Abstract
In coastal waters worldwide, an increase in frequency and intensity of algal blooms has been attributed to eutrophication, with further increases predicted because of climate change. Yet, the cellular-level changes that occur in blooming algae remain largely unknown. Comparative metatranscriptomics was used to investigate the underlying molecular mechanisms associated with a dinoflagellate bloom in a eutrophied estuary. Here we show that under bloom conditions, there is increased expression of metabolic pathways indicative of rapidly growing cells, including energy production, carbon metabolism, transporters and synthesis of cellular membrane components. In addition, there is a prominence of highly expressed genes involved in the synthesis of membrane-associated molecules, including those for the production of glycosaminoglycans (GAGs), which may serve roles in nutrient acquisition and/or cell surface adhesion. Biotin and thiamine synthesis genes also increased expression along with several cobalamin biosynthesis-associated genes, suggesting processing of B12 intermediates by dinoflagellates. The patterns in gene expression observed are consistent with bloom-forming dinoflagellates eliciting a cellular response to elevated nutrient demands and to promote interactions with their surrounding bacterial consortia, possibly in an effort to cultivate for enhancement of vitamin and nutrient exchanges and/or direct consumption. Our findings provide potential molecular targets for bloom characterization and management efforts.
Similar content being viewed by others
Introduction
Algal blooms have become a major nuisance in many estuarine and coastal marine systems worldwide (Hallegraeff, 1992; Anderson et al., 2008). Toxins produced by some harmful algal bloom species may be detrimental to fish, birds, marine mammals and even humans (Anderson et al., 2008). Just as important, blooms of nontoxic microalgae that result in bottom-water hypoxia are detrimental and potentially fatal to sessile macrofauna. Efforts made to manage nutrient inputs to estuaries and coastal waterways have achieved a limited degree of success in reducing bloom outbreaks. Most estuaries are under the influence of both nutrient-rich river runoff and climatic perturbations where sources of pollutants continue to steadily increase with growth in agriculture, industry and urbanization (Paerl, 2006). In addition to these sources, increased frequency and intensity of storms as a result of climate change have reduced the efficacy of both non-point and point-source nutrient reduction programs (Paerl, 2006).
The abundance and distribution of phytoplankton in shallow estuarine systems are greatly affected by physical, chemical and biological factors (Sellner et al., 2003). Optimal temperature, salinity and light conditions may vary among bloom-forming algal species (Smayda, 1997; Litaker et al., 2002). Similarly, nutrient supply rates, concentrations and ratios can play significant roles in algal bloom dynamics in coastal waters (Piehler et al., 2004; Paerl, 2006). Heterotrophic bacteria may also facilitate the growth of phytoplankton through remineralization of nutrients and biosynthesis of essential growth factors such as vitamins (Cole, 1982). Some phytoplankton produce polysaccharides that render them ‘sticky’, especially under nutrient-limited conditions. This promotes bacterial–phytoplankton interactions by trapping bacteria close to their cell surface (Kiørboe et al., 1990; Myklestad, 1999). Many phytoplankton also have the ability to switch trophic modes of energy acquisition by transitioning from autotrophy to heterotrophy (Smayda, 1997). This phenomenon, known as mixotrophy, is common among certain groups of phytoplankton, especially the dinoflagellates.
A complicating factor to deciphering the triggers of algal blooms is our rather limited capacity to predict when and where they will occur, and a lack of knowledge of the underlying mechanisms involved in the promotion and propagation of algal blooms impedes our ability to formulate preventative measures. Also, recent realization of there being a high degree of inter- and intraspecific species interactions during rapidly changing bloom microenvironments contributes to the difficulty in identifying the physiological processes involved in bloom formation and longevity (Cole, 1982; Amin et al., 2015; Lima-mendez et al., 2015; Zhuang et al., 2015). Primary methods for examining phytoplankton dynamics within water quality monitoring programs typically have used bulk plankton community analyses (for example, chlorophyll a, primary productivity, nutrient uptake rates, etc.). Sequencing of mRNA transcripts from the environment and subsequent analysis of gene expression patterns (commonly referred to as metatranscriptomics) has been shown to be a powerful tool for identifying the distinct cellular-level processes conducted by bloom-forming taxa within the background of all metabolic activities performed within a mixed plankton assemblage (Cooper et al., 2014). Recent applications of metatranscriptomic approaches have provided new insights into microalgal processes in natural marine environments and include characterization of the phytoplankton response to iron enrichment (Marchetti et al., 2012) and resource partitioning among distinct phytoplankton species in nutrient-rich coastal environments (Alexander et al., 2015).
Here we present a comparative metatranscriptomic analysis of a dinoflagellate bloom that was opportunistically sampled and characterized in the Neuse River Estuary (NRE), North Carolina in the early fall of 2012. Sequences in combination with environmental data collected before, during and following the bloom event are used to examine the conditions that may have triggered the bloom and the organismal responses at the molecular level. Transcripts obtained from eukaryotic plankton within the bloom were compared with those from surrounding non-bloom environments. Normalization techniques provided us the ability to distinguish between potential shifts in taxonomic cell abundance and gene expression patterns. We focus on the changes in genes grouped in metabolic modules that were assigned to dinoflagellates as a means to elucidate the cellular-level shifts occurring during the bloom.
Materials and methods
Study area and sampling collection
Sampling for metatranscriptomic sequencing took place in 2012 in conjunction with the NRE Modeling and Monitoring (ModMon) Program (http://www.unc.edu/ims/neuse/modmon); we sampled on five occasions throughout the year at four Modmon stations: 20, 70, 120 and 180 (Figure 1a), resulting in 20 routine metatranscriptomic samples. On 24 September 2012, the ModMon biweekly sampling survey reported a significant algal bloom near station 30, associated with bottom-water hypoxia and fish kills (Supplementary Figure S1). On 26 September 2012, metatranscriptomic sampling of a dinoflagellate bloom located near station 50 along with two non-bloom surrounding stations (stations 20 and 70) was conducted between 0930 and noon. In conjunction with metatranscriptomic sampling, physical and chemical parameters obtained from vertical YSI (Yellow Springs Instrument) profiles and Lugol’s preserved samples from the surface waters were also collected at each station (see Supplementary File 1). In this study, our primary analysis is focused on samples collected during this bloom event.
Map of the neuse river estuary and water quality parameters measured in 2012. (a) Location of ModMon stations (20–180) in the NRE. The bloom station is indicated with a red circle. (b) Salinity, phosphate (PO4) and nitrate (NO3) concentrations, (c) chlorophyll a (Chl a) concentrations and primary productivity (PP) and (d) primary photopigment concentrations of dinoflagellates (peridinin) and diatoms (fucoxanthin). Plotted are the annual averages and associated standard deviations of surface samples. Also provided are measurements obtained during the dinoflagellate bloom event in September of 2012 where the bloom station (red symbols) and the non-bloom stations (white and black symbols) are indicated.
