Abstract
Microbial interactions, including mutualistic nutrient exchange (cross-feeding), underpin the flow of energy and materials in all ecosystems. Metabolic exchanges are difficult to assess within natural systems. As such, the impact of exchange levels on ecosystem dynamics and function remains unclear. To assess how cross-feeding levels govern mutualism behavior, we developed a bacterial coculture amenable to both modeling and experimental manipulation. In this coculture, which resembles an anaerobic food web, fermentative Escherichia coli and photoheterotrophic Rhodopseudomonas palustris obligately cross-feed carbon (organic acids) and nitrogen (ammonium). This reciprocal exchange enforced immediate stable coexistence and coupled species growth. Genetic engineering of R. palustris to increase ammonium cross-feeding elicited increased reciprocal organic acid production from E. coli, resulting in culture acidification. Consequently, organic acid function shifted from that of a nutrient to an inhibitor, ultimately biasing species ratios and decreasing carbon transformation efficiency by the community; nonetheless, stable coexistence persisted at a new equilibrium. Thus, disrupting the symmetry of nutrient exchange can amplify alternative roles of an exchanged resource and thereby alter community function. These results have implications for our understanding of mutualistic interactions and the use of microbial consortia as biotechnology.
Similar content being viewed by others
Introduction
Ecosystems comprise a large, variable number of species that interact with both each other and their environment. Metabolic interactions between species, such as detoxification of metabolic waste and cross-feeding, shape microbial communities and regulate ecosystem processes (Schink, 2002; Fuhrman, 2009; Morris et al., 2013). Study of these interactions can be encumbered by the stochasticity and complexity of natural systems. As an alternative, synthetic microbial communities (cocultures) have been developed in which two or more species are cultivated together under laboratory conditions. Cocultures preserve core aspects of natural systems while offering greater practical experimental control (Shou et al., 2007; Hillesland and Stahl, 2010; Summers et al., 2010; Harcombe, 2010; Momeni et al., 2011; Hom and Murray, 2014; Mee et al., 2014). They are also more amenable to modeling than are natural systems and facilitate the development, experimental testing and refining of models for predicting community behavior (Zomorrodi and Segrè, 2015; Johns et al., 2016; Lindemann et al., 2016; Widder et al., 2016). As such, cocultures are very useful for defining the principles that underlie the stability and performance of microbial communities.
One factor that can promote cross-feeding in natural ecosystems is nutrient limitation, as cross-feeding can be a means by which to acquire scarce key nutrients (Shou et al., 2007; Klitgord and Segrè, 2010; Hom and Murray, 2014; Zelezniak et al., 2015). Limitation of bacterial growth is most commonly considered to result from nutrient deficiency; however, growth can also be influenced by diverse compounds that are inhibitory when abundant but serve as nutrients at lower concentrations (Abbott, 1973; Kunz et al., 1992; Alvarez et al., 2009; Barnhill et al., 2010). Organic acids, which are key intermediates in global carbon cycles (McInerney et al., 2009), exemplify this paradox in both natural and applied settings (Lee et al., 1976; Louis et al., 2007; Sousa et al., 2009; Huang et al., 2011). There is limited knowledge of how potentially toxic, cross-fed nutrients such as organic acids impact mutualism dynamics and stability. However, given the ubiquity of organic acid exchange, their impact may be profound, as the cross-feeding levels compatible with cooperation would be constrained between thresholds of inadequate exchange and toxicity.
Cocultures pairing fermentative bacteria with purple photoheterotrophic bacteria have been of interest for over 30 years (Odom and Wall, 1983; Fang et al., 2006; Ding et al., 2009; Sun et al., 2010; Jiao et al., 2012), primarily as a consolidated bioprocess for converting plant-derived sugars into H2. However, this combination of bacteria also resembles a natural anaerobic food web; fermentative bacteria consume plant-derived sugars and excrete products that serve as a carbon source for photoheterotrophs (Figure 1a). Despite decades of research, coculture instability has limited the utility of these cocultures for both practical applications, such as H2 production, and fundamental studies into anaerobic microbial cross-feeding.
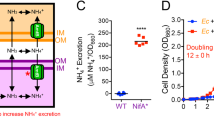
Imposing bidirectional cross-feeding changes coculture dynamics. (a, b) Cross-feeding under conditions compelling uni-directional transfer of carbon to R. palustris (a) or bidirectional transfer of carbon to R. palustris and nitrogen to E. coli (b). The latter requires a NH4+-excreting R. palustris strain. Filled bubbles indicate externally added compounds. (c) Representative growth curves for NH4+-supplied cocultures (15 mm NH4+ and 100% N2 headspace; open circles) and N2-supplied cocultures (100% N2 headspace; closed circles) containing either parental R. palustris (CGA4004; black), which does not share NH4+, or R. palustris Nx (CGA4005; green), which shares NH4+. Log transformed growth curves are in Supplementary Figure 3. All cocultures were inoculated at a 1:1 species ratio and contained 25 mm glucose. (d) Final E. coli percentage in NH4+-supplied (solid bars) or N2-supplied (hatched bars) cocultures with R. palustris Nx inoculated with different starting species ratios. A starting value of 1 corresponds to ~2.5 × 108 CFU ml−1. ND, not determined as E. coli dominated NH4+-supplied cocultures even when starting ratios did not favor E. coli. Ec, E. coli; Rp, R. palustris. Error bars, s.e.m., n=3.
Here, we use a bacterial coculture based on fermentative Escherichia coli and purple phototrophic Rhodopseudomonas palustris to assess how the dynamics and function of a mutualism are influenced by the degree of cross-feeding. In this coculture, E. coli and R. palustris stably coexist due to the obligate bidirectional exchange of two essential nutrients, carbon and nitrogen (Figure 1b). Using a combination of experimental and modeling approaches, we show that enhanced cross-feeding can disproportionately inhibit members of a mutualism by amplifying toxic attributes of an exchanged nutrient, with detrimental consequences for the community transformation of carbon into biomass and CO2. Nonetheless, obligate cross-feeding can uphold stable coexistence amid detrimental exchange levels.
