Abstract
The genus Bartonella comprises facultative intracellular bacteria with a unique lifestyle. After transmission by blood-sucking arthropods they colonize the erythrocytes of mammalian hosts causing acute and chronic infectious diseases. Although the pathogen–host interaction is well understood, little is known about the evolutionary origin of the infection strategy manifested by Bartonella species. Here we analyzed six genomes of Bartonella apis, a honey bee gut symbiont that to date represents the closest relative of pathogenic Bartonella species. Comparative genomics revealed that B. apis encodes a large set of vertically inherited genes for amino acid and cofactor biosynthesis and nitrogen metabolism. Most pathogenic bartonellae have lost these ancestral functions, but acquired specific virulence factors and expanded a vertically inherited gene family for harvesting cofactors from the blood. However, the deeply rooted pathogen Bartonella tamiae has retained many of the ancestral genome characteristics reflecting an evolutionary intermediate state toward a host-restricted intraerythrocytic lifestyle. Our findings suggest that the ancestor of the pathogen Bartonella was a gut symbiont of insects and that the adaptation to blood-feeding insects facilitated colonization of the mammalian bloodstream. This study highlights the importance of comparative genomics among pathogens and non-pathogenic relatives to understand disease emergence within an evolutionary-ecological framework.
Similar content being viewed by others
Introduction
Many host-restricted bacterial pathogens causing chronic infectious diseases have evolved from free-living environmental ancestors through a stepwise evolutionary process of host adaptation (Toft and Andersson, 2010; McCutcheon and Moran, 2012). Broadly, two major genomic changes accompany the evolution of pathogens: (i) genome reduction due to population bottlenecks and nutrient-rich host environments and (ii) acquisition of virulence factors for host invasion and persistence (Ochman and Moran, 2001; Pallen and Wren, 2007). Investigating the genomic changes associated with the transition of lifestyle is key to understand pathogen ecology and evolution (Parkhill et al., 2003; Rohmer et al., 2007; Langridge et al., 2015). To this end, comparative genome analysis of pathogens and closely related non-pathogenic environmental bacteria can provide valuable insights (Reuter et al., 2014; Bentley and Parkhill, 2015).
The alphaproteobacteria provide excellent subjects for comparative studies due to the large diversity of lifestyles within this class (Ettema and Andersson, 2009). Intracellular animal-associated pathogens emerged in several alphaproteobacterial lineages, each time associated with an overall reduction of genomic content and the acquisition of genes for the adoption of specific host infection strategies (Batut et al., 2004; Ettema and Andersson, 2009).
A striking example represents the genus Bartonella belonging to the Rhizobiales. This alphaproteobacterial order is dominated by plant and soil-associated bacteria. In contrast, Bartonella species include host-restricted facultative intracellular pathogens that have evolved a unique stealth infection strategy (Harms and Dehio, 2012): They colonize the erythrocytes of mammalian reservoir hosts resulting in acute or chronic, often asymptomatic, bloodstream infections. Transmission between hosts is ensured by blood-sucking arthropods. The evolutionary success of this life cycle is reflected by the adaptive radiation of this pathogen (Engel et al., 2011; Guy et al., 2013): More than 30 pathogenic Bartonella species have been described to date (Kešnerová et al., 2016), infecting a wide variety of mammalian host species.
The divergent adaptation of the genus Bartonella to distinct hosts has been linked to the horizontal gene transfer of several factors for host interaction (Saenz et al., 2007; Engel et al., 2011; Guy et al., 2013), including Type IV secretion systems, autotransporters and adhesins (Saenz et al., 2007; Engel et al., 2011; Guy et al., 2013). Moreover, bartonellae evolved a gene transfer agent, which mediates the transfer of genomic DNA between strains thereby facilitating diversification of genes for host interaction and enabling host adaptation (Guy et al., 2013).
Despite the acquisition of these genetic elements, the overall genomic evolution of Bartonella has been dominated by gene loss (Boussau et al., 2004). Bartonella genomes range in size from 1.4 to 2.6 Mb, which is small compared with soil or plant-associated Rhizobiales (Batut et al., 2004; Ettema and Andersson, 2009). The closest related family, the Brucellaceae, is phylogenetically already quite distant from the Bartonellaceae. Therefore, previous comparative analyses (Alsmark et al., 2004) provided limited insights into the evolutionary history of the genus Bartonella, especially in regard of the genomic makeup and ecology of its last common ancestor (LCA). However, microbial community analyses have recently indicated that honey bees and diverse ant species possess gut bacteria that are more closely related to the genus Bartonella than the family Brucellaceae (Cox-Foster et al., 2007; Martinson et al., 2011; Hu et al., 2014; Sanders et al., 2014). These insect-associated bacteria form deeply rooted sister lineages of the pathogenic Bartonella species (Kešnerová et al., 2016). Their symbiotic functions have so far remained elusive. However, several lines of evidence suggest that in ants these bacteria might play a role in nitrogen uptake by either fixing atmospheric nitrogen or recycling excreted insect waste products (Feldhaar et al., 2007; Russell et al., 2009; Anderson et al., 2012; Sapountzis et al., 2015). The symbiotic relationship between the Rhizobiales bacteria and the honey bee is so far unexplored.
We have recently cultivated several strains of the Rhizobiales gut symbiont of honey bees. On the basis of 16S rRNA sequence similarity (>95%), this bacterium belongs to the genus Bartonella and accordingly was named Bartonella apis (Kešnerová et al., 2016). The closest related Bartonella species to date is Bartonella tamiae, a pathogen that was isolated from the blood of three human patients in Thailand (Kosoy et al., 2008) and since then, B. tamiae-like DNA has been detected in various blood-sucking arthropods (Billeter et al., 2008; Kabeya et al., 2010; Leulmi et al., 2016). The close phylogenetic relationship with pathogenic Bartonella species renders B. apis promising to elucidate the evolutionary history of the unique infection strategy of the genus Bartonella. Here we sequenced the genomes of six divergent strains of B. apis. Our phylogenomic and comparative analyses shed light on the ancestral genomic state of the genus Bartonella providing novel insights into the evolutionary origin of these pathogens.
Materials and methods
Genome sequencing, assembly and annotation
Six cultured strains of B. apis were chosen for sequencing. Three strains (PEB0122, PEB0149 and PEB0150) originated from a honey bee colony (Apis mellifera) in West Haven, CT, US. Three other strains (BBC0122, BBC0178 and BBC0244) were isolated from a colony in Lausanne, Switzerland. The US strains were sequenced with Illumina MiSeq technology (2 × 250 bp reads) and assembled with Spades v3.6 (Bankevich et al., 2012). The Swiss strains were sequenced with SMRT technology (Pacific Biosciences, Menlo Park, CA, USA) and assembled with HGAP (Chin et al., 2013). All six genomes were annotated with the Integrated Microbial Genomes system (Markowitz et al., 2014). Additional details regarding DNA extraction and genome assembly are available in the Supplementary Methods and in Supplementary Table 1.
