Abstract
Chaperonins are protein-folding machinery found in all cellular life. Chaperonin genes have been documented within a few viruses, yet, surprisingly, analysis of metagenome sequence data indicated that chaperonin-carrying viruses are common and geographically widespread in marine ecosystems. Also unexpected was the discovery of viral chaperonin sequences related to thermosome proteins of archaea, indicating the presence of virioplankton populations infecting marine archaeal hosts. Virioplankton large subunit chaperonin sequences (GroELs) were divergent from bacterial sequences, indicating that viruses have carried this gene over long evolutionary time. Analysis of viral metagenome contigs indicated that: the order of large and small subunit genes was linked to the phylogeny of GroEL; both lytic and temperate phages may carry group I chaperonin genes; and viruses carrying a GroEL gene likely have large double-stranded DNA (dsDNA) genomes (>70 kb). Given these connections, it is likely that chaperonins are critical to the biology and ecology of virioplankton populations that carry these genes. Moreover, these discoveries raise the intriguing possibility that viral chaperonins may more broadly alter the structure and function of viral and cellular proteins in infected host cells.
Similar content being viewed by others
Introduction
Viruses are the most numerous biological entity on Earth, influencing nutrient cycling and the evolution of host organisms (Suttle, 2007). For successful replication, many viruses rely on proteins involved in nucleotide and amino acid metabolism encoded within their genomes. One such group of viral proteins that has heretofore rarely been observed, and thus poorly characterized, are chaperonins. Chaperonins are an ancient protein family that mediates the folding of nascent or misfolded polypeptides. Chaperonins occur in two main classes: Group I chaperonins, which comprise GroEL and its co-chaperonin GroES that function as a complex and are generally found in Bacteria and eukaryotic organelles, and Group II chaperonins that are found in Archaea (thermosomes) and Eukaryotes (TRiC/CCT) (Boisvert et al., 1996; Hartl et al., 2011; Leitner et al., 2012). Similarities in domain structure and primary sequence identity support an ancient evolutionary link between GroEL and thermosomes (Willison and Kubota, 1994).
Mutant strains of Escherichia coli unable to support the growth of bacteriophage led to the initial discovery and early work on chaperonins (Georgopoulos et al., 1972; Takano and Kakefuda, 1972; Georgopoulos, 2006; Murray and Gann, 2007). Protein-folding activity increases during infection due to host stress and production of viral proteins (Poranen et al., 2006). Host chaperonins are essential for capsid and/or tail assembly for several phages having lytic, temperate and transposable lifestyles (Takano and Kakefuda, 1972; Coppo et al., 1973; Georgopoulos et al., 1973; Zweig and Cummings, 1973; Hänninen et al., 1997; Andreadis and Black, 1998; Grimaud and Toussaint, 1998; Ang et al., 2001). For T4 and RB49 bacteriophages, two large genome myoviruses, the viral-encoded GroES homologs, gp31 and CocO, respectively, are necessary for folding the major capsid protein (van der Vies et al., 1994; Ang et al., 2001). Given the importance of chaperonins in phage assembly, it is surprising so few phages are known to encode chaperonin genes. Out of the 2178 phage genomes in GenBank, only 175 carry a chaperonin gene, with most of these (165 phages) carrying the small subunit gene (GroES or functional homolog such as gp31 (van der Vies et al., 1994; Supplementary Tables 1 and 2). Only ten phages are known to carry the large subunit chaperonin gene, GroEL, with only two having both the GroEL and ES genes for the Group I chaperonin complex (Hertveldt et al., 2005; Kurochkina et al., 2012; Supplementary Table 1). The need for protein-folding systems during infection likely selects for the acquisition and evolution of viral chaperonins and other protein-folding machinery. The low frequency of viral chaperonins in sequence databases may be due to the fact that cultivated phages are not representative of abundant phages in nature (Wommack et al., 2015) and that there may be proteins functioning as chaperones that we do not yet recognize within viral genomes (Ang and Georgopoulos, 2012).
For both bacteriophage T4 and bacteriophage EL, virally-encoded chaperonins are essential for productive infection (van der Vies et al., 1994; Kurochkina et al., 2012). However, the broader distribution of chaperonins among phages infecting a more diverse range of bacterial hosts is poorly known. The deep evolutionary history of chaperonins makes them excellent phylogenetic markers, and may reveal important insights into the ecological and biological features of unknown viruses in marine ecosystems (Georgopoulos, 2006). The prevalence and diversity of chaperonin genes in viral metagenomes from distinct marine ecosystems was investigated to ascertain the importance of chaperonins within the virioplankton. These data were interpreted with the goal of discovering how these ancient and essential genes may shape the biological features of virioplankton populations.
Materials and methods
Viral metagenome libraries
We examined sequences from the Chesapeake Bay (CFA-CFH) (Schmidt et al., 2014; Sakowski et al., 2014), Dry Tortugas (Sakowski et al., 2014; DTF), Gulf of Maine (Sakowski et al., 2014; GMF) and Pacific Ocean (Hurwitz and Sullivan, 2013; POF/STCS) viral metagenomes (viromes), available on the Viral Informatics Resource for Metagenome Exploration (VIROME, virome.dbi.udel.edu) (Wommack et al., 2012). Virome libraries from Raunefjordern, Norway (DYM) and the North Sea (SDO, YBW) are available on the Metagenomes Online (MgOL) database (metagenomesonline.org).
Isolation and sequencing of the SERC virome
In December 2012, fifty liters of surface water was collected from the Rhode River near the Smithsonian Environmental Research Center (SERC) in Edgewater, MD, USA. Viruses were concentrated using the FeCl3 flocculation method (John et al., 2010) and further purified by 0.2 μm filtration, ultrafiltration (Amicon Ultra 100 kDa, Millipore, Millerica, MA, USA) and three rounds of Ambion DNase treatment (Invitrogen, Carlsbad, CA, USA). DNA was isolated using phenol-chloroform extraction and ethanol precipitation.