Materials and methods
Strains, plasmids and growth conditions
Strains, plasmids and primers are listed in Supplementary Table 1. E. coli and R. palustris were cultivated on Luria–Burtani (LB) agar or defined mineral (PM) (Kim and Harwood, 1991) agar with 10 mm succinate, respectively. These media were also used to selectively plate for each species after coculturing, except that (NH4)2SO4 was excluded from PM plates to select for R. palustris. E. coli plates were incubated aerobically at 30 °C in the dark, whereas R. palustris plates were incubated anaerobically at 30 °C in a jar with a GasPak sachet (BD, Franklin Lakes, NJ, USA) in front of a 60 W light bulb. Cloning was performed using E. coli NEB10β (New England Biolabs, Ipswich, MA, USA) or E. coli XL10Gold (Stratagene, La Jolla, CA, USA). When necessary, gentamicin (Gm) was included at 15 μg ml−1 for E. coli or 100 μg ml−1 for R. palustris.
Cultures were grown in 10 ml of defined coculture medium (MDC) in 27-ml anaerobic test tubes except for time-course analyses, which used 60 ml MDC in 160-ml serum vials. MDC contained Na2HPO4 (42.5 mm), KH2PO4 (22 mm), Na2S2O3 (0.1 mm), p-aminobenzoic acid (14.6 μm) and trace elements solution (0.1% v/v; Kremer et al., 2015). MDC was made anaerobic by bubbling with 100% N2 and then sealing with rubber stoppers and aluminum crimps (100% N2 headspace). After autoclaving, MDC was supplemented with cation solution (1% v/v; 100 mm MgSO4 and 10 mm CaCl2) and glucose (25 mm) if used for cocultures. MDC also received either NH4Cl (15 mm, conditions which we refer to as NH4+-supplied cocultures) or NaCl (15 mm, conditions which we refer to as N2-supplied cocultures). For MOPS-supplemented cocultures, MOPS buffer (100 mm final concentration), pH 7 was added to MDC before autoclaving. For spent E. coli supernatant experiments, E. coli cultures were grown to stationary phase in MDC supplemented with cation solution, 25 mm glucose and 15 mm NH4Cl. Following removal of E. coli cells by filtration, spent supernatants were either supplemented with 100 mm MOPS buffer, pH 7 (+MOPS) or an equivalent volume of MDC (−MOPS). Spent supernatants were then injected into sterile anoxic tubes and flushed with 100% N2 before inoculation with R. palustris cells.
Final cell densities in starter monocultures were controlled using carbon limitation (MDC with 3 mm acetate) for R. palustris or nitrogen limitation (MDC with 1% v/v cation solution, 25 mm glucose and 2 mm NH4Cl) for E. coli. For non-growing E. coli suspensions, nitrogen-limited E. coli cultures were grown to stationary phase. Cells were then collected, washed and resuspended in 1 ml MDC, injected into fresh MDC with glucose but lacking NH4+, and then flushed with 100% N2. All cultures and cell suspensions were incubated at 30 °C laying flat under a 60 W incandescent bulb with shaking at 150 r.p.m.
Generation of R. palustris mutants
R. palustris mutants were derived from wild-type CGA009 (Larimer et al., 2004). The vector used to introduce the nifA* mutation was described previously (McKinlay and Harwood, 2010). Vectors used to delete other genes were generated by PCR amplifying regions flanking the gene to be deleted using primers in Supplementary Table 1. Product pairs were fused by overlap extension PCR and subsequently cloned into pJQ200SK. Vectors were introduced into R. palustris by conjugation with E. coli S17-1 (Rey et al., 2006) or by electroporation (Pelletier et al., 2008). Mutants were generated using sequential selection and screening as described (Rey et al., 2006). Mutant genotypes were confirmed by PCR and sequencing.
Generation of amtB2 complementation vector
A 2.2 kb region encompassing glnK2, amtB2 and their shared promoter was amplified using primers BL457 and BL498 (Supplementary Table 1) and cloned into pBBR1MCS-5 to generate pBBRGlnkAmtB2. To prevent GnlK2 over-expression, the gnlK2 start codon in pBBRGlnkAmtB2 was mutated to GGG by single-primer site-directed mutagenesis using primer BL499, resulting in pBBRAmtB2. The desired mutation was confirmed by sequencing.
Coculture inoculation
Cocultures were inoculated using either specific densities, single colonies or subculturing as indicated in figure legends. Cocultures inoculated with specific densities were generated by first growing monocultures in MDC with limiting nitrogen or carbon for E. coli or R. palustris, respectively. Monocultures were then diluted with MDC until the appropriate cell densities were achieved, as estimated from experimentally determined species-specific OD660/CFU per ml standard curves. For cocultures started from colonies, single colonies of each species were combined in 100 μl MDC for use as the inoculum. For subculturing, stationary phase cocultures were subcultured 1:100 into fresh medium. Growth and metabolic trends from cocultures inoculated by each of these methods were comparable.
Analytical procedures
Cell density was assayed by optical density at 660 nm (OD660) using a Genesys 20 visible spectrophotometer (Thermo-Fisher, Waltham, MA, USA). Most growth curve readings were taken in culture tubes without sampling. Specific growth rates were determined using tube OD measurements between 0.1 and 1.0 OD660 where there is linear correlation between cell density and OD660. Final cell densities and all serum vial readings were taken in cuvettes and samples were diluted as necessary to achieve values below 1 OD660. H2 was quantified using a Shimadzu (Kyoto, Japan) gas chromatograph as described (Huang et al., 2010). Glucose, organic acids, and ethanol were quantified using a Shimadzu high-performance liquid chromatograph (HPLC) as described (McKinlay et al., 2005). NH4+ was quantified using an indophenol colorimetric assay as follows. Cultures were sampled 15–30 h into stationary phase. Supernatant (550 μl) was mixed with 50 μl 1 m NaOH, 100 μl phenol nitroprusside (Sigma, St Louis, MO, USA), and 100 μl alkaline hypochlorite (Sigma), followed by incubation at room temperature for 15 min. The A630nm of each sample was converted into μM NH4+ using a standard curve and then normalized to cell density (OD660).