Determining orthologous gene families
For phylogenomic analyses, we determined orthologous gene families between the six strains of B. apis, two strains of B. tamiae (Bartonella initiative, Broad Institute) and 14 species representing the radiating lineage within the genus Bartonella (Supplementary Table 2). The latter group will be referred to as the eubartonellae in accordance with a previous study (Zhu et al., 2014). As outgroup taxa, we included the genome of the recently sequenced ant gut symbiont Ca. Tokpelaia hoelldoblerii (Neuvonen et al., 2016) and five other Rhizobiales. An all-against-all BLASTP analysis of the proteomes of these 28 strains identified candidate orthologs. BLAST hits with an e-value >10−5 and for which the query and the hit sequence had less than 50% overlap of their gene length were excluded. Clusters of orthologous gene families were created using OrthoMCL (Li et al., 2003) with recommended settings (—abc -I 1.5), resulting in a total of 7192 gene families.
Inferring phylogenetic trees
Genome-wide phylogenies were inferred from 589 orthologous gene families based on protein and DNA sequence alignments. Tree topologies and branch support values were inferred with maximum likelihood and Bayesian inference methods using RAxML v8.0.0 (Stamatakis, 2014) and PhyloBayes v4.1 (Lartillot et al., 2009), respectively. Additional details regarding the phylogenetic analyses are available in the Supplementary Methods.
Comparison of genome structure, genome divergence and gene content
To compare and visualize genomic regions we used the R-package genoPlotR (Guy et al., 2010). TBLASTX comparison files were created with DoubleACT (www.hpa-bioinfotools.org.uk). For comparison of specific regions, we generated pairwise comparison files with command-line TBLASTX. To estimate sequence divergence between genomes, we calculated pairwise average nucleotide identity (ANI) with JSpecies (Richter et al., 2015). We further estimated the number of synonymous substitutions per synonymous site (dS) with the Nei and Gojobori (1986) method in PAML-4.8 (Yang, 2007) for the 589 genes used in the phylogenetic analysis. The dS values were averaged over all genes to obtain genome-wide values for species comparisons. Pan genomes of B. apis, B. tamiae and the eubartonellae were based on the gene families identified with OrthoMCL.
Inferring gene gain and gene loss
To identify at which branch of the genome-wide phylogeny genes have been gained and lost, and to infer the gene content of the LCA of the genus Bartonella, we performed a gene flux analysis in PAUP v4.0 (Wilgenbusch and Swofford, 2003). We followed the method used in previous publications (Boussau et al., 2004; Guy et al., 2013): with generalized parsimony the 7192 protein families were mapped onto the phylogenetic tree. A cost-matrix was used which penalizes gene gain with 12, gene loss with 5, gene duplication with 1 and copy changes with 0.2 units. These parameters were based on previous publications (Guy et al., 2013; Tamarit et al., 2015), except that the gene gain penalty was increased from 10 to 12 based on empirical testing of a range of parameters.
Analysis of functional gene contents
Gene contents were categorized based on COG functions. We used the COG database, which was updated in 2014 (www.ncbi.nlm.nih.gov/COG). Analysis of amino acid and cofactor biosynthesis pathways were based on KEGG pathway maps and EC (Enzyme Commission) numbers.
Analysis of virulence factors
To analyze virulence factors in B. apis and B. tamiae, we compiled a list of 88 eubartonellae genes which were either experimentally identified to be essential for host colonization (Saenz et al., 2007; Harms and Dehio, 2012) or predicted to be involved in host interaction (Guy et al., 2013). We categorized these virulence factors into ‘conserved’ (n=69) and ‘Bartonella-specific’ (n=19) genes depending on their presence in the genomes of the outgroup. We then validated how many of these genes had orthologs in any of the eight genomes of B. apis and B. tamiae based on the OrthoMCL results.
Results and discussion
B. apis genomes reveal intra-species diversity, but a conserved genome structure
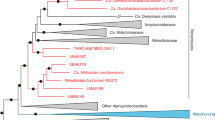
Assemblies of genomes sequenced with Illumina technology consisted of 7–16 contigs, whereas genomes sequenced with SMRT technology assembled into single circular chromosomes (Supplementary Table 1 and Supplementary Figure 1). The contigs of each draft genome were ordered according to the complete genome of strain BBC0122 (Supplementary Figure 2). The six B. apis genomes range in size between 2.53 and 2.91 Mb (Table 1), which is larger than the genomes of other bartonellae (1.39–2.38 Mb), except for B. tribocorum (2.64 Mb). The GC content (45–46%) is slightly elevated compared with other bartonellae (36–42%). ANI within B. apis ranges from 85% to 98% indicating substantial sequence divergence between strains, which is consistent with previous findings of intra-species diversity in other bee gut symbionts (Engel et al., 2012; Engel et al., 2014; Ellegaard et al., 2015). Despite the marked degree of sequence divergence, the genomic organization between the B. apis strains is conserved (Figure 1). A high degree of genome structure conservation also exists between B. apis and B. tamiae, with a few larger rearrangements present around the terminus of replication. In contrast, eubartonellae display a lower degree of structural conservation and many more rearrangements (Figure 1).
Comparative genome alignments of different Bartonella strains. Bartonella genomes are shown as gray horizontal lines. Genes specific to B. apis are shown in blue. Genes shared between B. apis and B. tamiae, B. apis and eubartonellae and B. tamiae and eubartonellae are shown in green, black and red, respectively. TBLASTX hits between genomes are shown in gray (bit score cutoff=600).
B. apis and B. tamiae are sister clades that diverged before the radiation of the eubartonellae
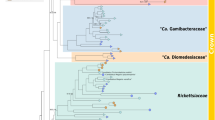
The phylogenetic analysis of the concatenated protein alignments using maximum likelihood and Bayesian methods grouped B. apis and B. tamiae into a monophyletic clade that diverged before the radiation of the eubartonellae (Figure 2 and Supplementary Figure 3a). In contrast, maximum likelihood and Bayesian trees inferred from aligned DNA sequences suggests that B. tamiae is monophyletic with the eubartonellae, and that B. apis branched off earlier (Supplementary Figures 3 and 4). The DNA analysis, however, was strongly affected by the similar GC content of B. tamiae and the eubartonellae. When only considering the first and second codon position for the maximum likelihood analysis (Supplementary Figure 4), the monophyletic group of B. tamiae and the eubartonellae was no longer supported by bootstrap analysis. On the basis of these findings and the fact that protein sequences are more reliable for divergent taxa (see high dS and low ANI values, Table 1), we decided to use the protein tree for further analyses. Sampling of additional taxa, closely related to B. apis or B. tamiae, will help to resolve the phylogenies further in the future. Independent of the slightly different topologies, both phylogenies show that B. apis and B. tamiae are sister clades that diverged before the radiation of the eubartonellae. This suggests that B. tamiae has a different evolutionary history than the eubartonellae despite the fact that both colonize the bloodstream of mammals and can cause human illness. Moreover, monophyly of B. apis and B. tamiae would suggest that pathogenicity in the genus Bartonella has evolved twice, once in an ancestor of the eubartonellae and independently in the lineage of B. tamiae.
