019)