Determination of species ratios and cell densities
All final species ratios were determined using fluorescence in situ hybridization and microscopy (FISH). Cells were fixed, washed, stored and dehydrated for hybridization as described (Oda et al., 2000). Dried cell pellets were resuspended in 50 μl hybridization buffer (Oda et al., 2000) incubated at 37 °C for 30 min, and then hybridized for 2.5 h at 60 °C with an E. coli specific probe (ALM-Ec3, 4.5 ng μl−1, ATTO550-5′–TGCTCTCGCGAGGTCGCTTC–3′; ATTO-TEC, Siegen, Germany). Probe specificity was empirically confirmed using E. coli and R. palustris monocultures. Hybridized samples were washed three times with hybridization buffer and then resuspended in 5 μm SYTO9 (Life Technologies, Carlsbad, CA, USA) to stain both species. At least 2000 cells were visualized per sample by epifluorescence microscopy (Ti-E; Nikon, Tokyo, Japan) within 72 h of hybridization. Cell counting was partially automated using Microbe J (Ducret et al., 2016). E. coli frequency was determined from the number of ATTO550-positive cells per total number of Syto9-positive cells counted in the sample. Species ratios determined using FISH were periodically confirmed by CFU enumeration for corresponding live cocultures. Intermediate species ratios as well as monoculture and coculture cell densities were determined by selective plating and CFU enumeration.
Mathematical modeling
Ecological models A and B were based on Monod models used to describe microbial mutualism (Meyer et al., 1975; Lee et al., 1976) but were modified as follows: (i) the dilution parameter was omitted to describe batch cultures rather than continuous cultures; (ii) growth-independent fermentation parameters were included (‘r’, Supplementary Table 2); (iii) a parameter to modulate E. coli product excretion between growth-dependent and growth-independent levels was included (‘ng’ used in the term ‘ng/(μ+ng)’, Supplementary Table 2); and (iv) model A also includes parameters to describe the growth-inhibiting pH effects of organic acids on both species (‘b’ used in the term ‘b/(b+10(f+C))’ Supplementary Table 2); this inhibition was omitted in model B. Growth inhibition by ethanol was not included as ethanol levels never reach those we determined to inhibit growth in monoculture. The models were developed in R studio and are available for download at https://github.com/McKinlab/Coculture-Mutualism. Default parameter values and descriptions are in Supplementary Table 2. The differential equations used in the models are as follows:


Equations (3–12) describe the changes in cell densities and extracellular compounds through time. Only net accumulation of formate, ethanol, CO2 and H2 are described in accordance with observed trends. Numerical constants in product excretion equations are used to account for the moles of product carbon relative to glucose. All R and r parameters are expressed in stoichiometric terms of glucose consumed except for RA, which is the amount of NH4+ produced per R. palustris cell (Supplementary Table S2).










where,
μ is the specific growth rate of the indicated species (h−1).
μMAX is the maximum specific growth rate of the indicated species (h−1), based on growth rates measured in monocultures in MDC with acetate and 100% N2 for R. palustris or glucose and NH4Cl for E. coli.
G, A, C, N, f, e and CO2 are the concentrations (mm) of glucose, NH4+, consumable organic acids, N2 (assumed to be fully dissolved), formate, ethanol and CO2, respectively. Consumable organic acids are those that R. palustris consumes, namely, lactate (3 carbons), acetate (2 carbons) and succinate (4 carbons). All consumable organic acids were simulated to have three carbons for convenience.
k is the half saturation constant for the indicated substrate (mm).
Ec and Rp are the cell densities (cells ml−1) of E. coli and R. palustris, respectively.
ng is used to modulate the transition between values used to describe product excretion by E. coli in the presence and absence of growth (h−1).
b is used to modulate the extent to which an indicated species resists growth-inhibiting effects of organic acids (mm).
Y is the E. coli or R. palustris cell yield from the indicated substrate (cells μmol−1 glucose). Values were determined for each species in MDC with the indicated substrate as the limiting nutrient.
R is the fraction of glucose converted into the indicated compound per E. coli cell during growth (μmol cell−1), except for RA. Values were based on product yields measured in monocultures in MDC with acetate and 100% N2 for R. palustris or glucose and NH4Cl for E. coli.
RA is the ratio of NH4+ produced per R. palustris cell during growth (μmol cell−1). The default value was based on that which led to simulations resembling observed trends.
r is the growth-independent rate of glucose converted into the indicated compound by E. coli (μmol cell−1 h−1). Specific production rates for non-growing E. coli (r) were determined in nitrogen-free cell suspensions by linear regression between days 1–7 (Supplementary Figure 1). E. coli product excretion rates were increased twofold from those measured in cell suspensions as we assumed that higher formate yield observed in coculture was true of other products as well. Ethanol and H2 production rates were unchanged from monoculture observations to match coculture observations.
Results
Obligate mutualism enforces stable coexistence in coculture by means of coupled growth
Cocultures pairing fermentative and purple photoheterotrophic bacteria resemble natural anaerobic foodwebs where organic acid fermentation products serve as a carbon source for the phototroph (Figure 1a). Over the last 30 years, traditional growth conditions for these cocultures promoted interactions in which phototrophs relied on fermentative bacteria but provided no reciprocal benefit (Figure 1a; Odom and Wall, 1983; Fang et al., 2006; Ding et al., 2009; Sun et al., 2010). Consequently, these systems commonly suffered from instability and variable product yields (Fang et al., 2006; Ding et al., 2009; Sun et al., 2010).
We overcame this long-standing stability issue by exploiting two metabolic features of the phototrophic purple nonsulfur bacterium, R. palustris. First, R. palustris can consume fermentation products but not glucose (Larimer et al., 2004). This means that R. palustris cannot compete against E. coli for glucose in coculture but is instead reliant on excreted E. coli fermentation products. Second, R. palustris is a diazotroph, and thus can use the enzyme, nitrogenase, to convert N2 into bioavailable ammonium (NH4+) in a process known as N2 fixation. We found that a NifA* mutant exhibiting constitutive nitrogenase activity (McKinlay and Harwood, 2010) excreted measurable NH4+ when fixing N2 (Supplementary Figure 2). This R. palustris strain, hereon referred to as Nx, has a NifA* mutation to allow for NH4+ excretion and ΔhupS and Δrpa2750 mutations to prevent H2 uptake (Rey et al., 2006) and cell aggregation (Fritts et al., in preparation), respectively. In contrast to R. palustris, E. coli cannot fix N2. Thus, we reasoned that NH4+ excreted by R. palustris could cross-feed E. coli and establish an obligate mutualism (that is, syntrophy; Figure 1b). Indeed, we observed robust growth when we paired E. coli with R. palustris Nx in a defined medium (MDC) supplemented with glucose as the sole carbon source and with N2 gas as the sole nitrogen source (N2-supplied coculture). In contrast, growth was negligible when E. coli was paired with the parent R. palustris strain that does not share NH4+ (Figure 1c). Our results demonstrate that under these nutrient conditions bidirectional cross-feeding is necessary for cooperative coculture growth; R. palustris relies on E. coli for essential carbon in the form of fermentation products, whereas E. coli relies on R. palustris for essential nitrogen in the form of NH4+ (Figure 1b).