Extracted DNA was prepared for Illumina sequencing using linker amplification. DNA was sheared using adaptive focused acoustics (Covaris, Woburn, MA, USA) and purified using Agencourt Ampure XP beads (Beckman Coulter, Brea, CA, USA) and BluePippin DNA size selection (Sage Sciences, Beverly, MA, USA). End repair, dA tailing and adapter ligation was performed using NEBNext DNA sample Prep Reagent Set (New England Biolabs, Ipswich, MA, USA) and barcoded NEXTflex Illumina-compatible adapters (Bioo Scientific, Austin, TX, USA). Eight cycles of PCR amplification were preformed following manufacturer recommendations (NEBNext, New England Biolabs) utilizing the adapters as priming sequences. The amplified library was sequenced on the Illumina HiSeq platform at the University of Delaware Sequencing and Genotyping Center.
SERC virioplankton DNA was also prepared for sequencing on the PacBio platform using the standard SMRTbell library preparation and terminal deoxynucleotidyl transferase (TdT) methods with a 10 kb target fragment size. In the standard library preparation, DNA was sheared to 10 kb using a Covaris g-Tube (Woburn, MA, USA) followed by DNA end repair, SMRTbell adapter ligation and sequence primer annealing performed as outlined in the PacBio 10 kb template preparation and sequencing protocol (http://www.pacb.com/support/documentation/). In the TdT method, DNA was sheared, followed by BluePippin size selection. PolyA tails were added to DNA fragments to facilitate MagBead loading of DNA templates into sequencing wells and as priming sites for sequencing. This method was optimized for smaller starting amounts of starting DNA (~300 ng sheared DNA). Libraries were sequenced at Pacific Biosciences (Menlo Park, CA, USA).
Bioinformatic assembly of SERC, Norway and North Sea viromes
Both Illumina and PacBio reads comprised the SERC data set. Before assembly, low quality bases and adapter sequences were trimmed from the Illumina data using CLC Genomics Workbench version 6.0.2 (https://www.w.qiagenbioinformatics.com). High accuracy unitigs were generated from the assembly of over 150 million paired-end 150 bp Illumina reads using Celera Assembler (CA) version 8.1 with recommended settings for Illumina reads (unitigger=bogart) (Myers et al., 2000). Subsequently, PacBio reads were error-corrected with the unitigs. Around 15 000 corrected PacBio reads were combined with the Illumina-only unitigs and assembled together using CA (unitigger=bogart). Only contigs ⩾600 bp were analyzed in this study.
Pyrosequencing reads from the Norway and North Sea viromes were first filtered to remove sequences with ⩾7% ambiguous bases and duplicate reads using CD-HIT-454 at 99% identity (Niu et al., 2010). Residual adapter sequences were trimmed from the reads, and any reads showing homology to sequences in the UniVec database (e-value cutoff ⩽10−75) were removed. Filtered reads were assembled using Celera with recommended settings for 454 reads (unitigger=bog, merSize=14; Myers et al., 2000). Assembled contigs with extreme GC skew (⩽5% or ⩾95% GC) or repetitive DNA sequences were filtered before analysis.
Open reading frames (ORFs) were predicted from the contigs using MetaGeneAnnotator (Noguchi et al., 2008) and compared against a custom database of chaperonin sequences (downloaded from UniRef (The UniProt Consortium, 2015)) using BLASTp (cutoff e-value <10−3; Altschul, 1997). For gene neighbor analyses, the function of ORFs adjacent to chaperonin genes on contigs from the DYM, SDO, YBW and SERC libraries was assigned by BLASTx comparison against the NCBI nr database with an e-value cutoff of 10−3. Chaperonin-encoding contigs from SERC, DYM, SDO and YBW libraries have been deposited in Genbank (LSRR00000000; KU756931-KU756933).
Identification and assembly of chaperonin genes
For unassembled libraries, partial chaperonin genes were identified through two methods. Partial chaperonin genes from MgOL viromes (DYM, SDO and YBW) were identified by BLASTx of predicted nucleotide ORFs against a custom database of bacterial, archaeal and viral chaperonins (cutoff e-value <10−3). For metagenomes available on VIROME (CFA-CFH, DTF, GMF and POF), ORFs with a hit to GroEL or thermosome genes in the SEED database (Overbeek et al., 2005) were used for analysis. To generate full-length chaperonin genes, partial nucleotide ORFs identified as GroEL or thermosomes were assembled for each library using Geneious 6.1.8 (Kearse et al., 2012) (http://www.geneious.com) with the following parameters: word length, 10; index word length, 10; maximum gap size, 5; maximum gaps per read, 2%; maximum mismatches per read, 10%; maximum ambiguity 16.
For all libraries, putative viral co-chaperonin genes (GroES) were identified by BLASTp comparison of predicted amino acid ORFs to a custom GroES database (e-value <10−3) and screened using NCBI conserved domain search (e-value <10−5) (Marchler-Bauer et al., 2015). Any sequences that did not meet these criteria were manually inspected. Sequences missing key regions necessary for function were discarded.
Alignment and phylogenetic analysis
Putative full-length viral metagenomic GroES protein sequences were clustered at 60% identity using USEARCH (UCLUST algorithm, v7.0.1090; Edgar, 2010). Co-chaperonin sequences were considered full-length if they contained the cpn10 domain equivalent to E. coli GroES amino acid residues 4–95 (GenBank Acc. AAN83648.1). The proportion of co-chaperonins from each library recruiting to clusters with at least 15 sequences was visualized using Cytoscape (Shannon et al., 2003). The 15 sequence cutoff for cluster size was selected as each of these clusters comprised ⩾1% of the total sequences analyzed. Representative sequences from the GroES clusters were aligned to evaluate the amino acid composition of conserved domains. Sequence logos (Crooks et al., 2004) for the GroES mobile loop region were constructed from alignments of 1479 full-length viral metagenomic co-chaperonins and 4363 bacterial co-chaperonins within the UniRef100 database (The UniProt Consortium, 2015). Full-length virioplankton GroEL sequences (spanning amino acid 8–514 of E. coli GroEL, GenBank Acc. AIZ93089) were aligned to known viral and cellular sequences. Putative viral metagenomic thermosome sequences were aligned to known archaeal sequences and trimmed to region 56–467 of the Thermoplasma acidophilum alpha gene (Genbank Acc. 1A6D_A). DNA polymerase A and B genes from SERC contigs were verified by conserved domain BLAST, aligned to known phage and bacterial sequences, and trimmed to include only the polymerase region (residue 631–882 in E. coli for polA, GenBank Acc. CDN84700; residue 410–626 in E. coli for polB, GenBank Acc. AIZ92772). All alignments were performed using MAFFT (Katoh et al., 2002) (FFT-NS-i X1000 algorithm). Phylogenetic trees were constructed using RAxML (Stamatakis, 2006) version 7.6.2 with the PROTGAMMAILG or PROTGAMMALG model with 100 bootstrap replicates. Viral GroEL, GroES, Thermosome and Polymerase A sequences are deposited in Genbank (GroEL: KU595435-KU595540; GroES: KU970419-KU971234; Thermosome: KU595566-KU595571; PolA: KU595541-KU595565).