Metatranscriptomic sample preparation and sequencing
RNA extraction and polyadenosine tail (poly(A)+) selection were conducted as described in Supplementary File 1. Briefly, mRNA samples from multiple filters collected at each station were extracted and then pooled to achieve a minimum mRNA concentration of 80 ng and provided to the UNC-CH High-throughput Sequencing Facility for Illumina sequence library synthesis using the TruSeq mRNA Library Preparation Kit (San Diego, CA, USA). mRNA from each of the stations collected during the bloom period was bar-coded and pooled. The combined sample was then sequenced on one lane using the Illumina HiSeq2000 platform (San Diego, CA, USA), generating between 56 million and 91 million pairs 100 bp paired-end reads per sample.
Differential expression analysis
Reads from each sample were assembled and functionally and taxonomically annotated. Details on assembly and annotation procedures are provided in Supplementary File 1. Differential expression of dinoflagellate-associated genes from the bloom versus the two non-bloom samples was performed on all genes that were annotated as belonging to dinoflagellates based on sequence similarity. Taxonomic grouping of genes was performed to the class level for expression analysis because of the scarcity of genomic or transcriptomic sequences for the blooming genus and incomplete transcriptomes for other closely related dinoflagellate genera (for example, Gyrodinium and Gymnodinium). Genes were examined at the KEGG Orthology (KO) level and reads for each KO were normalized for differences in dinoflagellate-specific transcript abundance among sequence libraries using the ‘TMM’ normalization procedure in the edgeR Bioconductor package, which normalize reads by computing a scaling factor after excluding genes that have high average counts and/or have large expression differences between samples with the assumption that most of the genes are not differentially expressed (Robinson and Oshlack, 2010). edgeR was then used to detect differentially expressed KOs between bloom and non-bloom samples as described in Marchetti et al. (2012), and significance of differentially expressed KOs were assigned with the ‘exactTest’ program in edgeR. Normalized reads for each KO were then grouped into modules using their KO-associated ‘Module ID’. To measure whether each module is significantly enriched with over- or underrepresented KOs between the bloom sample and each of the non-bloom samples, a Mann–Whitney U-test was conducted for each module (Nielsen et al., 2005). Fold change, counts per million (c.p.m.) and P-values for each module are provided in Data set 2.
Results and discussion
Bloom environmental conditions
The NRE is a tributary of North Carolina’s Albemarle-Pamlico Estuarine System, the USA’s largest lagoonal estuarine system (Figure 1a). The NRE is a partially mixed estuary, with riverine runoff and wind as the primary drivers of circulation. Strong vertical stratification is especially common in summer when the mild, cross-channel, southwest winds prevail (Luettich et al., 2000). Over the past several decades, increasing agricultural and industrial activities along with coastal urbanization have accelerated nutrient inputs to the NRE (Paerl and Whitall, 1999; Whitall and Paerl, 2001). Combined with a long residence time and strong seasonal stratification, the NRE suffers from eutrophication (Paerl, 2006). Under the influence of both anthropogenic and climatic activities, algal blooms occur frequently (Paerl et al., 1998, 2004).
During 2012, mean annual salinity along the estuary increased gradually from close to 0 at the head of the NRE (station 20) to about 18 at the mouth (station 180; Figure 1b). Phosphate concentrations were relatively consistent throughout the estuary, whereas nitrate concentrations were more variable, but, in general, were high at the head of the estuary and decreased down-estuary from an annual mean of 34 μmol l−1 at station 20 to commonly being below the detection limit by station 120. High variability in nitrate along the transect, particularly at the mid-estuary stations, is primarily due to fluctuations in river flow (Paerl et al., 2010). Chlorophyll a concentrations were consistently lower at station 20 and peaked at mid-estuary stations with an annual average of 25 μg l−1 before steadily declining down-estuary (Figure 1c). The low biomass at the head of the NRE likely results from short residence times in combination with the ‘geochemical filter’, where flocculation and aggregation of clay minerals and metal ions increase the turbidity within the water column (Sharp et al., 1984; Peierls et al., 2012). Thus, in combination with rapid flushing rates, light as well as nutrient supplies are likely limiting factors for phytoplankton growth in this section of the NRE (Peierls et al., 2012). Down-estuary of station 30, the NRE widens, resulting in reduced flushing rates, a decrease in turbidity and subsequent increase in primary production and phytoplankton biomass, which results in a rapid depletion of nutrients from the upper water column (Pinckney et al., 1997). At station 70 onward, low N supplies in surface waters tend to limit primary productivity, resulting in reduced phytoplankton biomass (Twomey et al., 2005).
During the bloom event, there were significant increases in chlorophyll a (Student’s t-test, P-value <0.001) at the bloom station relative to surrounding stations and the annual mean values (Figure 1c). Primary productivity was also slightly higher at the bloom station compared with the annual average, although not statistically significant (Student’s t-test, P-value=0.12). Concentrations of accessory pigments were used to distinguish the presence of different phytoplankton groups (Pinckney et al., 2001; Peierls et al., 2003). Among the accessory pigments measured, peridinin and fucoxanthin were used as proxies of dinoflagellate and diatom relative abundances, respectively. The peridinin concentration was significantly higher at the bloom station compared with other accessory pigments (Student’s t-test, P-value <0.001), indicating the blooming algae were primarily that of a single or multiple dinoflagellate species (Figure 1d). Consistent with the pigment data, cell densities of dinoflagellates at the bloom station were significantly higher compared with the surrounding non-bloom stations (one-way analysis of variance; Figure 2a). Blooming dinoflagellates were morphologically identified to be predominantly Levanderina fissa (formerly known as Gyrodinium instriatum; Moestrup et al., 2014).
Comparison of dinoflagellate abundances between the bloom and non-bloom samples. (a) Dinoflagellate mean cell counts in the bloom sample and stations 20 and 70. Error bars represent standard deviations (n=3). (b) Taxonomic proportions of read counts in the bloom sample and the average of 22 other samples obtained in 2012. (c) Community-wide TMM normalized transcript counts (counts per million reads, c.p.m.) of major taxonomic groups in the bloom sample and stations 20 and 70 (Supplementary File 1).
Transcriptome sequencing of the dinoflagellate bloom
One hundred and twelve million paired-end raw reads from the bloom station along with 182 million and 122 million paired-end raw reads from the two non-bloom samples located at stations 20 and 70, respectively, were sequenced (Supplementary Table S1). Assembly of reads yielded 1–1.6 million assembled contigs from each sample. For the bloom sample, 53% of contigs had sequence similarity to the reference sequences contained in the taxonomic sequence database, MarineRefII, and 39% of contigs had sequence similarity to genes in the functional KEGG (Kyoto Encyclopedia of Genes and Genomes) database. Of these KEGG matches, 25% had KEGG module annotations and were used for differential expression analysis. From stations 20 and 70, 58% and 59% of contigs were assigned a taxonomic identification, with 44% and 46% of these being assigned a functional annotation, in which 23% and 26% had KEGG module annotations, respectively (Supplementary Table S1).