Stable coexistence was interrogated by altering starting species ratios and by serially transferring cocultures. N2-supplied cocultures converged to a common species composition (~10% E. coli) from a range of starting species ratios spanning 12 orders of magnitude (Figure 1d). Once achieved, this ratio was maintained throughout exponential growth and over multiple transfers (⩾30 generations; Supplementary Figure 4). Furthermore, cocultures were readily initiated from single colonies grown on nutrient-replete agar (Supplementary Figure 4), demonstrating that metabolic cooperation was immediate and did not require metabolic priming or coevolution. Thus, in this system metabolic interdependency drives ratio convergence and enforces stable coexistence by means of coupled growth.
Stable coexistence was accompanied by reproducible fermentation product yields over serial transfers (Supplementary Figure 4). The soluble fermentation products excreted by E. coli are ethanol, formate, lactate, acetate and succinate (Figure 2a). There was no detectable lactate, acetate, or succinate remaining in N2-supplied cocultures, suggesting that these fermentation products were consumed by R. palustris whereas formate and ethanol were not (Figure 2a). Indeed, R. palustris only consumed lactate, acetate and succinate when fed supernatants from fermentative E. coli monocultures (Supplementary Figure 5). Hereon, we will collectively refer to lactate, acetate and succinate as consumable organic acids. In addition to soluble products, E. coli also produces H2 and CO2. As noted above, R. palustris Nx is genetically incapable of H2 consumption. CO2 fixation by R. palustris is likely negligible in cocultures as Calvin cycle transcript levels are low during N2 fixation (McKinlay and Harwood, 2010; 2011). Corroborating this expectation, cocultures containing an R. palustris Nx Calvin cycle mutant, incapable of CO2 fixation, behaved comparably to cocultures containing R. palustris Nx (Supplementary Figure 6).
Growth and metabolic trends of NH4+-supplied cocultures resemble those of E. coli monocultures. Metabolic profiles (a), specific growth rates (b), final cell densities (c) and final pH (d) of NH4+-supplied or N2-supplied cocultures compared with NH4+-supplied E. coli monocultures. All cocultures contained R. palustris Nx. Data shown is combined from experiments using cultures inoculated using single colonies, subculturing, and/or 1:1 species ratios (cocultures only). Ec, E. coli; Rp, R. palustris. Error bars, s.e.m., n⩾3.
To test the importance of metabolic interdependence for coculture stability, we initiated cocultures where we externally supplied either acetate, a carbon source for R. palustris, or NH4+, a nitrogen source that both species can use (Figure 1a). When acetate was externally supplied, coculture growth rate mirrored that of cocultures supplied glucose alone, even over serial transfers (Supplementary Figure 7). In addition, in acetate-supplied cocultures E. coli grew to final cell densities comparable to those in N2-supplied cocultures with glucose alone (Supplementary Figure 7). Within our coculture system, organic acids serve as both the essential carbon source for R. palustris growth as well as the electron source for N2 fixation. Hence, these data signify that growth and NH4+ cross-feeding by R. palustris in N2-supplied cocultures are not limited by either carbon or electron availability.
In contrast to acetate, external supply of NH4+ drastically altered coculture behavior. When NH4+ was supplied, we observed rapid growth regardless of the R. palustris genotype (Figures 1c and 2b). NH4+-supplied cocultures were white, unlike N2-supplied cocultures, which were red (Supplementary Figure 8), suggesting that NH4+-supplied cocultures were dominated by E. coli rather than R. palustris. E. coli dominance was confirmed by both FISH and selective plate counts (Figures 1d and 2c). Growth, fermentation profiles, and final pH of NH4+-supplied cocultures mirrored those of E. coli monocultures (Figure 2), indicating negligible metabolic contribution by R. palustris in these cocultures. Specifically, we observed the accumulation consumable organic acids (Figure 2a). The lack of conversion of consumable organic acids into R. palustris biomass in NH4+-supplied cocultures resulted in final cell densities that were much lower than those of N2-supplied cocultures (Figure 1c). Overall, these data demonstrate that the external supply of NH4+ breaks the reciprocal dependency of this syntrophy and allows E. coli to rapidly overtake the coculture due to its lower intrinsic generation time relative to that of R. palustris (2.5 h versus 9.9 h), and ultimately prevent R. palustris growth. These results, in combination with the acetate results discussed above, also corroborate the notion that stable coexistence relies on the metabolic dependence of the faster-growing species on its slower-growing partner and not the converse (Hom and Murray, 2014).
Organic acids have both positive and negative roles within the mutualism
It initially seemed paradoxical that there was negligible R. palustris growth in NH4+-supplied cocultures despite the presence of consumable organic acids (Figure 2). However, we noticed that the final pH of NH4+-supplied cocultures was much lower than that of N2-supplied cocultures (Figure 2d). On the basis of this observation, we reasoned that organic acids could have two roles in our system: a positive role as a carbon source for R. palustris and a negative role when abundant through culture acidification. To test this hypothesis, we monitored growth and fermentation product yields in NH4+-supplied cocultures that were additionally buffered with 100 mm MOPS, pH 7. We expected that the additional buffer would keep the pH sufficiently neutral to allow R. palustris growth. In agreement with this hypothesis, NH4+-supplied cocultures were indeed able to support R. palustris growth when supplemented with MOPS (Figure 3a; Supplementary Figure 8). Under these sufficiently buffered conditions, E. coli still grew rapidly and fermentation products accumulated (Figure 3); however, slower-growing R. palustris eventually assimilated the accumulated consumable organic acids and reached cell densities comparable to those seen in N2-supplied cocultures (Figure 3c; compare with Figure 2c). In a complementary approach, we also grew R. palustris in spent E. coli monoculture supernatants that were either unmodified (−MOPS; pH 4.9) or were additionally buffered with 100 mm MOPS, pH 7 (+MOPS; pH 6.2) before inoculation with R. palustris. No growth was observed in spent supernatants without MOPS. In contrast, in MOPS-buffered E. coli supernatants R. palustris assimilated consumable organic acids and grew to a similar cell density as that observed in N2-supplied cocultures (Supplementary Figure 5). The results from these two experiments corroborated that in NH4+-supplied cocultures without MOPS, the rapid accumulation of organic acids quickly shifts the role of organic acids from being a nutrient to being harmful to R. palustris.