Gene clustering and rank abundance of chaperonin sequences
All complete and partial peptide ORFs from the CFA-CFH, DTF, DYM, GMF, POF, SDO and YBW libraries, the SERC contigs, and 136 129 phage proteins in the NCBI database were clustered at 50% identity using USEARCH (UCLUST algorithm, v7.0.1090; Edgar, 2010). Predicted genes were sorted by length before clustering so that full-length genes from sequenced viruses and the SERC contigs had the best chance of acting as seed sequences for recruiting full and partial ORFs from the other viromes. Because each virome library differed in its total number of ORFs, it was necessary to normalize ORF counts within each cluster (ORFc) according to seed sequence length and library size using the following (Sakowski et al., 2014):

where C is the total number of ORFs from a given library recruiting to a given cluster,  is the average ORF length and
is the average ORF length and  is the average number of ORFs within all libraries considered, S is the seed ORF length and Lind is the total number of ORFs within a given library. Clusters containing putative chaperonin ORFs were identified through significant homology (BLASTp, e-value ⩽10−3) to annotated chaperonins in the NCBI nr database. Chaperonin rank abundance curves and heatmaps were visualized using R (http://www.r-project.org/).
is the average number of ORFs within all libraries considered, S is the seed ORF length and Lind is the total number of ORFs within a given library. Clusters containing putative chaperonin ORFs were identified through significant homology (BLASTp, e-value ⩽10−3) to annotated chaperonins in the NCBI nr database. Chaperonin rank abundance curves and heatmaps were visualized using R (http://www.r-project.org/).
All viral chaperonins (GroEL, GroES, thermosomes), polymerase genes and contigs analyzed in this study are available on GenBank (BioProject PRJNA305521).
Results
Identification and sequence conservation of virioplankton group I and II chaperonins
Sequence reads with homology to GroES and GroEL (Group I chaperonins) were detected in all viral metagenomic libraries (Supplementary Table 3). At least one full-length GroEL gene was assembled from each location, with the Smithsonian Environmental Research Center (SERC) assembly producing 79 GroEL genes. Interestingly, thermosome reads (Group II chaperonins) were identified in all libraries, and were abundant enough to assemble full-length genes from the Chesapeake Bay libraries (CFA-CFH) and the SERC library, and a nearly complete thermosome gene for the Gulf of Maine (GMF) data set. Viral GroEL and thermosome genes contained the conserved features that distinguish Group I and II chaperonins, as well as amino acid residues essential to folding activities (Supplementary Figure 1). ATP/ADP Mg+2 binding sites were the most conserved positions between viral and bacterial chaperonins (Supplementary Figure 2), with identical or synonymous amino acids at 21 out of 25 residues for viral GroEL genes and 22 out of 24 residues for viral thermosome genes. Glycine residues at hinge positions were also highly conserved, with 78.3 and 76.5% pairwise identity for putative viral GroEL and thermosome genes, respectively. The most variable sites included residues lining the interior of the GroEL complex, which may reflect evolution of residues that interact with misfolded peptides (Supplementary Figure 2).
Phylogenetic analysis of virioplankton GroEL genes
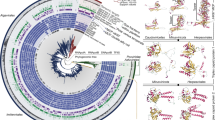
Most viral GroEL sequences fell into four clades corresponding with the presence and/or orientation of a GroES co-chaperonin gene (Figure 1; Supplementary Figure 3). Clade ‘GroEL’ was comprised exclusively of GroEL genes from SERC contigs that lacked a GroES gene. Presumably, these viruses either do not require a co-chaperonin, as observed for Pseudomonas aeruginosa bacteriophage EL (Kurochkina et al., 2012), or they utilize the host co-chaperonin. In clade ‘GroES>GroEL’ the GroES and GroEL genes are encoded in the canonical order typical within bacterial genomes (Lund, 2009). This clade was mostly comprised of GroEL genes from SERC contigs, and one GroEL gene from the Raunefjorden library (DYM). Most remaining virioplankton GroEL sequences fell within two clades labeled ‘GroEL>GroES-1’ and ‘GroEL>GroES-2’, as the canonical GroES-GroEL gene order on these contigs was reversed. All GroEL genes from the Chesapeake Bay (CFA-CFH), Dry Tortugas (DTF), Pacific Ocean (POF) and North Sea (SDO, YBW) viromes fell within these two clades. Two GroEL genes from SERC contigs claded closely with Cellulophaga phage phi38:1 (SERC_398908 and SERC_481644), indicating the presence of viral populations related to the Cellulophaga group of phages within the Chesapeake. Similar to Cellulophaga phage phi38:1, the GroES and GroEL genes on SERC_398908 and 481644 were found in the canonical GroES-EL gene order but were separated by a short ORF.
Gene order and content are defining features of viroplankton large subunit chaperonin phylogeny. Gray shaded ellipses indicate major clades of GroEL sequences that are labeled according to the presence and orientation of the GroES gene. The ‘GroEL’ clade did not show evidence of a neighboring GroES gene. For the ‘GroES>GroEL’ or ‘GroEL>GroES-1 or -2’ clades the GroES gene preceded or succeeded the GroEL gene, respectively. Sequenced viral chaperonins are indicated in italics. Nodes are colored by library location for the Chesapeake Bay (green), Dry Tortugas (pink), Norway (red), Gulf of Maine (yellow), North Sea (purple) and Pacific Ocean (blue) chaperonin sequences. For simplification, some of the SERC GroEL sequence labels were removed from the tree. Nodes with bootstrap values ⩾50, 70 and 90% are indicated by white, gray and black circles, respectively. The scale bar represents amino acid substitutions per site.