In the bloom sample, ~42% of the taxonomically annotated sequence reads were from dinoflagellates (Figure 2b), whereas from stations 20 and 70, only 22% and 17%, respectively, of these reads were from dinoflagellates. In all samples collected for sequencing in 2012, the average proportion of sequences assigned to dinoflagellates was 20% (Figure 2b). There was an approximate fivefold higher abundance of normalized transcripts assigned to dinoflagellates in the bloom sample relative to the non-bloom samples (Figure 2c), further confirming pigment and microscopic determinations of elevated dinoflagellate biomass and cell density at the bloom station. According to the taxonomic assignment of dinoflagellate sequences from the bloom sample, greatest sequence similarity was to transcriptomes of isolates from the genus Karenia (Supplementary Figure S2), a genus formerly part of Gymnodinium and closely related to Levanderina based on 18 S rDNA phylogeny (Daugbjerg et al., 2000; Heimann, 2012).
Of the total dinoflagellate reads, 20%, 22% and 15% from the bloom, station 20 and 70, respectively, obtained a KEGG module annotation. Sequences in the bloom sample were assigned to six major functional gene categories. Generally, about three-quarters of annotated reads were assigned to metabolic pathways, including synthesis of carbohydrates, fatty acids, amino acids, proteins, cofactors and vitamins, nucleotides and secondary metabolites, carbon fixation and energy-related processes such as ATP synthesis and photosynthesis; about one-fifth were related to genetic information processing, including processes involved in RNA degradation and other genetic information processing; and <1% reflected intracellular responses to the environment, including regulatory systems and transportation of macro/micronutrients, carbohydrates and amino acids.
Dinoflagellate-associated genes were grouped into a total of 399 KEGG modules, of which 352, 378 and 314 modules were expressed in the bloom sample and stations 20 and 70, respectively. Differential expression analysis was conducted at the module level to investigate the regulation of functional units within different biochemical processes. These modules represent a broad range of processes, including nutrient metabolism and transportation, carbohydrate production and transportation, and vitamin metabolism and transportation.
Differential expression analysis was performed on dinoflagellate-associated modules between the bloom sample versus station 20 and 70 to compare metabolic profiles under bloom conditions to those under non-bloom conditions. Modules were detected as over- or underrepresented in the bloom sample according to their transcripts’ fold change and relative abundance (c.p.m.) compared with the upstream and downstream non-bloom stations (Figure 3). Unless noted, the dinoflagellate-associated module expression patterns described herein are those that were differentially expressed between the bloom sample versus both stations 20 and 70.
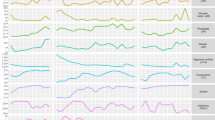
Metabolic pathway color-coded scatter plot of log fold change in transcript abundance of dinoflagellates between the bloom sample and the non-bloom sample for (a) bloom versus station 20, and (b) bloom versus station 70. Plotted are the log fold-change of module transcripts over average sequence abundance (log c.p.m.). Modules are grouped into different categories based on their metabolic functions. Circle size indicates the percentage of enzymes/proteins in each KEGG module with reads mapping to the underlying KEGG genes in dinoflagellates. The largest circles are 100% complete modules, whereas the smallest have at least a single gene mapped. The circle size legend indicates the proportion of genes with mapped reads for a given module. Both axes are in log scale (base 2). Selected modules are annotated as follows: ABC, putative ABC transport system (M00211); B1, thiamin biosynthesis (M00127); B7, biotin biosynthesis (M00123); B12, cobalamin biosynthesis (M00122); C4, C4 carbon fixation (M00172); Cho-Syn, cholesterol biosynthesis (M00101); Cho-Deg, cholesterol degradation (M00104); COX1, cytochrome c oxidase (M00155); FeCTS, iron complex transport system (M00240); FtsE, cell division transport system (M00256); GAG, glycosaminoglycan biosynthesis (M00059); GOGAT2, glutamate synthase(ferrodoxin) (K00284); GSL, glycosphingolipid biosynthesis (M00068); MST, putative multiple sugar transporters (M00207); N-reg, GlnL-GlnG two-component regulatory system (M00497); NUC, nucleotide sugar biosynthesis (M00361); PC, phosphatidylcholine biosynthesis (M00090); Pro, proline biosynthesis (M00015); PS II, photosystem II (M00161); PSR, phosphate starvation response (M00434); SS, sphingosine biosynthesis (M00099); SST, putative simple sugar transport system (M00221); RBC, RuBisCo (W00008); Spd, spermidine biosynthesis (M00133); VicK-VicR, cell wall metabolism (M00459).
Growth-related metabolic pathways during the dinoflagellate bloom
An algal bloom is often characterized by rapid growth of cells within a relatively short period of time (Pinckney et al., 1997), although it is recognized that there are numerous alternative conditions conducive to algal blooms, including a release of grazing pressure (Litaker et al., 2002). The relatively high concentrations of peridinin and abundance of dinoflagellates in the bloom sample is synonymous with high growth rates anticipated during the bloom event (Figure 2). Consistent with these biomass trends, putative genes encoding for a cell division transport system ATP-binding protein (FtsE; de Leeuw et al., 1999; Arends et al., 2009) as well as genes encompassing a two-component regulatory system for cell wall metabolism (VicK-VicR system; Bisicchia et al., 2007) were identified in dinoflagellates and highly overrepresented in the bloom sample, suggesting high rates of cell division (Figure 3). Although FtsE and VicK-VicR genes were previously only thought to be present in bacteria, our taxonomic analyses as well as evidence from other studies (For example, genomes of Thalassiosira pseudonana, Phaeodactylum tricornutum, Grateloupia taiwanensis and Paramecium tetraurelia) demonstrates their common presence in many eukaryotic phytoplankton.
Transcripts for components of C4/CAM carbon fixation were highly expressed in the bloom sample (Figures 3 and 4). C4 and CAM are CO2 concentrating mechanisms acquired by algae and plants to cope with the higher atmospheric O2 concentrations relative to CO2 (Reinfelder et al., 2000; Giordano et al., 2005). Transcripts for both pyruvate orthophosphate dikinase, which converts pyruvate to phosphoenolpyruvate, and NADP-malate decarboxylase, which catalyzes the decarboxylation of malate to pyruvate, releasing CO2 in the process, were overrepresented in the bloom sample compared with the non-bloom stations (Figure 4). Interestingly, transcripts for phosphoenolpyruvate carboxylase, which has the important role of catalyzing bicarbonate attachment to phosphoenolpyruvate, forming the four-carbon compound oxaloacetate, were underrepresented in the bloom sample relative to station 20 (Figure 4). Thus, although the high expression of genes encoding pyruvate orthophosphate dikinase and NADP-malate decarboxylase may be in response to higher O2 and lower CO2 concentrations and/or increased energy demands, the lack of corresponding overrepresentation of phosphoenolpyruvate carboxylase transcripts draws into question as to whether dinoflagellates use the full C4/CO2 concentrating mechanism pathway and how this pathway is regulated under bloom conditions (Haimovich-Dayan et al., 2013).