Rapid fermentation by E. coli in NH4+-supplied cocultures results in a growth-inhibiting pH. (a) Growth curves with corresponding final pH values of NH4+-supplied cocultures containing R. palustris Nx grown in MDC without or with additional 100 mm MOPS buffer, pH 7 (−/+ MOPS). Cocultures were inoculated by subculturing. Log transformed growth curves are in Supplementary Figure 3. (b, c) Fermentation product yields (b) and cell densities (c) at the intermediate and final time points indicated in a. Error bars, s.e.m., n=3. Some error bars are too small to visualize.
Development of an ecological model capable of predicting coculture behavior
Mathematical modeling of synthetic communities is a powerful tool for generating experimentally-testable predictions of what controls mutualism dynamics and stability (Klitgord and Segrè, 2010; Mee et al., 2014; Zomorrodi and Segrè, 2015; Zelezniak et al., 2015). We therefore developed an ecological model (Figure 4) based on previously described interactions for mutualistic systems with Monod uptake kinetics (Meyer et al., 1975; Lee et al., 1976). We modified the model to describe batch culturing and to include growth-independent fermentation by E. coli (Supplementary Table 2; Supplementary Figure 1; Wanner and Egli, 1990). Our model simulates NH4+ excretion in a growth-dependent manner because the essential carbon source for R. palustris growth (organic acids) is also the electron source for N2 fixation.
Schematic of the ecological model. Ec and Rp represent E. coli and R. palustris. Orange arrows indicate that production can be growth-independent. Red oval arrows represent inhibitory effects of organic acids; this parameter is included in model A but not in model B. Bracketed letters indicate model code designations (see ‘Materials and methods’ section, Supplementary Table 2). EtOH, ethanol; For, formate; Glc, glucose; OAcs, consumable organic acids (lactate, acetate and succinate).
We also modified our model to describe acid inhibition of both R. palustris and E. coli growth by organic acids (model A). This modification was necessary to correctly predict resource and population dynamics in NH4+-supplied cocultures, specifically domination of the coculture by E. coli, organic acid accumulation and inhibition of R. palustris growth (Figure 5b, left panel; compare with Figure 5a left panel and Figure 2). When acid inhibition was omitted from the model (model B), the model predicted growth and metabolic trends in NH4+-supplied cocultures that were inconsistent with our experimental observations. Specifically, model B incorrectly predicted that rapid E. coli growth would be accompanied by slower R. palustris Nx growth in NH4+-supplied cocultures and that final metabolic profiles and species densities would be comparable to N2-supplied cocultures (Figure 5c, left panel; compare with Figure 5c, right panel). These predictions were, however, consistent with the trends observed in MOPS-buffered NH4+-supplied cocultures (compare Figure 5c left panel with Figure 3), further substantiating both the impact of acid inhibition in NH4+-supplied cocultures and the utility of the ecological model in predicting how specific factors influence coculture behavior.
Ecological modeling accurately predicts coculture behavior when growth-inhibiting effects of organic acids are included. Experimental (a) and simulated (b, c) growth and metabolic profiles of E. coli+R. palustris Nx NH4+-supplied (left) or N2-supplied (right) cocultures. (a) All cocultures were inoculated by subculturing. Most error bars (s.e.m., n=3) are too small to visualize. (b, c) Simulations were run using model A (b) or model B (c) with default parameters (Supplementary Table 2) except that for simulations with NH4+ (left panels) the initial NH4+ concentration was set to 15 mm. Ec, E. coli; Rp, R. palustris; OAcs, consumable organic acids.
Predictions for N2-supplied cocultures were near identical using the two models (Figures 5b and c, right panels). Both models predicted that consumable organic acids would only briefly accumulate, and that the coculture would be primarily composed of R. palustris Nx (14% E. coli simulated versus ~10% experimental; Figure 5, right panels). These simulations were corroborated by our empirical data showing that additional buffer had no observable effect on growth or metabolic trends in N2-supplied cocultures (Supplementary Figure 9). Thus, the inhibitory effects of organic acids are not observed in N2-supplied cocultures as consumption by R. palustris keeps pace with production. In other words, balanced metabolic interactions in N2-supplied cocultures self-buffered the system.
High cross-feeding levels can be detrimental to a mutualism
Cross-feeding levels are inherently difficult to measure and yet are hypothesized to be a major determinant of mutualism dynamics and stability (Shou et al., 2007; Kim et al., 2008; Estrela et al., 2012; Hom and Murray, 2014). We therefore used our model to address the effect of NH4+-cross-feeding levels on mutualism dynamics. By varying the NH4+ excretion parameter we estimated the level of NH4+ excretion by R. palustris Nx that would result in observed coculture trends (Figures 6a, 1 ×). A large range of NH4+ excretion, both higher and lower than that of R. palustris Nx and spanning over 3 orders of magnitude, was predicted to support cooperative growth (Figure 6a). Surprisingly, at levels above that excreted by R. palustris Nx, the model predicted that increasing the amount of NH4+ excreted per R. palustris cell would decrease the R. palustris carrying capacity (the maximum population size allowed by the culture conditions; Figure 6a). More NH4+ excretion would result in faster E. coli growth and organic acid excretion, outpacing consumption by R. palustris. Consequently, organic acids would accumulate, acidify the medium and decrease the R. palustris carrying capacity (Figure 6a). The acid inhibition of R. palustris growth predicted to result from higher NH4+ excretion is analogous to trends observed when cocultures were supplemented with NH4+, though the effects from the NH4+ supplement are predicted to be more severe (compare Figure 6a with Figure 5b).