Viral GroEL genes from GMF_Contig_4 and contig SERC_383728 fell on a long branch that was part of a larger clade of GroEL genes from sequenced viral genomes (Figure 1; Supplementary Figure 3). Interestingly, contig SERC_383728 also encoded a DNA Polymerase B (polB) gene and an ATPase AAA gene homologous to archaeal genes from Methanoregula and Aciduliprofundum, respectively (Supplementary Figure 4). PolB has been used previously as a marker for phycodnaviruses and cyanoviruses (Chen and Suttle, 1996; Short, 2012; Ma et al., 2014) and can provide clues about viral morphology and host preference as demonstrated for Haloviruses HF1 and HF2 (Filée et al., 2002). Phylogenetic analysis of the SERC_383728 polB placed this peptide with the polB from thermosome-encoding viruses (SERC_352405) along with Thermoplasmata, methanogens and Marine Group II Archaea (Supplementary Figure 5). As Group I chaperonins have been detected in the genomes of Methanosarcina spp. (Klunker et al., 2003; Supplementary Table 4), this raises an intriguing possibility that SERC_383728 may represent a virus that infects archaea but carries a Group I GroEL chaperonin instead of a Group II chaperonin that is typically seen in archaea.
Most viral metagenomic GroELs were phylogenetically distant from bacterial GroELs, with four exceptions (Figure 1). The GroEL gene on contig SERC_344857 claded near Methanoregula boonei, a methanogen that presumably acquired GroEL through lateral gene transfer similar to Methanosarcina (Deppenmeier et al., 2002). In addition, GroEL genes from three SERC contigs formed a tight clade near Fluviicola taffensis (Figure 1) and shared close identity at the nucleotide level to GroEL genes from Bacteriodetes (Supplementary Figure 6). These contigs did not encode a co-chaperonin gene and putative genes flanking GroEL were not similar to other Bacteriodetes genes. In the case of contig SERC_344857, phage genes were also present. Because sequenced Bacteriodetes genomes rarely encode multiple GroEL genes separate from a GroES gene (Lund, 2009), these contigs likely represent viruses rather than bacterial contamination. The strong similarity of the viral GroEL genes to Bacteriodetes GroEL (~70% identity over ⩾92% of the gene) may indicate a gene transfer event from host to phage (Shi et al., 2005).
Phylogenetic analysis of virioplankton thermosome genes
Assembled viral thermosome genes were distantly related to archaeal thermosomes, with sequences in the Euryarchaeota lineage being closer than the Crenarchaeota lineage (Figure 2). Thermosomes, like the Group I GroES-EL system, have been transferred across kingdoms and are found primarily in Firmicutes (classes Bacilli and Clostridia; Williams et al., 2010). However, viral thermosome genes in this study were not related to bacterial thermosomes (Supplementary Figure 7). The best-supported phylogeny placed viral thermosome genes closer to Halobacteria than to other Euryarcheota (Figure 2). However, phylogenetic analysis using a larger region of the thermosome sequences placed them closer to Marine Group II Archaea (Supplementary Figure 8). This disagreement notwithstanding, it is clear that the virioplankton thermosomes were distantly related to the Euryarcheaota.
Thermosome sequence phylogeny indicates the presence of archaeal phages within the virioplankton. Branch colors correspond to the archaeal taxa noted in the tree. Thermosomes assembled from viral metagenomic reads are in bold. Nodes with bootstrap values ⩾50% are indicated. The scale bar represents amino acid substitutions per site.
Two of the eleven SERC contigs encoding a putative thermosome gene were long (55 and 99.3 Kb), allowing for a linkage-based assessment of the biological capabilities and potential hosts of these viruses. Conserved blocks of syntenous genes were observed between contigs SERC_272027 and 352405 (Supplementary Figure 9). Predicted capsid and portal proteins were most similar to genes within a haloarchaeal siphovirus (Supplementary Figures 10 and 11). A predicted polB on Contig 352405 shared the greatest homology to a Methanomicrobiales polymerase (Supplementary Figure 11). Phylogenetic analysis placed this viral polymerase gene near the polB gene on contig SERC_383728, along with methanogen, Thermoplasmata and Marine Group II polymerase genes (Supplementary Figure 4). Many genes on contigs SERC_272027 and 352405, including primase, terminase, ATPase AAA and various nucleases, were similar to euryarcheal proteins. The preponderance of evidence indicates these thermosome-encoding virioplankton populations are an unknown group of tailed archaeal viruses.
Lifestyle of viruses encoding group I chaperonins
Polymerase A peptide sequences on chaperonin-encoding SERC contigs were aligned with phage and bacterial polymerases, and polymerases from viral metagenomic libraries. The residue at the 762 position (E. coli numbering) was identified as this residue can be an indicator of phage lifecycle (Schmidt et al., 2014). Polymerases encoding leucine at position 762 were the most abundant (18 out of 26 contigs), suggesting that temperate phages in aquatic environments may carry co-chaperonin genes (Figure 3). Leucine-encoding polAs formed three distinct clades. Clades II and III contained virioplankton contigs encoding only GroES while Clade IV sequences derived from GroES>GroEL (GroESL) contigs (Figure 3). The largest clade of polA (GroES-encoding viruses in Clade II) was closely related to other virioplankton polA sequences (for example, ctg_DTF_polA_1102; Schmidt et al., 2014). Polymerases encoding phenylalanine and tyrosine at position 762, corresponding to clades I and V, respectively, are indicative of lytic virioplankton populations (Schmidt et al., 2014; Figure 3). Both clades were composed from polymerase sequences from SERC contigs encoding only GroES. Clade I contained two polymerase genes distantly related to polA from Enterobacteria phage T5, while clade V sequences formed a well-supported clade related to T7 bacteriophage. The polA from SERC_GroESL_Contig 398908 (Tyr762) claded closely to the polA from Cellulophaga phage phi38:1. This phylogenetic placement matched the GroEL phylogeny seen for the putative GroEL sequence on this contig (Figure 1), again suggesting contig SERC_398908 represents a virus related to Cellulophaga phages.
Viruses having presumptively lytic or temperate lifecycles carry chaperonin genes. Contig names reflect whether the polymerase sequence was found on a contig encoding a GroES gene or GroES>GroEL (GroESL) operon. Sequences from this study are in bold. Reference viral sequences and metagenomic viral polymerases from the Schmidt et al. (2014) study 23 are colored by the amino acid present at the 762 position. The scale bar represents amino acid substitutions per site.