Genome-wide phylogeny of the genus Bartonella and related alphaproteobacteria. Tree topology and bootstrap support values were inferred with RAxML (model: JTT+G+I). The three Bartonella clades (B. apis, B. tamiae and eubartonellae) are highlighted in blue, green and yellow, respectively. Ca. Tokpelaia hoelldoblerii is highlighted in orange. Values above branches show bootstrap support values (⩾80%). Numbers below certain branches indicate percentage of single gene trees with congruent topology at this branch. The branch toward the outgroup species Bradyrhizobium japonicum was shortened. A list of all strains with accession numbers is given in Supplementary Table 2. Scale bar indicates number of substitutions per site.
B. apis and B. tamiae share a large number of genes which are absent from the eubartonellae
The close phylogenetic position and conserved genomic synteny of B. apis and B. tamiae prompted us to analyze the gene content which they have in common with the eubartonellae (Figure 3a). We found that the three groups share a relatively large fraction of their pan genomes (1081 genes) of which 805 genes were present in all 22 analyzed genomes, thus representing the core genome of the genus Bartonella. Surprisingly, B. apis and B. tamiae share an additional 551 genes, which are absent from the eubartonellae. This represents a large fraction of their total genetic content, for example, 28% of the genome of B. tamiae strain Th239. In contrast, the eubartonellae only share 111 and 46 genes with B. apis and B. tamiae, respectively (Figure 3a). These results show that in terms of gene content, B. apis is more similar to B. tamiae than the eubartonellae, providing further evidence that the two pathogenic groups of the genus Bartonella have distinct evolutionary histories.
B. apis and B. tamiae share a large number of ancestral genes that have been lost in the eubartonellae. (a) Numbers of pan genome genes shared between B. apis (six genomes), B. tamiae (two genomes) and the eubartonellae (14 genomes). The number of core genome genes is shown in gray (that is, genes present in all 22 genomes). (b) Loss and gain of gene families mapped onto the phylogeny of Figure 2. Blue and red numbers indicate number of genes gained and lost at each branch, respectively. Black numbers below branches indicate number of gene families present in the ancestral genome at a given branch. Colored shading highlights the three groups of interest: B. apis (blue), B. tamiae (green) and eubartonellae (yellow). (c) Distribution of the 737 genes that were lost on the ancestral branch of the eubartonellae into subsets of shared genes according to the pan genome analysis in a.
The accessory gene content of B. tamiae and B. apis was vertically inherited from the LCA of the genus Bartonella
Analysis of the genomic structure revealed no clustering of the 551 genes exclusively shared by B. apis and B. tamiae providing little evidence for horizontal gene transfer en bloc (Figure 1). Using a gene flux analysis, we inferred when genes were gained and lost along the species tree shown in Figure 2. In agreement with previous studies (Boussau et al., 2004; Guy et al., 2013), this analysis revealed an extensive gene loss before and after the split of the families of Brucellaceae (Brucella melitensis and Ochrobactrum anthropi) and Bartonellaceae (Ca. T. hoelldoblerii and genus Bartonella) (Figure 3b). This general trend of genome reduction continued within the genus Bartonella. Our analysis predicted that an additional 737 gene families were lost after the divergence of B. apis and B. tamiae, on the ancestral branch toward the eubartonellae (Figure 3b, Supplementary Table 3). Strikingly, 456 of the 551 shared genes by B. tamiae and B. apis belong to these 737 gene families (Figure 3c), indicating that a large fraction (83%) of the genes exclusively shared by these two species were vertically inherited from the LCA of the genus Bartonella. A BLASTP analysis of these 551 genes against the non-redundant database corroborated these findings yielding mainly hits to other Rhizobiales (Supplementary Figure 5).
Among the remaining genes that were lost by the eubartonellae are 214 genes present only in B. apis (Figure 3c). Apparently, these vertically inherited genes were also lost by B. tamiae and only retained by B. apis. Conversely, only 60 genes were retained in B. tamiae, but lost by B. apis (Figure 3c). Moreover, only 31 genes were lost by B. tamiae and B. apis, but retained by the eubartonellae (Figure 3b). These findings show that B. tamiae and B. apis harbor a markedly larger set of ancestral gene functions than the eubartonellae and that B. apis has retained the largest number of such ancestral gene families. Consequently, based on its functional potential, B. apis seems to resemble most the LCA of this genus. Considering that all bartonellae colonize insect hosts, it is conceivable that the LCA of the genus Bartonella was already an insect-associated gut symbiont, from which mammalian pathogens evolved. In support of this scenario is the existence of a deeply rooted sister lineage comprising gut symbionts of diverse ant species (including Ca. Tokpelaia hoelldoblerii, Figure 2) (Stoll et al., 2007; Russell et al., 2009). Similar to this suggested evolutionary trajectory for the eubartonellae, vertebrate pathogens of the genus Rickettsia are thought to have emerged from insect symbionts. Pathogenic rickettsiae are transmitted by blood-feeding arthropods. However, the largest part of the currently known diversity of this genus consists of non-pathogenic arthropod associated strains (Perlman et al., 2006; Weinert et al., 2009). Also like B. apis, rickettsiae seem to be mainly facultative symbionts of their invertebrate hosts (Perlman et al., 2006). Possibly, a larger diversity of Bartonella-like bacteria is still to be discovered among insects.
The LCA of the genus Bartonella encoded amino acid and cofactor biosynthesis pathways
One hundred ten of the 456 ancestral gene families that were retained in the genomes of B. apis and B. tamiae, but lost in the eubartonellae belong to the COG categories ‘Amino acid transport and metabolism’, and ‘Coenzyme transport and metabolism’ (Figure 4a; Supplementary Table 3), suggesting that core metabolic functions were lost by the eubartonellae. For example, B. tamiae and B. apis both encode complete biosynthesis pathways for all amino acids except Asn and Ala (Supplementary Table 4). According to our gene flux analysis, these pathways were vertically inherited from the LCA of Bartonella. The eubartonellae, however, have experienced substantial gene loss in these pathways (Figure 4b). Only six pathways were found to be complete in all analyzed genomes of the eubartonellae (Lys, Asp, Glu, Gln, Pro and Gly) (Supplementary Table 4). Although most genes were lost on the ancestral branch of the eubartonellae, we noted that further gene loss must have occurred within sublineages, as some species were missing more genes than others (Figure 4b and Supplementary Table 4).