Schematic plot of select components in photosynthesis and carbon fixation pathways and the predicted flow of nitrogen into select dinoflagellate biosynthetic compounds. Metabolic reactions are illustrated as pie charts. Within each pie chart, the upper half represents log fold change (log FC) of transcripts between the bloom and station 20 samples, and the bottom half represents log FC between the bloom and station 70 samples, with radius indicative of average sequence abundance (log c.p.m.). Pie chart color gradients: cooler colors indicate underrepresented in bloom sequence library versus stations 20 or 70; warmer colors, indicate overrepresented in bloom sequence library versus stations 20 or 70; and gray, no change between samples. C4 pathway is incomplete and shown is the predicted localization of proteins based on Giordano et al. (2005).
Dinoflagellate-associated genes involved in N metabolism, including nitrate/ammonium transporters, nitrate/nitrite reductases, urease and glutamine and glutamate synthases were expressed in all sequence libraries, indicating that dinoflagellates are likely using multiple N sources (Zhuang et al., 2015; Figure 4). However, expression patterns revealed discrepancies in N metabolism between the bloom and the non-bloom samples. Ammonium transporters, ferredoxin-dependent glutamate synthase (GOGAT2) and several components of the urea cycle were overrepresented relative to both non-bloom stations, whereas nitrite reductase was overrepresented relative to station 20 (Figure 4). GOGAT2 has a significant role in ammonium uptake and assimilation (Okuhara et al., 1999). Carbamoyl-phosphate synthase, argininosuccinate synthase and argininosuccinate lyase are three (out of five) key enzymes in the urea cycle and were also overrepresented in the bloom sample. Overrepresentation of genes in urea cycle along with those of genes involved in other nitrogen metabolisms infer high N compound cycling by blooming dinoflagellates because of high N demands (Coschigano et al., 1998; Okuhara et al., 1999; Maheswari et al., 2010; Allen et al., 2011; Kang et al., 2015). Genes involved in proline (Pro), glutamate (GOGAT) and spermidine (Spd) were also overrepresented (Figures 3 and 4). These N-rich metabolites are critical components for protein biosynthesis and cell wall formation, as well as being influential in regulating cellular N status (Allen et al., 2011). Spermidine synthesis has also been found to increase under rapid growth (Nishibori and Nishio, 1997; Nishibori and Nishijima, 2004) consistent with high growth rates in the bloom sample. In addition, a regulatory system for N (GlnL-GlnG two-component regulatory system), where expression increases under N limitation (Reitzer, 2003), was overrepresented in the bloom sample (Figure 3). Noticeably, porphyrin and chlorophyll metabolism (HemA-HemH, HemL, EARS, Chl, Chlp, Chlm), which are downstream biochemical processes of glutamate biosynthesis, were also primarily overrepresented in the bloom condition (Figure 4). The high production of chlorophyll is indicative of high growth rates and probably higher photosynthetic activities (Bollivar, 2006). Taken together, despite the steep gradients in NO3 concentrations and variations in N sources observed during our sampling effort, the biological processes in N-requiring metabolic pathways that are overrepresented in the bloom sample suggest a higher metabolic demand for N likely stemming from rapid cell growth within an increasingly N-deprived environment. Another differential expression analysis was conducted between station 20 and station 70 samples. Station 70 was characterized with module expression profiles consistent with low nitrogen availability (Figure 1, Data set 2). Overrepresentation of nitrogen assimilation modules at the bloom station compared with both non-bloom samples suggests that blooming dinoflagellates had higher nitrogen demand/nitrogen stress compared with that at station 70 where a high nitrogen demand was also inferred by changes in expression of nitrogen-associated modules relative to station 20.
Signals of higher metabolic demands for phosphorous (P) and iron (Fe) were also detected. Transcripts for a phosphate starvation response system and a Fe complex transport system were overrepresented in the bloom sample (Figure 3). Phosphorous is a major component of nucleic acids and phospholipids (Van Mooy et al., 2006). The high expression of the phosphate starvation response suggests an increased P demand of dinoflagellates, which was possibly caused by higher growth rates. Iron serves as an electron carrier in various protein complexes and has a central role in photosynthesis, cellular respiration and N metabolism, as well as other biological reactions (Marchetti and Maldonado, 2016). Expression of Fe complex transport system has been found to increase under Fe starvation conditions (Katoh et al., 2001; Yuan et al., 2005, 2008). Thus, the increased expression of genes involved in this system can also serve as a signal of increased Fe demands in dinoflagellates during the bloom. Thus, our study highlights how cellular nutrient demands associated with rapid growth in specific members of a phytoplankton community may elicit nutrient limitation-like responses regardless of the external nutrient concentrations.
Under the bloom condition, production and transportation of various polysaccharides were overrepresented, including metabolism of N-glycans, GAGs, glycosphingolipids, sphingosines and phosphatidylcholines (PCs), as well as putative single/multiple sugar transport systems (SST/MST; Figure 3). The highly represented polysaccharides are structural components within cell membranes and surface-bound proteins, involved in cell interactions, nutrient acquisition, growth factors and acquisition of numerous other substances from the surrounding environment (Sarrazin et al., 2011; Delbarre-Ladrat et al., 2014). With higher metabolic rates and growth rates, dinoflagellates would likely need to produce more polysaccharides to meet the increased demand for cell membrane formation and other polysaccharide-involved biological processes.
A putative multiple sugar transporter system and another polysaccharide-associated ABC transporter system (ABC) were overrepresented (Figure 3), both of which have been shown to export polysaccharides outside of cells (Delbarre-Ladrat et al., 2014). Secretion of polysaccharides has previously been observed in blooming phytoplankton in response to elevated nutrient demands (Myklestad, 1999; Decho, 2000). Exopolysaccharides are made of a large number of exported polysaccharides that attach to cellular membranes or are dispersed into the environment. They may have important roles in relieving environmental stress, mostly through trapping nutrients to alleviate nutrient shortages and enhancing cell membrane integrity under harsh physical environments (Delbarre-Ladrat et al., 2014). In particular, genes involved in the synthesis of GAGs, a major group of mucopolysaccharides present in almost all domains of life, were overrepresented in the bloom sample (Figure 3), suggesting their elevated production under bloom conditions. Although GAGs have multiple suspected roles, they are often involved in cell adhesion processes. In the planktonic diatom Bacteriastrum jadranum, the formation of GAG-like exopolysaccharide particles forms a fibrillar network that can extend large distances from the cell surface, facilitating colony formation and other interactions with their surrounding environment (Bosak et al., 2012).
Cholesterol biosynthesis in dinoflagellates was overrepresented, whereas cholesterol degradation was underrepresented in the bloom sample relative to the surrounding non-bloom stations (Figure 3). Dinoflagellate sterols (also referred to as dinosterols) have been shown to increase during resting cyst formation, possibly for use as long-term organic carbon reserves (Amo et al., 2010). Chemically reduced carbon allocated to the synthesis of cholesterols would then be used to support later growth when environmental conditions are more favorable to proliferate. Because of the high biomass of dinoflagellates along with an increased nutrient demand within an environment containing a reduced nutrient supply, the infered accumulation of dinosterols in dinoflagellates at the peak of the bloom could be a response to the suboptimal environmental conditions where both intra- and interspecies competition for limiting growth resources is intense.