Prevalent gene neighbor associations on chaperonin-encoding contigs
Several recurring gene neighbor associations were observed for particular chaperonin gene arrangements on contigs assembled from the SERC, North Sea and Norway libraries (Figure 4). Forty percent of SERC contigs with GroEL genes also carried a co-chaperonin gene (133 out of 337 contigs). The orientation of the GroES co-chaperonin gene was about equally split between the GroES>GroEL (63 out of 133) and GroEL>GroES gene order (70 out of 133). For GroES>GroEL contigs, small heat shock proteins (sHSP), which bind misfolded proteins and prevent aggregation, were predicted upstream of the GroES>GroEL operon for 25 SERC contigs (Figure 4) and were largely distinct from sHSP genes in sequenced marine phage (Maaroufi and Tanguay, 2013; Supplementary Figure 12). These sHSP presumably work in concert with the viral chaperonin complex to fold viral proteins. Forty-four SERC contigs containing a GroEL>GroES operon encoded an ORF downstream with a significant BLAST hit to a predicted phage protein from a viral fosmid sequence (Oxic3_1) from the Saanich Inlet, British Columbia, that also encodes a GroEL>GroES operon immediately upstream of this protein (Chow et al., 2015; for example, SERC Contig 314715, Figure 4). BLAST hits to Cellulophaga phages having Podoviridae morphology were common for structural proteins predicted on contigs with GroEL>GroES operons (data not shown), suggesting these viruses may also belong to the Podoviridae. Among SERC contigs encoding only a co-chaperonin gene, 523 (53.6%) of the 976 contigs encoded a kinesin-like protein downstream of GroES (Figure 4). The homologous region between known kinesins and the kinesin-like phage proteins on GroES virioplankton contigs did not include the region responsible for motor function. Thus, this protein may not function as a kinesin. This GroES and kinesin-like protein association was also reported in a study examining carbon metabolism genes in phage, albeit in the reverse orientation (Hurwitz et al., 2013). The consistent syntenic order of these two genes suggests these genes may be under similar genetic regulation and have allied functions during infection. Longer contigs also encoded a putative phage head protein downstream of the kinesin-like protein (Figure 4), which may rely on the host large subunit chaperonin for folding in a similar manner to the capsid protein in T4 bacteriophage (van der Vies et al., 1994; Bakkes et al., 2005).
Sequence conservation and diversity of virioplankton co-chaperonin genes
Clustering and analysis of the 1479 full-length virioplankton GroES genes revealed only 28.8% average pairwise identity between GroES representative cluster sequences. Despite low amino acid conservation, key functional residues in the mobile loop region, consisting of a conserved glycine followed by three small hydrophobic amino acids (Xu et al., 1997), were usually conserved among virioplankton co-chaperonins (Supplementary Figures 1 and 13). However, 336 (23%) of the full-length virioplankton co-chaperonins encoded a charged or polar residue instead of a hydrophobic residue in the second or third position of the mobile loop region, including sequences representing the most abundant co-chaperonin clusters (for example, Clusters 4, 8, 10 and 12; Figure 5b; Supplementary Figure 13). GroES genes from GroES>GroEL contigs recruited to two of the predominant co-chaperonin clusters (Clusters 15 and 18, Figures 5a and b) and were composed exclusively of sequences from the DYM and SERC viromes. This grouping agreed with the GroEL phylogeny, where GroEL sequences within the GroES>GroEL clade were exclusively from the DYM and SERC libraries (Figure 1), suggesting GroES>GroEL-encoding viruses rather than GroEL>GroES were prevalent in the DYM library. For co-chaperonins in these clusters, the conserved glycine residue in the mobile loop was replaced with an asparagine (clusters 15 and 18, yellow box, Figure 5b) or serine residue in all but one of the GroES sequences. This modification may facilitate interaction with the viral GroEL instead of the host GroEL. Another well-defined domain in co-chaperonin proteins is the roof hairpin, which covers the dome of the GroEL/ES complex in bacterial co-chaperonins but is deleted in the viral co-chaperonin paralogs T4 gp31 and RB49 CocO (Ang et al., 2001). Interestingly, co-chaperonin sequences from all libraries (except DYM) fell into clusters without a dome loop structure and included GroES sequences from SERC contigs with a GroEL>GroES gene order (Clusters 6, 7 and 16 in the red box/outline, Figures 5a and b). Deletion of the roof hairpin structure may provide a larger folding cavity, as observed in the T4 gp31-GroEL complex (Bakkes et al., 2005).
The co-chaperonin GroES is ubiquitous within the virioplankton and demonstrates unique structural features. (a) Network analysis of co-chaperonin clusters generated by clustering of full-length viral metagenomic GroES sequences at 60% amino acid identity. Blue ellipses represent cluster rank, with the width of the ellipse representing the cluster size. Clusters containing SERC GroES found in GroES>GroEL or GroEL>GroES operons are outlined in yellow and red, respectively. Viral metagenomic libraries are indicated by green boxes (Chesapeake Bay (ChesBay and SERC), Dry Tortugas (DTF), Gulf of Maine (GMF), Pacific Ocean (POF), Raunefjordern, Norway (DYM) and the North Sea (SDO, YBW)). The proportion of GroES genes recruiting to a given cluster is represented by the edge width. Only clusters with ⩾15 sequences are displayed. (b) Amino acid alignment of GroES cluster representatives for co-chaperonin clusters with ⩾15 sequences. Numbers in parentheses indicate the number of co-chaperonin sequences in the cluster. Clusters containing SERC GroES found in GroES>GroEL or GroEL>GroES operons are outlined in yellow and red, respectively. The roof hairpin deletion in three of the cluster representative sequences is outlined in black. The depicted alignment spans from E. coli GroES position 5–72.
Relative abundance of virioplankton chaperonins
Putative chaperonin genes were consistently among the most abundant genes in the viromes, particularly for DYM, GMF and POF viromes (Figure 6; Supplementary Table 5). Putative GroES clusters were ranked in the 100 most abundant peptide clusters for 4 out of 7 libraries (DYM, DTF, GMF and POF), and putative GroEL genes were ranked in the top 100 peptide clusters for two of the libraries (GMF and DYM). Viral chaperonin genes were most common (relative to other genes) for the GMF library while the POF library had the most chaperonin clusters ranked within the top 1000 peptide clusters. With the exception of DYM, most chaperonin clusters contained sequences from multiple viromes, suggesting chaperonin-encoding virioplankton populations are widely distributed across marine environments. Chaperonin genes from DYM were related to SERC but were not closely related to chaperonin-encoding viruses from other locations (Figures 1, 5a and 6) due to the predominance of GroES>GroEL phage in this data set.