Functional classification of ancestral gene families that were lost in the eubartonellae, but retained in B. apis and B. tamiae. (a) COG category distribution of B. apis and B. tamiae shared genes, which were lost in the eubartonellae. Of 456 gene families 387 that could be assigned to a COG category are shown in the bar graph. Categories were sorted according to the number of assigned gene families. Numbers indicate percentages. (b) Presence of amino acid and cofactor biosynthesis pathway genes in 22 Bartonella genomes and their inferred LCA. Only pathways for which genes were detected in the inferred genome of the LCA are shown (Supplementary Tables 4 and 5). Cofactors marked with asterisks indicate that some genes of the pathway were missing in the LCA and could not be identified in any of the analyzed genomes of contemporary Bartonella species. Intensity of blue coloring indicates number of genes present in each pathway according to the depicted scale.
A similar pattern of gene loss was found for cofactor biosynthesis (Figure 4b). All strains of B. apis and B. tamiae encode ancestral pathways for the biosynthesis of cofactors including heme, vitamin B12, vitamin B6, molybdopterine and tetrahydrofolate (Supplementary Table 5). It remains elusive whether B. apis and B. tamiae are completely autonomous in producing these cofactors, because some necessary enzymes could not be identified in the analyzed genomes. However, in the eubartonellae, these pathways are practically absent with most genes predicted to have been lost on the ancestral branch preceding the radiation (Supplementary Table 5).
The eubartonellae likely exploit the host for amino acid and cofactor acquisition, which makes the corresponding biosynthetic pathways superfluous. Indeed, genome-wide experimental screens have previously revealed that amino acid transporters and heme-binding proteins and transporters are essential for eubartonellae to colonize and persist in the mammalian bloodstream (Mavris et al., 2005; Saenz et al., 2007). These findings are in line with the absence of biosynthesis capabilities in other (facultative) intracellular pathogens (Zhang and Rubin, 2013).
In contrast, the honey bee gut symbiont B. apis does not seem to rely on amino acids and cofactors from the host. Its extracellular lifestyle, the competition with other gut bacteria and the nitrogen-limited plant diet of the host may impose strong selective pressure to retain the vertically inherited biosynthetic capabilities. Accordingly, these pathways were also found to be conserved in other honey bee gut symbionts (Kwong and Moran, 2016).
The conservation of the vitamin B12 biosynthesis pathway is particularly interesting. This pathway involves over 30 different steps, but only a few enzymes require vitamin B12 as cofactor, including methylmalonyl CoA mutase (MCM, EC 5.4.99.2) and methionine synthase H (MetH, EC 2.1.1.13). Both enzymes are encoded in the genomes of B. apis and B. tamiae, but are absent from the eubartonellae. MetH catalyzes the last step of the methionine biosynthesis. Thus, the conservation of the vitamin B12 and the methionine biosynthesis pathways is likely coupled. The second vitamin B12-dependent enzyme, MCM, catalyzes the last step in the degradation of propionate. This pathway is present in all six B. apis strains, but in none of the other bartonellae (Supplementary Figure 6). As in mammals or termites (den Besten et al., 2013; Brune, 2014), propionate may be an end product of bacterial fermentation in the honey bee hindgut, which could be utilized by B. apis for energy production.
The presence of ancestral biosynthetic pathways in B. apis suggests that the LCA of the genus Bartonella was a metabolically self-reliant bacterium. However, it was rather surprising to find that the pathogen B. tamiae has retained most of these ancestral pathways. In line with the evolution of the eubartonellae, adaptation to the mammalian bloodstream should have resulted in the loss of these pathways. Possibly, the primary ecological niche of B. tamiae is in the gut of hematophageous insects and colonization of mammals only happens incidentally. Thus, the genome of B. tamiae may not be streamlined to a host-restricted intraerythrocytic lifestyle as in the case of the eubartonellae. Indeed, to date little is known about a possible mammalian reservoir host of B. tamiae. Only three studies have so far detected B. tamiae in mammals, two in humans (Kosoy et al., 2008; 2010) and one in bats (Leulmi et al., 2016).
Bartonella apis-specific gene contents
We analyzed the B. apis-specific gene content to obtain insights into possible functional roles of this symbiont in the honey bee gut, but also to better understand the functional capabilities of the inferred ancestor of the entire genus. Out of 1241 B. apis-specific genes (Figure 3a), 289 are present in all six strains suggesting key functions for the ecology of this bacterium. On the basis of the gene flux analysis, 158 genes were vertically inherited from the LCA and 131 genes were acquired by horizontal gene transfer.
Anaerobic respiration via nitrate reduction
Interestingly, a large fraction of the B. apis-specific core genes are predicted to be involved in nitrogen metabolism. All six strains of B. apis encode a nitrate reductase gene cluster for anaerobic respiration (Supplementary Figure 7a). Two nitrate/nitrite antiporter genes are located upstream of the reductase genes and four to five ABC-type nitrate/taurine transporters are encoded elsewhere in the six genomes. According to the gene flux analysis and phylogenetic trees, the reductase genes were vertically inherited, whereas the ABC-type transporters were acquired by horizontal gene transfer (Supplementary Figure 7b and 7c). The acquisition of these additional transporters suggests that B. apis has a high demand for nitrate, possibly as an electron acceptor to acquire energy via anaerobic respiration. Previous experiments have indeed shown that B. apis, in contrast to other bartonellae, reduces nitrate to nitrite in vitro (Kešnerová et al., 2016).
Recycling of nitrogenous waste products
For several herbivorous insects, it was proposed that gut bacteria may recycle nitrogenous waste products into amino acids (for example, Anderson et al., 2012). Strikingly, all six strains of B. apis encode a vertically inherited urease gene cluster to degrade urea into ammonia, which in turn can be converted into glutamine and glutamate (Supplementary Figure 8). Urea may derive from uric acid, the major waste product released by insects into the hindgut (McNally et al., 1965; Gullan and Cranston, 2009). Accordingly, a gene cluster for uric acid degradation is encoded in all six strains of B. apis. However, genes for two key enzymes are missing, suggesting that B. apis is not capable of converting uric acid on its own (Supplementary Figure 9). It is interesting to note that also the related ant gut symbiont Ca. Tokpelaia hoelldoblerii has retained the urease gene cluster suggesting that recycling of urea may be an important function of rhizobial gut symbionts that was also present in the LCA of the genus Bartonella. In contrast, neither the analyzed eubartonellae, nor the two B. tamiae strains encode the urease gene cluster. Thus, it was recently hypothesized that the loss of this function may have been a critical step toward the adaptation to hematophageous insect hosts (Neuvonen et al., 2016).