Vitamin synthesis
Phytoplankton require multiple B vitamins, including vitamin B12 (cobalamin), vitamin B1 (thiamine) and vitamin B7 (biotin), as growth factors that have been shown to be in varying supply in marine environments (Sañudo-Wilhelmy et al., 2014). Combinations of these B vitamins are required for different phytoplankton (Croft et al., 2006). B vitamin auxotrophy is present in several unrelated phyla, suggesting that it has arisen independently in the evolution of phytoplankton, and variability in B vitamin requirements is observed at even the species level (Croft et al., 2006). For auxotrophic eukaryotic phytoplankton, bacteria have pivotal roles in vitamin supply to support phytoplankton growth (Croft et al., 2006). Expression of biotin and thiamine biosynthesis genes was detected in all of our sequence libraries (Figure 3), indicating the ability of at least some of the dinoflagellates in our samples to biosynthesize both of these vitamins. Although eukaryotic phytoplankton are thought to be unable to synthesize cobalamin (Croft et al., 2005), unexpectedly, several dinoflagellate-associated genes encoding enzymes involved in the biosynthesis of cobalamin were detected in our sequence libraries, as discussed below.
Biotin is a cofactor for several fundamental carboxylase enzymes and is involved in central pathways in both prokaryotic and eukaryotic cell metabolisms (Streit and Entcheva, 2003). Four enzymes, BioF, BioA, BioD and BioB are required to convert pimeloyl-CoA, which is the precursor for biotin biosynthesis, to biotin (Croft et al., 2006). Expression of BioF, BioA and BioB were detected in both the bloom and non-bloom samples, whereas BioD was not detected in either sample. However, the absence of BioD transcripts does not necessarily negate dinoflagellates’ ability to synthesize BioD, as BioD is not present in the genomes of many biotin-producing phytoplankton and plant species (for example, Thalassiosira pseudonana and Arabidopsis thaliana). The absence of BioD suggests that the function of BioD might be carried out by another unidentified enzyme (Croft et al., 2006). Under the bloom condition, genes for biotin biosynthesis were overrepresented, consistent with higher biotin production corresponding with the high metabolic rates of the blooming dinoflagellates.
Thiamine is another important cofactor for carbohydrate and branched-chain amino-acid metabolisms. Biosynthesis of thiamine requires the coupling of thiazole and pyrimidine to form thiamine monophosphate, which is then catalyzed by thiamine pyrophosphokinase to form thiamine pyrophosphate, the active form of thiamine (Croft et al., 2006). Similar to biotin, genes involved in the biosynthesis of thiamine were also overrepresented in the bloom condition. All enzymes involved in pyrimidine metabolism in dinoflagellates were detected in each sample in addition to genes encoding enzymes for catalyzing the coupling of pyrimidine and thiazole. However, genes involved in thiazole metabolism were not detected in the bloom sample nor was thiamine pyrophosphokinase (Data set 2, M00127). The absence of genes encoding for some of these essential enzymes in thiamine biosynthesis may be a result of our current incomplete understanding of this pathway in phytoplankton. However, the high expression of most of the thiamine biosynthesis genes in the blooming dinoflagellates suggests an increased production of thiamine.
Cobalamin is a cofactor catalyzing either rearrangement–reduction reactions or methyl transfer reactions, and is essential for cobalamin-dependent enzymes (Croft et al., 2006). Some phytoplankton require cobalamin for cobalamin-dependent methionine synthase (MetH), whereas an alternative form of methionine synthase (MetE) exists in some microalgae that do not require B12, alleviating them of most of their B12 requirement (Helliwell et al., 2011; Bertrand et al., 2012). Eukaryotic phytoplankton must obtain B12 from their environment and/or close bacterial associations (Croft et al., 2005). Surprisingly, two putative genes assigned to dinoflagellates, CobC1 and CobC2, encoding for part of the cobalamin biosynthesis pathway were detected in the bloom sample (CobC2 only) as well as station 20 (both genes; Supplementary Figure S3). α-Ribazole phosphatase (CobC2) is an enzyme that transforms α-ribazole 5'-phosphate to α-ribazole, and threonine-phosphate decarboxylase (CobC1) catalyzes a step in the pathway of adenosylcobalamin biosynthesis, both of which are necessary steps in the formation of the final product, cobalamin coenzyme (Scott and Roessner, 2002). Besides CobC1 and CobC2, other putative genes involved in B12 biosynthesis were also detected in NRE samples collected at different time points (data not shown); however, we did not detect the expression of the entire suite of genes required for B12 synthesis in a single sample of our eukaryotic sequence libraries. The high expression of genes for putative dinoflagellate-associated CobC2 in the bloom sample is consistent with dinoflagellates being able to at least partially biosynthesize vitamin B12 following the acquisition of B12 intermediates. MetH was highly expressed at station 20 but was not detected at the bloom station, suggesting low B12 availability during the bloom (Data set 4; Bertrand et al., 2013). Given bacteria are major B12 suppliers to dinoflagellates, we thus hypothesize that bacterially derived B12 intermediates may be acquired by dinoflagellates and processed to biosynthesize B12. Similar to our findings, Zhang et al. (2013) detected the expression of the cobalamin biosynthesis gene, CobW, in dinoflagellates, before which there was only limited support for full or partial cobalamin synthesis within any algal lineage.
Implications for dinoflagellate-bacterial interactions
Phytoplankton secrete dissolved organic matter, especially when nutrient stressed (Wear et al., 2015). Similarly, previous studies have demonstrated that under bloom conditions, phytoplankton can release large amounts of extracellular polysaccharides into their surrounding environment, forming large sticky aggregates (Myklestad, 1995). The mucilage material coagulates both phytoplankton and surrounding bacteria cells, possibly serving as protection for phytoplankton against predation and a means to deal with turbulence within the water column (Smayda, 2002). Some of the mucilage secreted by phytoplankton can also be used as an organic carbon source for heterotrophic bacteria (Decho, 2000; Croft et al., 2005). Production and transportation of simple sugars (ABC, SST), which are readily assimilated by heterotrophic bacteria, were also overrepresented during the bloom (Figure 3, Data set 2). Correspondingly, previous studies have shown that organic carbon uptake increases in bacteria during algal blooms, as well as adhesiveness of bacterial cell surfaces that may also be associated with phytoplankton mucilage formation (Rinta-Kanto et al., 2012). The inferred increased production of both simple sugar and polysaccharide exudates suggests that dinoflagellates may actively promote the growth and interaction with their surrounding heterotrophic bacteria community. Notably, bacterial communities have also been shown to have higher biomass and more efficient remineralization of organic materials when in aggregates, producing more inorganic nutrients that could then be made available to phytoplankton (Müller-MNiklas et al., 1994; Decho, 2000; Cooper and Smith, 2015).