Biogeography indicates both broad and narrow distribution of chaperonin-carrying virioplankton populations. (a) Stacked bar plot of GroES clusters. (b) Stacked bar plot of GroEL clusters. The raw number of reads recruiting to a given cluster is indicated by the number above the stacked bar. Each bar represents the normalized abundance of reads recruiting to the cluster. Each location is indicated by color. Bars with an asterisk underneath indicate that the seed sequence for that cluster was a less than full-length GroEL gene (⩽300 amino acids). (c) Rank abundance of predominant chaperonin clusters per library. Each row represents a co-chaperonin or chaperonin cluster and columns represent individual libraries. Rank abundance of chaperonin clusters for a particular library is indicated by color. Numbers within the heatmap report the rank of chaperonin clusters that were in the top 500 peptide clusters for a given library.
Discussion
This survey of virioplankton chaperonin genes was facilitated by virome sequence data from a variety of marine locations, coupled with the SERC virome that was deeply sequenced and included long PacBio sequence reads. These data provided long contigs for examining the diversity and gene content of chaperonin-encoding viruses. Our analyses revealed a surprising abundance and diversity of Group I and II viral chaperonins that were evolutionarily distinct from cellular chaperonins. The higher prevalence of putative GroES genes compared with GroEL genes (Figure 6; Supplementary Table 3) indicated that encoding only a co-chaperonin is common within the virioplankton. However, for viruses encoding GroEL, the majority of genes fell into clades corresponding with GroES>GroEL or GroEL>GroES operons (Figure 1), suggesting aquatic viruses carrying a GroEL gene are more likely to encode their own co-chaperonin gene. Also surprising was the occurrence of thermosome sequences, revealing the presence of euryarchaeal viruses in surface waters. Previous research has suggested that Euryarcheaota populations are infected by tailed viruses which share a common ancestry with tailed bacteriophage (Krupovic et al., 2010, 2011). The deep branching of viral thermosomes from this study supports an ancient association between Archaea and tailed viruses.
Encoding thermosomes and complete GroES and GroEL operons is likely a defining feature of larger genome viruses (Holmfeldt et al., 2013), which are hypothesized to represent low burst size, large genome viruses infecting abundant microbial populations (Suttle, 2007). The full chaperonin complex may be necessary for folding the major capsid protein (MCP), as modified chaperonins are essential for folding the MCP of T4-like bacteriophage (van der Vies et al., 1994; Ang et al., 2000, 2001), along with the fact that encoding a greater number of viral proteins may lead to increased reliance on chaperonins. In general, carrying chaperonins may allow a virus to fold a more diverse suite of proteins not accommodated by the host GroES-EL complex (Andreadis and Black, 1998) or expand its host range (Ang et al., 2000). There is also the potential that viral chaperonins may play a more global role in folding or modulation of both viral and host proteins during infection. Previous studies describing multiple moonlighting functions for chaperonins (Henderson et al., 2013), and the ability of viral chaperonins to fold host proteins in vivo (van der Vies et al., 1994; Ang et al., 2001), support this notion.
The observed links between GroEL phylogeny, chaperonin gene order and associations with nearby genes demonstrated that large subunit chaperonins are a defining feature of certain virioplankton populations. Similar to findings for DNA polymerase A (Schmidt et al., 2014), ribonucleotide reductase (Dwivedi et al., 2013; Sakowski et al., 2014) and photosystem genes (Mann et al., 2003; Sullivan et al., 2006; Roitman et al., 2015), important biological and ecological characteristics may be linked with the type and organization of chaperonin genes a virus carries. Given the common observation of chaperonin genes within viromes, it is surprising that so few known viruses carry chaperonin genes. This discrepancy is likely because most genome-sequenced phages infect a narrow taxonomic group of hosts. For example, 83% of known phages infect hosts within only three phyla (Wommack et al., 2015) and archaeal viruses, infecting mostly Haloarchaea or hyperthermophillic Crenarchaea (Pina et al., 2011), account for only 65 sequenced viral genomes. The fact that chaperonin genes are relatively common in the virioplankton, and are known to demonstrate vertical inheritance patterns within cellular life (Chaban and Hill, 2011; Links et al., 2012) makes these targets ideal for follow-up studies examining the ecology of chaperonin-encoding viruses.
References
Altschul S . (1997). Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25: 3389–3402.
Andreadis JD, Black LW . (1998). Substrate mutations that bypass a specific Cpn10 chaperonin requirement for protein folding. J Biol Chem 273: 34075–34086.
Ang D, Georgopoulos C . (2012). An ORFan no more: the bacteriophage T4 39.2 gene product, NwgI, modulates GroEL chaperone function. Genetics 190: 989–1000.
Ang D, Keppel F, Klein G, Richardson A, Georgopoulos C . (2000). Genetic analysis of bacteriophage-encoded cochaperonins. Annu Rev Genet 34: 439–456.
Ang D, Richardson A, Mayer MP, Keppel F, Krisch H, Georgopoulos C . (2001). Pseudo-T-even bacteriophage RB49 encodes CocO, a cochaperonin for GroEL, which can substitute for Escherichia coli’s GroES and bacteriophage T4’s Gp31. J Biol Chem 276: 8720–8726.
Bakkes PJ, Faber BW, van Heerikhuizen H, van der Vies SM . (2005). The T4-encoded cochaperonin, gp31, has unique properties that explain its requirement for the folding of the T4 major capsid protein. Proc Natl Acad Sci USA 102: 8144–8149.
Boisvert DC, Wang J, Otwinowski Z, Horwich AL, Sigler PB . (1996). The 2.4 A crystal structure of the bacterial chaperonin GroEL complexed with ATP gamma S. Nat Struct Biol 3: 170–177.
Chaban B, Hill JE . (2011). A ‘universal’ type II chaperonin PCR detection system for the investigation of Archaea in complex microbial communities. ISME J 6: 430–439.
Chen F, Suttle CA . (1996). Evolutionary relationships among large double-stranded DNA viruses that infect microalgae and other organisms as inferred from DNA polymerase genes. Virology 219: 170–178.