Degradation of plant secondary metabolites
Among the B. apis-specific gene content were also genes for the degradation of secondary plant metabolites. Four of the six strains encode the complete protocatechuate pathway for the degradation of 4-hydroxybenzoate, a common plant-derived aromatic compound (Figure 5). In addition, all six B. apis strains encode two quinone-dependent quinate dehydrogenases. One of these genes and the genes for the biosynthesis of the cofactor pyrroloquinoline quinone are located upstream of the gene cluster of the protocatechuate pathway (Figure 5). Quinate is a plant-derived cyclic polyol that is degraded by bacteria into shikimate and hippuric acid and subsequently used for amino acid biosynthesis (Teramoto et al., 2009). The protocatechuate pathway and the two quinate dehydrogenase genes were likely acquired by horizontal gene transfer, as the gene trees are incongruent with the species phylogeny (Supplementary Figures 10 and 11).
Genomic region encoding genes for the degradation of aromatic compounds in B. apis. The genomic regions of the six B. apis strains are compared with the corresponding region in B. tamiae strain Th239. Genes are depicted as arrows in different colors according to the legend. TBLASTX hits between genomes are shown as bands, with gray intensity reflecting the percentage identity of a given hit (e-value cutoff=10−15). In strain BBC0122 and PEB0150, the gene cluster of the protocatechuate pathway (that is, 4-hydroxybenzoate degradation) is interrupted by the insertion of a gene cluster coding for several dehydrogenase genes.
As plant secondary metabolites are present in pollen and nectar, they could be used by bee gut symbionts for energy and amino acid production. Likewise, these functions may have been beneficial for a herbivorous insect-associated ancestor of the genus Bartonella. However, at which point in evolution these genes were acquired, whether in a common ancestor of the entire genus or specifically in the lineage of B. apis, cannot be concluded from the current data.
Acquisition and expansion of virulence factors in the eubartonellae after divergence from B. apis and B. tamiae
To learn more about the evolutionary origin of the genus’ pathogenicity, we analyzed which virulence factors of the eubartonellae are present in B. apis and B. tamiae. We searched for 88 virulence factors (that is, genes important for host interaction or establishment of an intraerythrocytic infection) of which 69 were classified as conserved (that is, present in alphaproteobacterial outgroup species) and 19 as Bartonella-specific (Supplementary Table 6). Except for two genes of unknown functions, none of the Bartonella-specific virulence factors are present in the genomes of B. apis and B. tamiae (Figure 6a) including the well-characterized trw and virB T4SS and most autotransporter genes (Harms and Dehio, 2012). Consequently, these virulence factors must have been acquired after the divergence of B. apis and B. tamiae, consistent with the hypothesis that they were key innovations for the radiation of the eubartonellae, facilitating the adaptation to different mammalian host species (Engel et al., 2011; Guy et al., 2013).
Bartonella virulence factors in B. apis and B. tamiae. (a) Distribution of 88 virulence factors in B. apis and B. tamiae categorized according to their evolutionary history (conserved in Rhizobiales versus Bartonella-specific). (b) The YadA-like trimeric autotransporter gene badA of B. henselae Houston-1 (GenBank accession CAF26961) compared with three homologous genes or gene fragments identified on different contigs in the draft genome of B. tamiae Th239. Protein domains are shown in different colors according to the legend and based on Riess et al., (2004). Contig names are indicated below the gene representations. AIMB01000003 and AIMB01000004 are adjacent contigs suggesting that the two open reading frames may present the N- and C-terminal part of the gene. In strain Th239, but not in strain Th307, the gene on contig AIMB01000003 has a frameshift in a homopolymeric G stretch as indicated by an asterisk. Vertical lines show contig ends. Diagonal dashes indicate that the contig would continue. TBLASTX hits are shown between the homologs of B. henselae and B. tamiae, with gray intensity reflecting the percentage identity of a hit (e-value cutoff=10−3). (c) Maximum likelihood phylogeny (model: WAG) of the hemin-binding proteins (Hbps) of Bartonella and corresponding homologs identified in outgroup species. The tree is based on the protein alignment (273 aa) of the conserved C-terminal region. The alignment was stripped of alignment positions with less than 50% coverage. Colors indicate different Bartonella species. A tree including the Hbps identified in all analyzed Bartonella species is shown in Supplementary Figure 13a. Bar indicates number of substitutions per site.
The majority of the conserved virulence factors have orthologs in B. apis and B. tamiae (Figure 6a). Interestingly, both strains of B. tamiae encode orthologs of the YadA-like trimeric autotransporter family (TAA) (Supplementary Figure 12). Representatives of this adhesin gene family, for example, BadA of B. henselae, mediate important interactions of the eubartonellae with their mammalian hosts (Riess et al., 2004; Zhang et al., 2004; Saenz et al., 2007; Lu et al., 2013). Both homologs of B. tamiae have the conserved C-terminal anchor domain as well as the characteristic stalk repeats of TAAs (Figure 6b). However, the N-terminal YadA-like head domain, thought to mediate binding to host components (Riess et al., 2004; Szczesny et al., 2008), is not conserved. Instead, one of the two homologs of B. tamiae contains a 600-bp N-terminal extension including a signal peptide. Future experimental studies need to verify whether the TAAs of B. tamiae play similar roles for mammalian host interaction as their homologs in the eubartonellae.
Another group of conserved virulence factors, which show distinct patterns of evolution within the three major lineages of Bartonellae are hemin-binding proteins (Hbp). Although both B. apis and B. tamiae encode two ancestrally duplicated paralogs (one is pseudogenized in B. tamiae), this gene family has substantially expanded and diversified in the eubartonellae. Certain strains encode up to eight copies of hbp genes. Phylogenetic analysis suggests that this expansion occurred after the divergence of B. apis and B. tamiae via repeated duplication events (Figure 6c). Moreover, paralogs of this gene family show higher levels of sequence divergence in the eubartonellae than in B. tamiae and B. apis (Supplementary Figure 13b). Hbps are outer membrane proteins involved in the acquisition of heme and iron from the environment (Lee, 1992). Thus, the expansion of this family may have complemented the loss of the heme biosynthesis genes in the eubartonellae (Figure 4b). Expression of a diverse set of Hbps may increase the efficiency to absorb and uptake heme from the blood. Alternatively, the binding properties of the Hbps may lead to an extracellular heme coat that could serve as a nutritive reservoir for Bartonella during passage through the arthropod host, as shown for Y. pestis (Hinnebusch et al., 1996), or may act as antioxidant barrier against reactive oxygen species (Harms and Dehio, 2012). Independent of the precise role of the Hbps, our data strongly suggests that the expansion of this gene family is an adaptation to the specific infection strategy of Bartonella.