Higher NH4+ excretion levels lead to cocultures with less R. palustris and less carbon assimilation. (a) Simulated effect of the R. palustris NH4+ excretion level on growth and organic acid accumulation in N2-supplied cocultures. Concentration of 1 × is the default NH4+ excretion level (0.25 fmol NH4+/cell) and is thought to represent that excreted by R. palustris Nx, based on model approximation of experimental trends. Concentration of 3 × (0.75 fmol NH4+/cell) indicates an NH4+ excretion level thought to represent that by R. palustris NxΔAmtB (CGA4021), based on monoculture NH4+ levels compared with those of R. palustris Nx (Supplementary Figure 2). OAcs, consumable organic acids; For, formate; Eci and Rpi, initial E. coli and R. palustris cell densities (dashed lines); Ecf and Rpf, final E. coli and R. palustris cell densities (solid lines). (b–d) Experimental data from R. palustris Nx-based and R. palustris NxΔAmtB-based cocultures, including growth curves and final pH values (b), final cell densities (c) and final E. coli percentages over serial transfers (d). Log transformed growth curves are in Supplementary Figure 3. All cocultures were inoculated at a 1:1 species ratio. Error bars, s.e.m., n=3.
These predictions of altered community composition highlighted a crucial aspect of this mutualism: the inhibitory activity of organic acids is directly proportional to their net excretion level. Consequently, the level of cross-feeding determines the equilibrium ratio and carrying capacity for each species. Moreover, contradictory as it may seem, enhanced cross-feeding is predicted to actually be detrimental to this mutualism (lower R. palustris carrying capacity and less carbon assimilated by the community).
To experimentally test these predictions, we engineered R. palustris to excrete more NH4+ than the Nx strain. In several diazotroph species, disruption of the NH4+ transporter, AmtB, results in extracellular NH4+ accumulation during N2 fixation (Yakunin and Hallenbeck, 2002; Zhang et al., 2012; Barney et al., 2015). We deleted the genes for both AmtB homologs in R. palustris Nx, resulting in R. palustris NxΔAmtB, and found that NH4+ accumulated to nearly three times to that of R. palustris Nx in monocultures (Supplementary Figure 2). The increased NH4+ accumulation was due to the amtB2 deletion; the amtB1 deletion had no effect (Supplementary Figure 2). When we paired R. palustris NxΔAmtB with E. coli, coculture behavior matched that predicted by the modeled 3 × NH4+ excretion level (Figure 6a). The combined carrying capacity was lower than that of cocultures with R. palustris Nx (Figure 6b), R. palustris cell densities declined (Figure 6c), the final species ratio changes to approximately 1:1 (Figure 6c), and consumable organic acids accumulated and acidified the medium (Figure 6b; Supplementary Figure 10). Growth yield and organic acid assimilation levels in cocultures with R. palustris NxΔAmtB were restored to levels seen in cocultures with R. palustris Nx by adding MOPS buffer, confirming that acidification was prematurely inhibiting R. palustris NxΔAmtB growth (Supplementary Figure 10).
Despite the detrimental effect of increased NH4+ excretion on the R. palustris carrying capacity, metabolic and growth trends were reproducible and stable coexistence was maintained over multiple transfers (Figure 6c; Supplementary Figure 10). Thus, the obligate nature of cross-feeding within this mutualism ensured stable coexistence even at a new equilibrium state. The model predicted that different NH4+ excretion levels would establish a range of stable equilibria (Figure 6a). These distinct equilibria are likely all stabilized by negative frequency-dependent selection on E. coli (Morris, 2015), wherein the relative amount of NH4+ available per E. coli cell, which determines E. coli fitness, decreases as the E. coli frequency increases. The level of NH4+ exchange also influenced organic acid transformation by R. palustris into biomass and CO2 (Figure 6a; Supplementary Figure 10) and thus impacted the nature and efficiency with which this synthetic community transformed carbon.
Discussion
Organic acids are important drivers of anaerobic food web ecology, acting as a resource for some microbes while inhibiting others (Russell and Diez-Gonzalez, 1997; Schink, 2002; McInerney et al., 2009). In our synthetic community, the organic acids cross-fed by E. coli function as both a nutrient and an inhibitor to R. palustris. This attribute crucially impacts how cross-feeding levels govern mutualism dynamics. Specifically, excessive cooperation by R. palustris (that is, increased NH4+ cross-feeding) stimulates reciprocation by E. coli, resulting in organic acid production that outpaces consumption by R. palustris. Consequently, increased cooperation moves the nature of organic acids along a continuum from beneficial to detrimental, limiting total coculture growth and carbon transformation. Presumably, the influence of dose-dependent metabolite toxicity on a mutualism depends on at least three factors: (i) the rate of excretion, which depends on the producer population size; (ii) the rate of consumption or detoxification by the partner, which depends on the consumer population size; and (iii) the metabolite’s inhibitory activity. For example, a high-metabolite excretion level and/or a high-metabolite toxicity would more readily impair cooperation. In contrast, enhanced consumption by the partner would facilitate cooperation at higher exchange levels (Supplementary Figure 11). Thus, the detrimental influence exerted by a metabolite on the system will depend on the activity of both partners.
The closed nature of our experimental system circumvents two other factors that could also influence cross-feeding and dose-dependent nutrient toxicity in natural systems: diffusion of metabolites in an open system and metabolic contributions by additional species. Both of these factors would influence the accumulation of a metabolite and therefore determine its position on the continuum between beneficial nutrient and detrimental toxin. Synthetic ecosystems such as the one described here could potentially be modified to understand how such additional factors influence dose-dependent nutrient toxicity and thereby mutualism behavior.
Separately, we have also shown herein that NH4+ cross-feeding by R. palustris can result in an immediate and stable syntrophic relationship with fermentative E. coli under conditions requiring N2 fixation. Metabolite excretion is a mechanism by which cooperative interactions can be initiated and sustained (Sachs et al., 2004). There is accumulating evidence that some diazotrophs excrete NH4+ during N2 fixation (Adam et al., 2016) and that AmtB functions to reacquire NH4+ and limit its availability to nearby organisms (Supplementary Figure 2) (Yakunin and Hallenbeck, 2002; Zhang et al., 2012; Barney et al., 2015). As proposed in the Black Queen hypothesis (Morris et al., 2012), this NH4+ leakage makes diazotrophs well-suited to establish nascent mutualisms, both in natural and synthetic communities. Our results support this notion. Furthermore, the specific ability of enhanced NH4+ cross-feeding to elevate reciprocal exchange of potentially toxic compounds could have implications for the deployment of engineered NH4+-excreting diazotrophs as biofertilizers for agricultural and industrial applications (Ortiz-Marquez et al., 2012; Barney et al., 2015; Geddes et al., 2015): Increasing NH4+ excretion might not equate to optimizing NH4+ excretion. Overall, our results indicate that variations in cross-feeding levels have the potential to alter the nature of interspecies relationships and potentially ecosystem function.