Croft et al. (2005) also found that coculturing with phytoplankton significantly increases bacteria’s B12 production. Similarly, increased biosynthesis and secretion of biotin and thiamine by dinoflagellates would support further bacterial growth, as there are numerous bacteria that do not possess the metabolic pathways for synthesis of these vitamins (Myklestad, 2000; Sañudo-Wilhelmy et al., 2014). In addition to roles in nutrient trapping and cell adhesion, polysaccharide exudates can bind chemokines, which are used to facilitate cell-to-cell communications (Sarrazin et al., 2011). Remarkably, a two-component regulatory system for chemotaxis was overrepresented in dinoflagellates under bloom conditions (Figure 3). The expression of chemotaxis was shown to be a response of a marine heterotrophic bacterial assemblages to higher nutrient concentrations (Shi et al., 2012). Taken together, we hypothesize that under bloom conditions, dinoflagellates produce excess polysaccharides and vitamins in efforts to cultivate bacteria so that they may acquire essential macro- and micronutrients (N, P, B12, etc.) produced by bacteria to prolong the bloom. Alternatively, if the blooming dinoflagellates are mixotrophic, which many of them are (Jeong et al., 2010), bacterial cultivation may provide a source of organic matter when inorganic nutrients are depleted to levels where autotrophy is no longer favored.
Potential molecular indicators for bloom development
Characterized by rapid growth, blooming phytoplankton have high metabolic activity rates. Overrepresented transcripts in the nucleotide sugar biosynthesis and other DNA/RNA processing systems are consistent with the blooming phytoplankton experiencing high rates of energy-demanding transcription and translation (Figure 3). To meet this increase in energy demands, high proportions of transcripts were devoted to ATP synthesis, carbon fixation and photosynthesis in the bloom sample (Figures 3 and 5). As organic carbon synthesis and energy production are fundamental to phytoplankton growth, increased expression of photosynthesis and carbon fixation are crucial for the formation and propagation of an algal bloom. Thus, molecular activities of these processes could be used as markers for assessing bloom potential. However, their increased expression alone does not guarantee a bloom will develop. In addition to these metabolic processes, carbohydrate biosynthesis and transportation may also serve as molecular proxies for bloom formation. Increased production of carbohydrate exudates is common in intense algal blooms, which are consistent with the overrepresentation of transcripts for single/polysaccharide transportation and biosynthesis activities in our bloom sample (Figure 3). Lastly, as increased sterol synthesis may be associated with resting cysts, the accumulation of dinosterols may be indicative of bloom termination, providing a useful indicator of the bloom entering senescent phase. Although more work is needed, we suggest such genes as potential targets that can be used in conjunction with advancing probe technologies for bloom characterization and management efforts.
Summary
Metatranscriptomic analysis has provided a comprehensive examination into the molecular underpinnings of blooming dinoflagellates within a natural setting. When combined with environmental data, our study provides a unique and in-depth approach to coupling the transcriptome-level responses by dinoflagellates to changes in their surrounding environment under bloom-favorable conditions. Our findings indicate that dinoflagellates increased many components of their cellular metabolism and growth, including a possible reliance on various components of CO2-concentrating mechanisms. In response to increased cellular demands, dinoflagellates increased expression levels of genes involved in N, P and Fe acquisition, as well as the biosynthesis of B vitamins. Despite these efforts, a decreasing N supply in combination with gene expression patterns indicative of high N demand suggests that the blooming dinoflagellates were possibly experiencing the onset of N limitation. We speculate dinoflagellates were producing and exporting sugars, exopolysaccharides and vitamins to facilitate bacterial interactions, either to increase remineralization of nutrients by bacteria and promote nutrient acquisition or for their direct consumption. Thus, we speculate increased interactions between dinoflagellates and bacteria may be a common phenomenon that facilitates the propagation of dinoflagellate blooms. Our study is based on transcriptomes. Dinoflagellates have been shown to undergo widespread trans-splicing, which may complicate interpretations based on gene transcripts (Lidie and van Dolah, 2007; Lin et al., 2010). Thus, further research would be beneficial to assess whether changes in the gene module expression we have discussed are reflective of protein expression and activity. In addition, further investigation of our hypothesized phytoplankton and bacterial interactions under bloom conditions is warranted.
References
Alexander H, Jenkins BD, Rynearson TA, Dyhrman ST . (2015). Metatranscriptome analyses indicate resource partitioning between diatoms in the field. Proc Natl Acad Sci USA 112: 201421993.
Allen AE, Dupont CL, Oborník M, Horák A, Nunes-Nesi A, McCrow JP et al. (2011). Evolution and metabolic significance of the urea cycle in photosynthetic diatoms. Nature 473: 203–207.
Amin SA, Hmelo LR, van Tol HM, Durham BP, Carlson LT, Heal KR et al. (2015). Interaction and signalling between a cosmopolitan phytoplankton and associated bacteria. Nature 522: 98–101.
Amo M, Suzuki N, Kawamura H, Yamaguchi A, Takano Y, Horiguchi T . (2010). Sterol composition of dinoflagellates: different abundance and composition in heterotrophic species and resting cysts. Geochem J 44: 225–231.
Anderson DM, Burkholder JM, Cochlan WP, Glibert PM, Gobler CJ, Heil CA et al. (2008). Harmful algal blooms and eutrophication: examining linkages from selected coastal regions of the United States. Harmful Algae 8: 39–53.
Arends SJR, Kustusch RJ, Weiss DS . (2009). ATP-binding site lesions in FtsE impair cell division. J Bacteriol 191: 3772–3784.
Bertrand EM, Allen AE, Dupont CL, Norden-Krichmar TM, Bai J, Valas RE et al. (2012). PNAS Plus: influence of cobalamin scarcity on diatom molecular physiology and identification of a cobalamin acquisition protein. Proc Natl Acad Sci USA 109: E1762–E1771.
Bertrand EM, Moran DM, Mcilvin MR, Hoffman JM, Allen AE, Saito MA . (2013). Methionine synthase interreplacement in diatom cultures and communities: Implications for the persistence of B 12 use by eukaryotic phytoplankton. Limnol Oceanogr 58: 1431–1450.
Bisicchia P, Noone D, Lioliou E, Howell A, Quigley S, Jensen T et al. (2007). The essential YycFG two-component system controls cell wall metabolism in Bacillus subtilis. Mol Microbiol. 65: 180–200.
Bollivar DW . (2006). Recent advances in chlorophyll biosynthesis. Photosynth Res 90: 173–194.
Bosak S, Pletikapić G, Hozić A, Svetličić V, Sarno D, Viličić D . (2012). A novel type of colony formation in marine planktonic diatoms revealed by atomic force microscopy. PLoS One 7: e44851.
Cole JJ . (1982). Interactions between bacteria and algae in aquatic ecosystems. Annu Rev Ecol Syst 13: 291–314.