Conclusion
In this study, we sequenced the first six genomes of the honey bee gut symbiont B. apis, the closest known relative of the pathogenic members of the genus Bartonella. Our results show that B. apis has retained a large ancestral gene pool, which allowed us to reconstruct the metabolic capabilities of the Bartonella LCA. This provided compelling new insights into the genomic changes associated with the evolution of the characteristic intraerythrocytic infection strategy of Bartonella.
We conclude that the LCA of Bartonella was likely an amino acid and cofactor self-reliant gut symbiont that recycled nitrogenous waste products from its insect host. This is corroborated by the fact that a deeply rooted lineage of the genus Bartonella comprises a large group of ant-associated Rhizobiales with presumably similar functional capabilities (for example, Stoll et al., 2007; Russell et al., 2009; Neuvonen et al., 2016). Our hypothesis that the mammalian pathogens of the genus Bartonella derived from an insect-associated gut symbiont is in contrast to the evolution of other vector-borne pathogens. For example, Y. pestis is believed to have been adapted to the mammalian host before it evolved insect-vector transmission (Chain et al., 2004). It is similar, however, to the emergence of pathogens in the genera Rickettsia (Perlman et al., 2006; Weinert et al., 2009) and Coxiella (Duron et al., 2015). Bacterial strains of the genus Coxiella have so far only been found in ticks and among them are maternally inherited endosymbionts that form a basal lineage to the mammalian pathogen Coxiella burnetti (Duron et al., 2015). Genome analyses of these endosymbionts suggest that they provision their tick hosts with vitamins and cofactors (Gottlieb et al., 2015; Smith et al., 2015). A key step in the evolution of the pathogenic Bartonella, Coxiella and Rickettsia was probably the colonization of blood-sucking arthropods. In Bartonella, this transition could have happened twice, once in the ancestor of the eubartonellae, and another time in the lineage of B. tamiae. Because our phylogenetic analyses based on amino acid and DNA sequences disagree on the placement of B. tamiae, discovery of more Bartonella-like bacteria could further resolve the evolution of the genus.
In contrast to the eubartonellae, B. tamiae has retained ancestral amino acid and cofactor biosynthesis pathways and lacks most Bartonella-specific virulence factors. This indicates that B. tamiae may have a different ecology than the eubartonellae. Possibly, B. tamiae presents an intermediate state of the evolutionary transition from a gut symbiont to a mammalian parasite. Although it is adapted to colonize hematophageous insects, it may not be adapted to a mammalian reservoir host and only causes opportunistic infections. Future experimental studies focusing on the infection course of B. tamiae in mammals will shed light on the stepwise evolution of virulence in the genus Bartonella. For example, it is currently unknown whether B. tamiae can enter erythrocytes and persist in the bloodstream, as in the case of the eubartonellae (Harms and Dehio, 2012).
Finally, our study shows that genome reduction in host-restricted bacterial pathogens may be counteracted by the expansion of vertically inherited gene families. Besides the acquisition of dedicated virulence factors, we find that the eubartonellae have compensated the loss of the heme biosynthesis pathway by the expansion of an ancestral family of Hbps. This may allow pathogenic Bartonella to efficiently acquire heme and iron from the mammalian host and overcome nutritional shortages during passage through the insect vector.
Bartonellae are experimentally amenable and in vivo colonization models are established for both honey bee gut symbionts (Engel et al., 2015) and Bartonella mammalian pathogens (Harms and Dehio, 2012). Thus, our comparative genomic analysis sets the ground for future experimental studies to understand how changes in gene content affect host interactions during the evolutionary transition from an insect-associated gut symbiont to a mammalian pathogen.
References
Alsmark CM, Frank AC, Karlberg EO, Legault B-A, Ardell DH, Canbäck B et al. (2004). The louse-borne human pathogen Bartonella quintana is a genomic derivative of the zoonotic agent Bartonella henselae. Proc Natl Acad Sci USA 101: 9716–9721.
Anderson KE, Russell JA, Moreau CS, Kautz S, Sullam KE, Hu Y et al. (2012). Highly similar microbial communities are shared among related and trophically similar ant species. Mol Ecol 21: 2282–2296.
Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS et al. (2012). SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol 19: 455–477.
Batut J, Andersson SGE, O'Callaghan D . (2004). The evolution of chronic infection strategies in the alpha-proteobacteria. Nat Rev Microbiol 2: 933–945.
Bentley SD, Parkhill J . (2015). Genomic perspectives on the evolution and spread of bacterial pathogens. Proc Biol Sci 282: 20150488.
Besten, den G, van Eunen K, Groen AK, Venema K . (2013). The role of short-chain fatty acids in the interplay between diet, gut microbiota, and host energy metabolism. J Lipid Res 54: 2325–2340.
Billeter SA, Miller MK, Breitschwerdt EB, Levy MG . (2008). Detection of two Bartonella tamiae-like sequences in Amblyomma americanum Acari: Ixodidae using 16S-23S intergenic spacer region-specific primers. J Med Entomol 45: 176–179.
Boussau B, Karlberg EO, Frank AC, Legault B-A, Andersson SGE . (2004). Computational inference of scenarios for alpha-proteobacterial genome evolution. Proc Natl Acad Sci USA 101: 9722–9727.
Brune A . (2014). Symbiotic digestion of lignocellulose in termite guts. Nat Rev Microbiol 12: 168–180.
Chain PSG, Carniel E, Larimer FW, Lamerdin J, Stoutland PO, Regala WM et al. (2004). Insights into the evolution of Yersinia pestis through whole-genome comparison with Yersinia pseudotuberculosis. Proc Natl Acad Sci USA 101: 13826–13831.
Chin C-S, Alexander DH, Marks P, Klammer AA, Drake J, Heiner C et al. (2013). Nonhybrid, finished microbial genome assemblies from long-read SMRT sequencing data. Nat Methods 10: 563–569.
Cox-Foster DL, Conlan S, Holmes EC, Palacios G, Evans JD, Moran NA et al. (2007). A metagenomic survey of microbes in honey bee colony collapse disorder. Science 318: 283–287.
Duron O, Noël V, McCoy KD, Bonazzi M, Sidi-Boumedine K, Morel O et al. (2015). The recent evolution of a maternally-inherited endosymbiont of ticks led to the emergence of the Q fever pathogen, Coxiella burnetii. PLoS Pathog 11: e1004892.
Ellegaard KM, Tamarit D, Javelind E, Olofsson TC, Andersson SGE, Vásquez A . (2015). Extensive intra-phylotype diversity in lactobacilli and bifidobacteria from the honeybee gut. BMC Genomics 16: 284.
Engel P, Bartlett KD, Moran NA . (2015). The Bacterium Frischella perrara causes scab formation in the gut of its honeybee host. MBio 6: e00193–15.
Engel P, Martinson VG, Moran NA . (2012). Functional diversity within the simple gut microbiota of the honey bee. Proc Natl Acad Sci USA 109: 11002–11007.