References
Abbott BJ . (1973). Ethanol inhibition of a bacterium (Acinetobacter calcoaceticus in chemostat culture. J Gen Microbiol 75: 383–389.
Adam B, Klawonn I, Svedén JB, Bergkvist J, Nahar N, Walve J et al. (2016). N2-fixation, ammonium release and N-transfer to the microbial and classical food web within a plankton community. ISME J 10: 450–459.
Alvarez LA, Exton DA, Timmis KN, Suggett DJ, McGenity TJ . (2009). Characterization of marine isoprene-degrading communities. Environ Microbiol 11: 3280–3291.
Barney BM, Eberhart LJ, Ohlert JM, Knutson CM, Plunkett MH . (2015). Gene deletions resulting in increased nitrogen release by Azotobacter vinelandii: Application of a novel nitrogen biosensor. Appl Environ Microbiol 81: 4316–4328.
Barnhill AE, Weeks KE, Xiong N, Day TA, Carlson SA . (2010). Identification of multiresistant Salmonella isolates capable of subsisting on antibiotics. Appl Environ Microbiol 76: 2678–2680.
Ding J, Liu BF, Ren NQ, Xing DF, Guo WQ, Xu JF et al. (2009). Hydrogen production from glucose by co-culture of Clostridium butyricum and immobilized Rhodopseudomonas faecalis RLD-53. Int J Hydrogen Energy 34: 3647–3652.
Ducret A, Quardokus E, Brun Y . (2016). MicrobeJ, a high throughput tool for quantitative bacterial cell detection and analysis. Nat Microbiol 1: 16077.
Estrela S, Trisos CH, Brown SP . (2012). From metabolism to ecology: cross-feeding interactions shape the balance between polymicrobial conflict and mutualism. Am Nat 180: 566–576.
Fang HHP, Zhu H, Zhang T . (2006). Phototrophic hydrogen production from glucose by pure and co-cultures of Clostridium butyricum and Rhodobacter sphaeroides. Int J Hydrogen Energy 31: 2223–2230.
Fuhrman JA . (2009). Microbial community structure and its functional implications. Nature 459: 193–199.
Geddes BA, Ryu M-H, Mus F, Garcia Costas A, Peters JW, Voigt CA et al. (2015). Use of plant colonizing bacteria as chassis for transfer of N2-fixation to cereals. Curr Opin Biotechnol 32: 216–222.
Harcombe W . (2010). Novel cooperation experimentally evolved between species. Evolution 64: 2166–2172.
Hillesland KL, Stahl DA . (2010). Rapid evolution of stability and productivity at the origin of a microbial mutualism. Proc Natl Acad Sci USA 107: 2124–2129.
Hom EFY, Murray AW . (2014). Niche engineering demonstrates a latent capacity for fungal-algal mutualism. Science 345: 94–88.
Huang CB, Alimova Y, Myers TM, Ebersole JL . (2011). Short- and medium-chain fatty acids exhibit antimicrobial activity for oral microorganisms. Arch Oral Biol 56: 650–654.
Huang JJ, Heiniger EK, McKinlay JB, Harwood CS . (2010). Production of hydrogen gas from light and the inorganic electron donor thiosulfate by Rhodopseudomonas palustris. Appl Environ Microbiol 76: 7717–7722.
Jiao Y, Navid A, Stewart BJ, McKinlay JB, Thelen MP, Pett-Ridge J . (2012). Syntrophic metabolism of a co-culture containing Clostridium cellulolyticum and Rhodopseudomonas palustris for hydrogen production. Int J Hydrogen Energy 37: 11719–11726.
Johns NI, Blazejewski T, Gomes AL, Wang HH . (2016). Principles for designing synthetic microbial communities. Curr Opin Microbiol 31: 146–153.
Kim HJ, Boedicker JQ, Choi JW, Ismagilov RF . (2008). Defined spatial structure stabilizes a synthetic multispecies bacterial community. Proc Natl Acad Sci USA 105: 18188–18193.
Kim M, Harwood CS . (1991). Regulation of benzoate-CoA ligase in Rhodopseudomonas palustris. FEMS Micrbiol Lett 83: 199–203.
Klitgord N, Segrè D . (2010). Environments that induce synthetic microbial ecosystems. PLoS Comput Biol 6: e1001002.
Kremer TA, LaSarre B, Posto AL, McKinlay JB . (2015). N2 gas is an effective fertilizer for bioethanol production by Zymomonas mobilis. Proc Natl Acad Sci USA 112: 2222–2226.
Kunz DA, Nagappan O, Silva-Avalos J, Delong GT . (1992). Utilization of cyanide as nitrogenous substrate by Pseudomonas fluorescens NCIMB 11764: evidence for multiple pathways of metabolic conversion. Appl Environ Microbiol 58: 2022–2029.
Larimer FW, Chain P, Hauser L, Lamerdin J, Malfatti S, Do L et al. (2004). Complete genome sequence of the metabolically versatile photosynthetic bacterium Rhodopseudomonas palustris. Nat Biotechnol 22: 55–61.
Lee IH, Fredrickson AG, Tsuchiya HM . (1976). Dynamics of mixed cultures of Lactobacillus plantarum and Propionibacterium shermanii. Biotechnol Bioeng 18: 513–526.
Lindemann SR, Bernstein HC, Song H-S, Fredrickson JK, Fields MW, Shou W et al. (2016). Engineering microbial consortia for controllable outputs. ISME J 10: 2077–2084.
Louis P, Scott KP, Duncan SH, Flint HJ . (2007). Understanding the effects of diet on bacterial metabolism in the large intestine. J Appl Microbiol 102: 1197–1208.
McInerney MJ, Sieber JR, Gunsalus RP . (2009). Syntrophy in anaerobic global carbon cycles. Curr Opin Biotechnol 20: 623–632.
McKinlay JB, Harwood CS . (2011). Calvin cycle flux, pathway constraints, and substrate oxidation state together determine the H2 biofuel yield in photoheterotrophic bacteria. mBio 2: e00323–10.
McKinlay JB, Harwood CS . (2010). Carbon dioxide fixation as a central redox cofactor recycling mechanism in bacteria. Proc Natl Acad Sci USA 107: 11669–11675.