Cooper MB, Smith AG . (2015). Exploring mutualistic interactions between microalgae and bacteria in the omics age. Curr Opin Plant Biol 26: 147–153.
Cooper ED, Bentlage B, Gibbons TR, Bachvaroff TR, Delwiche CF . (2014). Metatranscriptome profiling of a harmful algal bloom. Harmful Algae 37: 75–83.
Coschigano KT, Melo-Oliveira R, Lim J, Coruzzi GM . (1998). Arabidopsis gls mutants and distinct Fd-GOGAT genes. Implications for photorespiration and primary nitrogen assimilation. Plant Cell 10: 741–752.
Croft MT, Lawrence AD, Raux-Deery E, Warren MJ, Smith AG . (2005). Algae acquire vitamin B12 through a symbiotic relationship with bacteria. Nature 438: 90–93.
Croft MT, Warren MJ, Smith AG . (2006). Algae need their vitamins. Eukaryot Cell 5: 1175–1183.
Daugbjerg N, Hansen G, Larsen J, Moestrup Ø . (2000). Phylogeny of some of the major genera of dinoflagellates based on ultrastructure and partial LSU rDNA sequence data, including the erection of three new genera of unarmoured dinoflagellates. Phycologia 39: 302–317.
Decho AW . (2000). Microbial biofilms in intertidal systems: an overview. Cont Shelf Res 20: 1257–1273.
Delbarre-Ladrat C, Sinquin C, Lebellenger L, Zykwinska A, Colliec-Jouault S . (2014). Exopolysaccharides produced by marine bacteria and their applications as glycosaminoglycan-like molecules. Front Chem 2: 1–15.
Giordano M, Beardall J, Raven JA . (2005). CO2 concentrating mechanisms in algae: mechanisms, environmental modulation, and evolution. Annu Rev Plant Biol 56: 99–131.
Haimovich-Dayan M, Garfinkel N, Ewe D, Marcus Y, Gruber A, Wagner H et al. (2013). The role of C4 metabolism in the marine diatom Phaeodactylum tricornutum. New Phytol 197: 177–185.
Hallegraeff GM . (1992). Harmful algal blooms in the Australian region. Mar Pollut Bull 25: 186–190.
Heimann K . (2012). Gymnodinium and related dinoflagellates. eLS Available at: http://www.els.net/WileyCDA/ElsArticle/refId-a0001967.html (last accessed 28 September 2015).
Helliwell KE, Wheeler GL, Leptos KC, Goldstein RE, Smith AG . (2011). Insights into the evolution of vitamin B12 auxotrophy from sequenced algal genomes. Mol Biol Evol 28: 2921–2933.
Jeong HJ, Yoo YD, Kim JS, Seong KA, Kang NS, Kim TH . (2010). Growth, feeding and ecological roles of the mixotrophic and heterotrophic dinoflagellates in marine planktonic food webs. Ocean Sci J 45: 65–91.
Kang L-K, Gong G-C, Wu Y-H, Chang J . (2015). The expression of nitrate transporter genes reveals different nitrogen statuses of dominant diatom groups in the southern East China Sea. Mol Ecol 24: 1374–1386.
Katoh H, Hagino N, Grossman AR, Ogawa T . (2001). Genes essential to iron transport in the cyanobacterium Synechocystis sp. strain PCC 6803. J Bacteriol 183: 2779–2784.
Kiørboe T, Andersen KP, Dam HG . (1990). Coagulation efficiency and aggregate formation in marine phytoplankton. Mar Biol 107: 235–245.
de Leeuw E, Graham B, Phillips GJ, ten Hagen-Jongman CM, Oudega B, Luirink J . (1999). Molecular characterization of Escherichia coli FtsE and FtsX. Mol Microbiol 31: 983–993.
Lidie KB, van Dolah FM . (2007). Spliced leader RNA-mediated trans-splicing in a dinoflagellate, Karenia brevis. J Eukaryot Microbiol 54: 427–435.
Lima-mendez G, Faust K, Henry N, Decelle J, Colin S, Carcillo F et al. (2015). Determinants of community structure in the global plankton interactome. Ocean Plankton 348: 1–10.
Lin S, Zhang H, Zhuang Y, Tran B, Gill J . (2010). Spliced leader-based metatranscriptomic analyses lead to recognition of hidden genomic features in dinoflagellates. Proc Natl Acad Sci USA 107: 20033–20038.
Litaker RW, Tester PA, Duke CS, Kenney BE, Pinckney JL, Ramus J . (2002). Seasonal niche strategy of the bloom-forming dinoflagellate Heterocapsa triquetra. Mar Ecol Prog Ser 232: 45–62.
Luettich RA Jr, McNinch JE, Paerl HW, Peterson CH, Wells JT, Alperin M et al. (2000). Neuse River Estuary modeling and monitoring project. Water Resources Research Institute of the University of North Carolina. Available at http://www.lib.ncsu.edu/resolver/1840.4/1891.
Maheswari U, Jabbari K, Petit J-L, Porcel BM, Allen AE, Cadoret J-P et al. (2010). Digital expression profiling of novel diatom transcripts provides insight into their biological functions. Genome Biol 11: R85.
Marchetti A, Maldonado MT . (2016) Iron. In: Borowitzka MA, Beardall J, Raven JA (eds), The Physiology of Microalgae. Springer International Publishing: Cham, Switzerland, pp 233–279.
Marchetti A, Schruth DM, Durkin CA, Parker MS, Kodner RB, Berthiaume CT et al. (2012). Comparative metatranscriptomics identifies molecular bases for the physiological responses of phytoplankton to varying iron availability. Proc Natl Acad Sci USA 109: E317–E325.
Moestrup Ø, Hakanen P, Hansen G, Daugbjerg N, Ellegaard M . (2014). On Levanderina fissa gen. & comb. nov. (Dinophyceae) (syn. Gymnodinium fissum Gyrodinium instriatum Gyr. uncatenum, a dinoflagellate with a very unusual sulcus. Phycologia 53: 265–292.
Van Mooy BAS, Rocap G, Fredricks HF, Evans CT, Devol AH . (2006). Sulfolipids dramatically decrease phosphorus demand by picocyanobacteria in oligotrophic marine environments. Proc Natl Acad Sci USA 103: 8607–8612.
Müller-MNiklas G, Stefan S, Kaltenböok E, Herndl GJ . (1994). Organic content and bacterial metabolism in amorphous aggregations of the northern Adriatic Sea. Limnol Oceanogr 39: 58–68.
Myklestad SM . (1995). Release of extracellular products by phytoplankton with special emphasis on polysaccharides. Sci Total Environ 165: 155–164.
Myklestad SM . (1999). Phytoplankton extracellular production and leakage with considerations on the polysaccharide accumulation. Ann Super Sanita 35: 401–404.
Myklestad SM . (2000). Dissolved organic carbon from phytoplankton. Mar Chem 5: 112–144.
Nielsen R, Bustamante C, Clark AG, Glanowski S, Sackton TB, Hubisz MJ et al. (2005). A scan for positively selected genes in the genomes of humans and chimpanzees. PLoS Biol 3: e170.