Engel P, Salzburger W, Liesch M, Chang C-C, Maruyama S, Lanz C et al. (2011). Parallel evolution of a type IV secretion system in radiating lineages of the host-restricted bacterial pathogen Bartonella. PLoS Genet 7: e1001296.
Engel P, Stepanauskas R, Moran NA . (2014). Hidden diversity in honey bee gut symbionts detected by single-cell genomics. PLoS Genet 10: e1004596.
Ettema TJG, Andersson SGE . (2009). The alpha-proteobacteria: the Darwin finches of the bacterial world. Biol Lett 5: 429–432.
Feldhaar H, Straka J, Krischke M, Berthold K, Stoll S, Mueller MJ et al. (2007). Nutritional upgrading for omnivorous carpenter ants by the endosymbiont Blochmannia. BMC Biol 5: 48.
Gottlieb Y, Lalzar I, Klasson L . (2015). Distinctive genome reduction rates revealed by genomic analyses of two Coxiella-like endosymbionts in ticks. Genome Biol Evol 7: 1779–1796.
Gullan PJ, Cranston PS . (2009) The Insects: An Outline of Entomology. Wiley-Blackwell: West Sussex, UK.
Guy L, Kultima JR, Andersson SGE . (2010). genoPlotR: comparative gene and genome visualization in R. Bioinformatics 26: 2334–2335.
Guy L, Nystedt B, Toft C, Zaremba-Niedzwiedzka K, Berglund EC, Granberg F et al. (2013). A gene transfer agent and a dynamic repertoire of secretion systems hold the keys to the explosive radiation of the emerging pathogen Bartonella. PLoS Genet 9: e1003393.
Harms A, Dehio C . (2012). Intruders below the radar: molecular pathogenesis of Bartonella spp. Clin Microbiol Rev 25: 42–78.
Hinnebusch JB, Perry RD, Schwan TG . (1996). Role of the Yersinia pestis hemin storage (hms) locus in the transmission of plague by fleas. Science 273: 367–370.
Hu Y, Łukasik P, Moreau CS, Russell JA . (2014). Correlates of gut community composition across an ant species (Cephalotes varians elucidate causes and consequences of symbiotic variability. Molecular Ecology 23: 1284–1300.
Kabeya H, Colborn JM, Bai Y, Lerdthusnee K, Richardson JH, Maruyama S et al. (2010). Detection of Bartonella tamiae DNA in ectoparasites from rodents in Thailand and their sequence similarity with bacterial cultures from Thai patients. Vector Borne Zoonotic Dis 10: 429–434.
Kešnerová L, Moritz R, Engel P . (2016). Bartonella apis sp. nov., a honey bee gut symbiont of the class Alphaproteobacteria. Int J Syst Evol Microbiol 66: 414–421.
Kosoy M, Bai Y, Sheff K, Morway C, Baggett H, Maloney SA et al. (2010). Identification of Bartonella infections in febrile human patients from Thailand and their potential animal reservoirs. Am J Trop Med Hyg 82: 1140–1145.
Kosoy M, Morway C, Sheff KW, Bai Y, Colborn J, Chalcraft L et al. (2008). Bartonella tamiae sp. nov., a newly recognized pathogen isolated from three human patients from Thailand. J Clin Microbiol 46: 772–775.
Kwong WK, Moran NA . (2016). Gut microbial communities of social bees. Nat Rev Microbiol 14: 374–384.
Langridge GC, Fookes M, Connor TR, Feltwell T, Feasey N, Parsons BN et al. (2015). Patterns of genome evolution that have accompanied host adaptation in Salmonella. Proc Natl Acad Sci USA 112: 863–868.
Lartillot N, Lepage T, Blanquart S . (2009). PhyloBayes 3: a Bayesian software package for phylogenetic reconstruction and molecular dating. Bioinformatics 25: 2286–2288.
Lee BC . (1992). Isolation of an outer membrane hemin-binding protein of Haemophilus influenzae type b. Infect Immun 60: 810–816.
Leulmi H, Aouadi A, Bitam I, Bessas A, Benakhla A, Raoult D et al. (2016). Detection of Bartonella tamiae Coxiella burnetii and rickettsiae in arthropods and tissues from wild and domestic animals in northeastern Algeria. Parasit Vectors 9: 27.
Li L, Stoeckert CJ, Roos DS . (2003). OrthoMCL: identification of ortholog groups for eukaryotic genomes. Genome Res 13: 2178–2189.
Lu Y-Y, Franz B, Truttmann MC, Riess T, Gay-Fraret J, Faustmann M et al. (2013). Bartonella henselae trimeric autotransporter adhesin BadA expression interferes with effector translocation by the VirB/D4 type IV secretion system. Cell Microbiol 15: 759–778.
Markowitz VM, Chen I-MA, Chu K, Szeto E, Palaniappan K, Pillay M et al. (2014). IMG/M 4 version of the integrated metagenome comparative analysis system. Nucl Acids Res 42: D568–D573.
Martinson VG, Danforth BN, Minckley RL, Rueppel O, Tingek S, Moran NA . (2011). A simple and distinctive microbiota associated with honey bees and bumble bees. Mol Ecol 20: 619–628.
Mavris M, Saenz H, Monteil M, Boulouis HJ, Dehio C, Vayssier-Taussat M . (2005). Characterization of genes involved in long-term bacteremia in mice by Bartonella birtlesii. Ann N Y Acad Sci 1063: 312–314.
McCutcheon JP, Moran NA . (2012). Extreme genome reduction in symbiotic bacteria. Nat Rev Microbiol 10: 13–26.
McNally JB, McCaughey WF, Standifer LN, Todd FE . (1965). Partition of excreted nitrogen from honey bees fed various proteins. J Nutr 85: 113–116.
Nei M, Gojobori T . (1986). Simple methods for estimating the numbers of synonymous and nonsynonymous nucleotide substitutions. Mol Biol Evol 3: 418–426.
Neuvonen M-M, Tamarit D, Näslund K, Liebig J, Feldhaar H, Moran NA et al. (2016). The genome of Rhizobiales bacteria in predatory ants reveals urease gene functions but no genes for nitrogen fixation. Sci Rep 6: 39197.
Ochman H, Moran NA . (2001). Genes lost and genes found: evolution of bacterial pathogenesis and symbiosis. Science 292: 1096–1099.
Pallen MJ, Wren BW . (2007). Bacterial pathogenomics. Nature 449: 835–842.
Parkhill J, Sebaihia M, Preston A, Murphy LD, Thomson N, Harris DE et al. (2003). Comparative analysis of the genome sequences of Bordetella pertussis Bordetella parapertussis and Bordetella bronchiseptica. Nat Genet 35: 32–40.