Chow C-ET, Winget DM, White RA, Hallam SJ, Suttle CA . (2015). Combining genomic sequencing methods to explore viral diversity and reveal potential virus-host interactions. Front Microbiol 6: 265.
Coppo A, Manzi A, Pulitzer JF, Takahashi H . (1973). Abortive bacteriophage T4 head assembly in mutants of Escherichia coli. J Mol Biol 76: 61–87.
Crooks GE, Hon G, Chandonia J-M, Brenner SE . (2004). WebLogo: a sequence logo generator. Genome Res 14: 1188–1190.
Deppenmeier U, Johann A, Hartsch T, Merkl R, Schmitz RA, Martinez-Arias R et al. (2002). The genome of Methanosarcina mazei: evidence for lateral gene transfer between bacteria and archaea. J Mol Microbiol Biotechnol 4: 453–461.
Dwivedi B, Xue B, Lundin D, Edwards RA, Breitbart M . (2013). A bioinformatic analysis of ribonucleotide reductase genes in phage genomes and metagenomes. BMC Evol Biol 13: 33.
Edgar RC . (2010). Search and clustering orders of magnitude faster than BLAST. Bioinforma Oxf Engl 26: 2460–2461.
Filée J, Forterre P, Sen-Lin T, Laurent J . (2002). Evolution of DNA polymerase families: evidences for multiple gene exchange between cellular and viral proteins. J Mol Evol 54: 763–773.
Georgopoulos C . (2006). Toothpicks, serendipity and the emergence of the Escherichia coli DnaK (Hsp70) and GroEL (Hsp60) chaperone machines. Genetics 174: 1699–1707.
Georgopoulos CP, Hendrix RW, Casjens SR, Kaiser AD . (1973). Host participation in bacteriophage lambda head assembly. J Mol Biol 76: 45–60.
Georgopoulos CP, Hendrix RW, Kaiser AD, Wood WB . (1972). Role of the host cell in bacteriophage morphogenesis: effects of a bacterial mutation on T4 head assembly. Nat New Biol 239: 38–41.
Grimaud R, Toussaint A . (1998). Assembly of both the head and tail of bacteriophage Mu is blocked in Escherichia coli groEL and groES mutants. J Bacteriol 180: 1148–1153.
Hartl FU, Bracher A, Hayer-Hartl M . (2011). Molecular chaperones in protein folding and proteostasis. Nature 475: 324–332.
Henderson B, Fares MA, Lund PA . (2013). Chaperonin 60: a paradoxical, evolutionarily conserved protein family with multiple moonlighting functions. Biol Rev Camb Philos Soc 88: 955–987.
Hertveldt K, Lavigne R, Pleteneva E, Sernova N, Kurochkina L, Korchevskii R et al. (2005). Genome comparison of Pseudomonas aeruginosa large phages. J Mol Biol 354: 536–545.
Holmfeldt K, Solonenko N, Shah M, Corrier K, Riemann L, Verberkmoes NC et al. (2013). Twelve previously unknown phage genera are ubiquitous in global oceans. Proc Natl Acad Sci USA 110: 12798–12803.
Hurwitz BL, Hallam SJ, Sullivan MB . (2013). Metabolic reprogramming by viruses in the sunlit and dark ocean. Genome Biol 14: R123.
Hurwitz BL, Sullivan MB . (2013). The Pacific Ocean virome (POV): a marine viral metagenomic cudataset and associated protein clusters for quantitative viral ecology. PloS One 8: e57355.
Hänninen AL, Bamford DH, Bamford JK . (1997). Assembly of membrane-containing bacteriophage PRD1 is dependent on GroEL and GroES. Virology 227: 207–210.
John SG, Mendez CB, Deng L, Poulos B, Kauffman AKM, Kern S et al. (2010). A simple and efficient method for concentration of ocean viruses by chemical flocculation. Environ Microbiol Rep 3: 195–202.
Katoh K, Misawa K, Kuma K, Miyata T . (2002). MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res 30: 3059–3066.
Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S et al. (2012). Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinforma Oxf Engl 28: 1647–1649.
Klunker D, Haas B, Hirtreiter A, Figueiredo L, Naylor DJ, Pfeifer G et al. (2003). Coexistence of group I and group II chaperonins in the archaeon Methanosarcina mazei. J Biol Chem 278: 33256–33267.
Krupovic M, Forterre P, Bamford DH . (2010). Comparative analysis of the mosaic genomes of tailed archaeal viruses and proviruses suggests common themes for virion architecture and assembly with tailed viruses of bacteria. J Mol Biol 397: 144–160.
Krupovic M, Spang A, Gribaldo S, Forterre P, Schleper C . (2011). A thaumarchaeal provirus testifies for an ancient association of tailed viruses with archaea. Biochem Soc Trans 39: 82–88.
Kurochkina LP, Semenyuk PI, Orlov VN, Robben J, Sykilinda NN, Mesyanzhinov VV . (2012). Expression and functional characterization of the first bacteriophage-encoded chaperonin. J Virol 86: 10103–10111.
Leitner A, Joachimiak LA, Bracher A, Mönkemeyer L, Walzthoeni T, Chen B et al. (2012). The molecular architecture of the eukaryotic chaperonin TRiC/CCT. Structure 20: 814–825.
Links MG, Dumonceaux TJ, Hemmingsen SM, Hill JE . (2012). The chaperonin-60 universal target is a barcode for bacteria that enables de novo assembly of metagenomic sequence data. PloS One 7: e49755.
Lund PA . (2009). Multiple chaperonins in bacteria–why so many? FEMS Microbiol Rev 33: 785–800.
Ma Y, Allen LZ, Palenik B . (2014). Diversity and genome dynamics of marine cyanophages using metagenomic analyses. Environ Microbiol Rep 6: 583–594.
Maaroufi H, Tanguay RM . (2013). Analysis and phylogeny of small heat shock proteins from marine viruses and their cyanobacteria host. PloS One 8: e81207.
Mann NH, Cook A, Millard A, Bailey S, Clokie M . (2003). Marine ecosystems: bacterial photosynthesis genes in a virus. Nature 424: 741.
Marchler-Bauer A, Derbyshire MK, Gonzales NR, Lu S, Chitsaz F, Geer LY et al. (2015). CDD: NCBI’s conserved domain database. Nucleic Acids Res 43: D222–D226.