McKinlay JB, Zeikus JG, Vieille C . (2005). Insights into Actinobacillus succinogenes fermentative metabolism in a chemically defined growth medium. Appl Environ Microbiol 71: 6651–6656.
Mee MT, Collins JJ, Church GM, Wang HH . (2014). Syntrophic exchange in synthetic microbial communities. Proc Natl Acad Sci USA 111: E2149–E2156.
Meyer JS, Tsuchiya HM, Fredrickson AG . (1975). Dynamics of mixed populations having complementary metabolism. Biotechnol Bioeng 17: 1065–1081.
Momeni B, Chen C-C, Hillesland KL, Waite A, Shou W . (2011). Using artificial systems to explore the ecology and evolution of symbioses. Cell Mol Life Sci 68: 1353–1368.
Morris BEL, Henneberger R, Huber H, Moissl-Eichinger C . (2013). Microbial syntrophy: interaction for the common good. FEMS Microbiol Rev 37: 384–406.
Morris JJ . (2015). Black Queen evolution: the role of leakiness in structuring microbial communities. Trends Genet 31: 475–482.
Morris JJ, Lenski RE, Zinser ER . (2012). The Black Queen Hypothesis: evolution of dependencies through adaptive gene loss. mBio 3: e00036–12.
Oda Y, Slagman S-J, Meijer WG, Forney LJ, Gottschal JC . (2000). Influence of growth rate and starvation on fluorescent in situ hybridization of Rhodopseudomonas palustris. FEMS Microbiol Ecol 32: 205–213.
Odom JM, Wall JD . (1983). Photoproduction of H2 from cellulose by an anaerobic bacterial coculture. Appl Environ Microbiol 45: 1300–1305.
Ortiz-Marquez JCF, Do Nascimento M, Dublan M de LA, Curatti L . (2012). Association with an ammonium-excreting bacterium allows diazotrophic culture of oil-rich eukaryotic microalgae. Appl Environ Microbiol 78: 2345–2352.
Pelletier DA, Hurst GB, Foote LJ, Lankford PK, McKeown CK, Lu TY et al. (2008). A general system for studying protein-protein interactions in Gram-negative bacteria. J Proteome Res 7: 3319–3328.
Rey FE, Oda Y, Harwood CS . (2006). Regulation of uptake hydrogenase and effects of hydrogen utilization on gene expression in Rhodopseudomonas palustris. J Bacteriol 188: 6143–6152.
Russell JB, Diez-Gonzalez F . (1997). The Effects of fermentation acids on bacterial growth. Adv Microb Physiol 39: 205–234.
Sachs JL, Mueller UG, Wilcox TP, Bull JJ . (2004). The evolution of cooperation. Q Rev Biol 79: 135–160.
Schink B . (2002). Synergistic interactions in the microbial world. Antonie Van Leeuwenhoek 81: 257–261.
Shou W, Ram S, Vilar JMG . (2007). Synthetic cooperation in engineered yeast populations. Proc Natl Acad Sci USA 104: 1877–1882.
Sousa DZ, Smidt H, Alves MM, Stams AJM . (2009). Ecophysiology of syntrophic communities that degrade saturated and unsaturated long-chain fatty acids. FEMS Microbiol Ecol 68: 257–272.
Summers ZM, Fogarty HE, Leang C, Franks AE, Malvankar NS, Lovley DR . (2010). Direct exchange of electrons within aggregates of an evolved syntrophic coculture of anaerobic bacteria. Science 330: 1413–1415.
Sun Q, Xiao W, Xi D, Shi JP, Yan X, Zhou ZH . (2010). Statistical optimization of biohydrogen production from sucrose by a co-culture of Clostridium acidisoli and Rhodobacter sphaeroides. Int J Hydrogen Energy 35: 4076–4084.
Wanner U, Egli T . (1990). Dynamics of microbial growth and cell composition in batch culture. FEMS Microbiol Lett 75: 19–43.
Widder S, Allen RJ, Pfeiffer T, Curtis TP, Wiuf C, Sloan WT et al. (2016). Challenges in microbial ecology: building predictive understanding of community function and dynamics. ISME J doi:10.1038/ismej.2016.45.
Yakunin AF, Hallenbeck PC . (2002). AmtB is necessary for NH4+-induced nitrogenase switch-off and ADP-ribosylation in Rhodobacter capsulatus. J Bacteriol 184: 4081–4088.
Zelezniak A, Andrejev S, Ponomarova O, Mende DR, Bork P, Patil KR . (2015). Metabolic dependencies drive species co-occurrence in diverse microbial communities. Proc Natl Acad Sci USA 112: 6449–6454.
Zhang T, Yan Y, He S, Ping S, Alam KM, Han Y et al. (2012). Involvement of the ammonium transporter AmtB in nitrogenase regulation and ammonium excretion in Pseudomonas stutzeri A1501. Res Microbiol 163: 332–339.
Zomorrodi AR, Segrè D . (2015). Synthetic ecology of microbes: mathematical models and applications. J Mol Biol 428: 837–861.
Acknowledgements
We thank A Posto for early contributions; Prof SR Hall for modeling discussions; Prof Y Brun for critical reading; and RK Fritts and the McKinlay lab for discussions. This work was supported by the US Department of Energy, Office of Science, Office of Biological and Environmental Research, under Award Number DE-SC0008131. Model development was supported in part by the US Army Research Office, Grant W911NF-14-1-0411.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies this paper on The ISME Journal website
Supplementary information
Rights and permissions
About this article
Cite this article
LaSarre, B., McCully, A., Lennon, J. et al. Microbial mutualism dynamics governed by dose-dependent toxicity of cross-fed nutrients. ISME J 11, 337–348 (2017). https://doi.org/10.1038/ismej.2016.141
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ismej.2016.141
This article is cited by
-
Two-tiered mutualism improves survival and competitiveness of cross-feeding soil bacteria
The ISME Journal (2023)
-
Microbial active functional modules derived from network analysis and metabolic interactions decipher the complex microbiome assembly in mangrove sediments
Microbiome (2022)
-
Priority effects in microbiome assembly
Nature Reviews Microbiology (2022)
-
A light tunable differentiation system for the creation and control of consortia in yeast
Nature Communications (2021)
-
The ecology of wine fermentation: a model for the study of complex microbial ecosystems
Applied Microbiology and Biotechnology (2021)