Perlman SJ, Hunter MS, Zchori-Fein E . (2006). The emerging diversity of Rickettsia. Proc Royal Soc London B 273: 2097–2106.
Reuter S, Connor TR, Barquist L, Walker D, Feltwell T, Harris SR et al. (2014). Parallel independent evolution of pathogenicity within the genus Yersinia. Proc Natl Acad Sci USA 111: 6768–6773.
Richter M, Rosselló-Móra R, Glöckner FO, Peplies J . (2015). JSpeciesWS: a web server for prokaryotic species circumscription based on pairwise genome comparison. Bioinformatics 32: btv681–btv931.
Riess T, Andersson SGE, Lupas A, Schaller M, Schäfer A, Kyme P et al. (2004). Bartonella adhesin a mediates a proangiogenic host cell response. J Exp Med 200: 1267–1278.
Rohmer L, Fong C, Abmayr S, Wasnick M, Freeman TJL, Radey M et al. (2007). Comparison of Francisella tularensis genomes reveals evolutionary events associated with the emergence of human pathogenic strains. Genome Biol 8: R102.
Russell JA, Moreau CS, Goldman-Huertas B, Fujiwara M, Lohman DJ, Peirce NE . (2009). Bacterial gut symbionts are tightly linked with the evolution of herbivory in ants. Proc Natl Acad Sci USA 106: 21236–21241.
Saenz HL, Engel P, Stoeckli MC, Lanz C, Raddatz G, Vayssier-Taussat M et al. (2007). Genomic analysis of Bartonella identifies type IV secretion systems as host adaptability factors. Nat Genet 39: 1469–1476.
Sanders JG, Powell S, Kronauer DJC, Vasconcelos HL, Frederickson ME, Pierce NE . (2014). Stability and phylogenetic correlation in gut microbiota: lessons from ants and apes. Mol Ecol 23: 1268–1283.
Sapountzis P, Zhukova M, Hansen LH, Sørensen SJ, Schiøtt M, Boomsma JJ . (2015). Acromyrmex leaf-cutting ants have simple gut microbiota with nitrogen-fixing potential. Appl Environ Microbiol 81: 5527–5537.
Smith TA, Driscoll T, Gillespie JJ, Raghavan R . (2015). A Coxiella-like endosymbiont is a potential vitamin source for the Lone Star tick. Genome Biol Evol 7: 831–838.
Stamatakis A . (2014). RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30: 1312–1313.
Stoll S, Gadau J, Gross R, Feldhaar H . (2007). Bacterial microbiota associated with ants of the genus Tetraponera. Biol J Linnean Soc 90: 399–412.
Szczesny P, Linke D, Ursinus A, Bär K, Schwarz H, Riess TM et al. (2008). Structure of the head of the Bartonella adhesin BadA. PLoS Pathog 4: e1000119.
Tamarit D, Ellegaard KM, Wikander J, Olofsson T, Vásquez A, Andersson SGE . (2015). Functionally structured genomes in Lactobacillus kunkeei colonizing the honey crop and food products of honeybees and stingless bees. Genome Biol Evol 7: 1455–1473.
Teramoto H, Inui M, Yukawa H . (2009). Regulation of expression of genes involved in quinate and shikimate utilization in Corynebacterium glutamicum. Appl Environ Microbiol 75: 3461–3468.
Toft C, Andersson SGE . (2010). Evolutionary microbial genomics: insights into bacterial host adaptation. Nat Rev Genet 11: 465–475.
Weinert LA, Werren JH, Aebi A, Stone GN, Jiggins FM . (2009). Evolution and diversity of Rickettsia bacteria. BMC Biol 7: 1.
Wilgenbusch JC, Swofford D . (2003). Inferring evolutionary trees with PAUP*. Curr Protoc Bioinformatics Chapter 6: Unit 6.4.1–6.4.28.
Yang Z . (2007). PAML 4: phylogenetic analysis by maximum likelihood. Mol Biol Evol 24: 1586–1591.
Zhang P, Chomel BB, Schau MK, Goo JS, Droz S, Kelminson KL et al. (2004). A family of variably expressed outer-membrane proteins (Vomp) mediates adhesion and autoaggregation in Bartonella quintana. Proc Natl Acad Sci USA 101: 13630–13635.
Zhang YJ, Rubin EJ . (2013). Feast or famine: the host-pathogen battle over amino acids. Cell Microbiol 15: 1079–1087.
Zhu Q, Kosoy M, Olival KJ, Dittmar K . (2014). Horizontal transfers and gene losses in the phospholipid pathway of bartonella reveal clues about early ecological niches. Genome Biol Evol 6: 2156–2169.
Acknowledgements
We thank Christoph Dehio, Alexander Harms, Fabienne Wichmann and Kirsten Ellegaard for critical reading of the manuscript. We wish to thank Michael Feldgarden and Paul Godfrey of the Broad Institute of Harvard and MIT for sequencing and annotating the genomes of B. tamiae and making the genome sequence publically accessible and to acknowledge the role of James Kirby of the Beth Israel Deaconess Medical Center in organizing the project on comparative genomics of multiple Bartonella species. We are very grateful to Nancy Moran for access to the genome of Ca. Tokpelaia hoelldoblerii before publication and for funding the sequencing of strains PEB0122, PEB0149 and PEB0150. This research was funded by the Swiss National Science foundation grants PZ00P3_148264 and 31003A_160345 (to P.E.) and by the NSF grant DEB 1415604 (to N.M.).
Sequence deposition
The genome sequences of Bartonella apis PEB0122, PEB0149, PEB0150, BBC0122, BBC0178, and BBC0244 are deposited in GenBank with accession numbers LXYU00000000, LXYT00000000, LXYS00000000 CP015625, CP015820 and CP015821.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies this paper on The ISME Journal website
Rights and permissions
This work is licensed under a Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-nd/4.0/
About this article
Cite this article
Segers, F., Kešnerová, L., Kosoy, M. et al. Genomic changes associated with the evolutionary transition of an insect gut symbiont into a blood-borne pathogen. ISME J 11, 1232–1244 (2017). https://doi.org/10.1038/ismej.2016.201
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ismej.2016.201
This article is cited by
-
Turnover of strain-level diversity modulates functional traits in the honeybee gut microbiome between nurses and foragers
Genome Biology (2023)
-
The Effect of Residual Pesticide Application on Microbiomes of the Storage Mite Tyrophagus putrescentiae
Microbial Ecology (2023)
-
First data on bacteria associated with bat ectoparasites collected in Kharkiv oblast, Northeastern Ukraine
Parasites & Vectors (2022)
-
Seed properties and bacterial communities are associated with feeding preferences of a seed-eating beetle
Plant and Soil (2022)
-
Global similarity, and some key differences, in the metagenomes of Swedish varroa-surviving and varroa-susceptible honeybees
Scientific Reports (2021)