Murray NE, Gann A . (2007). What has phage lambda ever done for us? Curr Biol 17: R305–R312.
Myers EW, Sutton GG, Delcher AL, Dew IM, Fasulo DP, Flanigan MJ et al. (2000). A whole-genome assembly of Drosophila. Science 287: 2196–2204.
Niu B, Fu L, Sun S, Li W . (2010). Artificial and natural duplicates in pyrosequencing reads of metagenomic data. BMC Bioinformatics 11: 187.
Noguchi H, Taniguchi T, Itoh T . (2008). MetaGeneAnnotator: detecting species-specific patterns of ribosomal binding site for precise gene prediction in anonymous prokaryotic and phage genomes. DNA Res 15: 387–396.
Overbeek R, Begley T, Butler RM, Choudhuri JV, Chuang H-Y, Cohoon M et al. (2005). The subsystems approach to genome annotation and its use in the project to annotate 1000 genomes. Nucleic Acids Res 33: 5691–5702.
Pina M, Bize A, Forterre P, Prangishvili D . (2011). The archeoviruses. FEMS Microbiol Rev 35: 1035–1054.
Poranen MM, Ravantti JJ, Grahn AM, Gupta R, Auvinen P, Bamford DH . (2006). Global changes in cellular gene expression during bacteriophage PRD1 infection. J Virol 80: 8081–8088.
Roitman S, Flores-Uribe J, Philosof A, Knowles B, Rohwer F, Ignacio-Espinoza JC et al. (2015). Closing the gaps on the viral photosystem-I psaDCAB gene organization. Environ Microbiol 17: 5100–5108.
Sakowski EG, Munsell EV, Hyatt M, Kress W, Williamson SJ, Nasko DJ et al. (2014). Ribonucleotide reductases reveal novel viral diversity and predict biological and ecological features of unknown marine viruses. Proc Natl Acad Sci USA 111: 15786–15791.
Schmidt HF, Sakowski EG, Williamson SJ, Polson SW, Wommack KE . (2014). Shotgun metagenomics indicates novel family A DNA polymerases predominate within marine virioplankton. ISME J 8: 103–114.
Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D et al. (2003). Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 13: 2498–2504.
Shi S-Y, Cai X-H, Ding D . (2005). Identification and categorization of horizontally transferred genes in prokaryotic genomes. Acta Biochim Biophys Sin 37: 561–566.
Short SM . (2012). The ecology of viruses that infect eukaryotic algae. Environ Microbiol 14: 2253–2271.
Stamatakis A . (2006). RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinforma Oxf Engl 22: 2688–2690.
Sullivan MB, Lindell D, Lee JA, Thompson LR, Bielawski JP, Chisholm SW . (2006). Prevalence and evolution of core photosystem II genes in marine cyanobacterial viruses and their hosts. PLoS Biol 4: e234.
Suttle CA . (2007). Marine viruses–major players in the global ecosystem. Nat Rev Microbiol 5: 801–812.
Takano T, Kakefuda T . (1972). Involvement of a bacterial factor in morphogenesis of bacteriophage capsid. Nature New Biol 239: 34–37.
The UniProt Consortium. (2015). UniProt: a hub for protein information. Nucleic Acids Res 43: D204–D212.
van der Vies SM, Gatenby AA, Georgopoulos C . (1994). Bacteriophage T4 encodes a co-chaperonin that can substitute for Escherichia coli GroES in protein folding. Nature 368: 654–656.
Williams TA, Codoñer FM, Toft C, Fares MA . (2010). Two chaperonin systems in bacterial genomes with distinct ecological roles. Trends Genet 26: 47–51.
Willison KR, Kubota H . (1994). The structure, function, and genetics of the chaperonin containing TCP-1 (CCT) in eukaryotic cytosol. Cold Spring Harb Monogr Arch 26: 299–312.
Wommack KE, Bhavsar J, Polson SW, Chen J, Dumas M, Srinivasiah S et al. (2012). VIROME: a standard operating procedure for analysis of viral metagenome sequences. Stand Genomic Sci 6: 421–433.
Wommack KE, Nasko DJ, Chopyk J, Sakowski EG . (2015). Counts and sequences, observations that continue to change our understanding of viruses in nature. J Microbiol Seoul Korea 53: 181–192.
Xu Z, Horwich AL, Sigler PB . (1997). The crystal structure of the asymmetric GroEL-GroES-(ADP)7 chaperonin complex. Nature 388: 741–750.
Zweig M, Cummings DJ . (1973). Cleavage of head and tail proteins during bacteriophage T5 assembly: selective host involvement in the cleavage of a tail protein. J Mol Biol 80: 505–518.
Acknowledgements
We thank Bruce Kingham and Olga Shevchenko of the University of Delaware Sequencing and Genotyping Facility for sequencing support. This research was supported by a National Science Foundation grant to KEW and SP (OCE-1148118). Computational analyses using the BioHen computing cluster were made possible through funding from Delaware INBRE (NIGMS GM103446), the State of Delaware, and the Delaware Biotechnology Institute. RM was supported through Graduate and Dissertation Fellowship awards from the University of Delaware Office of Graduate and Professional Education.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies this paper on The ISME Journal website
Supplementary information
Rights and permissions
This work is licensed under a Creative Commons Attribution-NonCommercial-ShareAlike 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-sa/4.0/
About this article
Cite this article
Marine, R., Nasko, D., Wray, J. et al. Novel chaperonins are prevalent in the virioplankton and demonstrate links to viral biology and ecology. ISME J 11, 2479–2491 (2017). https://doi.org/10.1038/ismej.2017.102
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ismej.2017.102
This article is cited by
-
Community ecology and functional potential of bacteria, archaea, eukarya and viruses in Guerrero Negro microbial mat
Scientific Reports (2024)
-
Towards an integrative view of virus phenotypes
Nature Reviews Microbiology (2022)
-
Moisture modulates soil reservoirs of active DNA and RNA viruses
Communications Biology (2021)
-
Viral elements and their potential influence on microbial processes along the permanently stratified Cariaco Basin redoxcline
The ISME Journal (2020)
-
Single-cell genomics uncover Pelagibacter as the putative host of the extremely abundant uncultured 37-F6 viral population in the ocean
The ISME Journal (2019)