Nishibori N, Nishijima T . (2004). Changes in polyamine levels during growth of a red-tide causing phytoplankton Chattonella antiqua (Raphidophyceae). Eur J Phycol 39: 51–55.
Nishibori N, Nishio S . (1997). Occurrence of polyamines in the broom forming toxic dinoflagellate Alexandrium tamarense Naoyoshi and Sachio Key words: Alexandrium late. The peaks corresponding to putrescine and spermidine benzoates in HPLC analysis were subjected to LC/FABMS analysis. Fish Sci 63: 319–320.
Okuhara H, Matsumura T, Fujita Y, Hase T . (1999). Cloning and inactivation of genes encoding ferredoxin- and NADH-dependent glutamate synthases in the cyanobacterium Plectonema boryanum. Imbalances in nitrogen and carbon assimilations caused by deficiency of the ferredoxin-dependent enzyme. Plant Physiol 120: 33–42.
Paerl HW . (2006). Assessing and managing nutrient-enhanced eutrophication in estuarine and coastal waters: interactive effects of human and climatic perturbations. Ecol Eng 26: 40–54.
Paerl HW, Pinckney JL, Fear J, Peierls BL . (1998). Ecosystem responses to internal and watershed organic matter loading: consequences for hypoxia in the eutrophying Neuse River Estuary, North Carolina, USA. Mar Ecol Prog Ser 166: 17–25.
Paerl HW, Rossignol KL, Hall NS, Peierls BL, Wetz MS . (2010). Phytoplankton community indicators of short- and long-term ecological change in the anthropogenically and climatically impacted neuse river estuary, North Carolina, USA. Estuar Coasts 33: 485–497.
Paerl HW, Valdes LM, Joyner AR, Piehler MF, Lebo ME . (2004). Solving problems resulting from solutions: evolution of a dual nutrient management strategy for the eutrophying Neuse River Estuary, North Carolina. Environ Sci Technol 38: 3068–3073.
Paerl HW, Whitall DR . (1999). Anthropogenically-derived atmospheric nitrogen deposition, marine eutrophication and harmful algal bloom expansion. Ambio 28. Available at: http://uncch.pure.elsevier.com/en/publications/anthropogenically-derived-atmospheric-nitrogen-deposition-marine (last accessed 19 January 2016).
Peierls BL, Christian RR, Paerl HW . (2003). Water quality and phytoplankton as indicators of hurricane impacts on a large estuarine ecosystem. Estuaries 26: 1329–1343.
Peierls BL, Hall NS, Paerl HW . (2012). Non-monotonic responses of phytoplankton biomass accumulation to hydrologic variability: a comparison of two coastal plain North Carolina Estuaries. Estuar Coasts 35: 1376–1392.
Piehler MF, Twomey LJ, Hall NS, Paerl HW . (2004). Impacts of inorganic nutrient enrichment on phytoplankton community structure and function in Pamlico Sound, NC, USA. Estuar Coast Shelf Sci 61: 197–209.
Pinckney JL, Millie DF, Vinyard BT, Paerl HW . (1997). Environmental controls of phytoplankton bloom dynamics in the Neuse River Estuary, North Carolina, U.S.A. Can J Fish Aquat Sci 54: 2491–2501.
Pinckney JL, Paerl HW, Tester P, Richardson TL . (2001). The role of nutrient loading and eutrophication in estuarine ecology. Environ Health Perspect 109: 699–706.
Reinfelder JR, Kraepiel AM, Morel FM . (2000). Unicellular C4 photosynthesis in a marine diatom. Nature 407: 996–999.
Reitzer L . (2003). Nitrogen assimilation and global regulation in Escherichia coli. Annu Rev Microbiol 57: 155–176.
Rinta-Kanto JM, Sun S, Sharma S, Kiene RP, Moran MA . (2012). Bacterial community transcription patterns during a marine phytoplankton bloom. Environ Microbiol 14: 228–239.
Robinson MD, Oshlack A . (2010). A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biol 11: R25.
Sañudo-Wilhelmy SA, Gómez-Consarnau L, Suffridge C, Webb EA . (2014). The role of B vitamins in marine biogeochemistry. Ann Rev Mar Sci 6: 339–367.
Sarrazin S, Lamanna WC, Esko JD . (2011). Heparan sulfate proteoglycans. Cold Spring Harb Perspect Biol 3: 1–33.
Scott AI, Roessner CA . (2002). Biosynthesis of cobalamin (vitamin B12 . Biochem Soc Trans 30: 613–620.
Sellner KG, Doucette GJ, Kirkpatrick GJ . (2003). Harmful algal blooms: causes, impacts and detection. J Ind Microbiol Biotechnol 30: 383–406.
Sharp JH, Pennock JR, Church TM, Tramontano JM, Cifuentes LA . ((1984)). Estuarine interaction of nutrients, organics, and metals: A Case Study in the Delaware Estuary. In: VS Kennedy (ed), Estuary as a Filter. Elsevier: Orlando, FL, USA, pp 241–258.
Shi Y, McCarren J, Delong EF . (2012). Transcriptional responses of surface water marine microbial assemblages to deep-sea water amendment. Environ Microbiol 14: 191–206.
Smayda TJ . (1997). Harmful algal blooms: their ecophysiology and general relevance to phytoplankton blooms in the sea. Limnol Oceanogr 42: 1137–1153.
Smayda TJ . (2002). Turbulence, watermass stratification and harmful algal blooms: an alternative view and frontal zones as ‘pelagic seed banks’. Harmful Algae 1: 95–112.
Streit WR, Entcheva P . (2003). Biotin in microbes, the genes involved in its biosynthesis, its biochemical role and perspectives for biotechnological production. Appl Microbiol Biotechnol 61: 21–31.
Twomey LJ, Piehler MF, Paerl HW . (2005). Phytoplankton uptake of ammonium, nitrate and urea in the Neuse River Estuary, NC, USA. Hydrobiologia 533: 123–134.
Wear EK, Carlson CA, Windecker LA, Brzezinski MA . (2015). Roles of diatom nutrient stress and species identity in determining the short- and long-term bioavailability of diatom exudates to bacterioplankton. Mar Chem 177: 335–348.
Whitall DR, Paerl HW . (2001). Spatiotemporal variability of wet atmospheric nitrogen deposition to the Neuse River Estuary, North Carolina. J Environ Qual 30: 1508.
Yuan Y, Wu H, Wang N, Li J, Zhao W, Du J et al. (2008). FIT interacts with AtbHLH38 and AtbHLH39 in regulating iron uptake gene expression for iron homeostasis in Arabidopsis. Cell Res 18: 385–397.
Yuan YX, Zhang J, Wang DW, Ling HQ . (2005). AtbHLH29 of Arabidopsis thaliana is a functional ortholog of tomato FER involved in controlling iron acquisition in strategy I plants. Cell Res 15: 613–621.
Zhang H, Zhuang Y, Gill J, Lin S . (2013). Proof that dinoflagellate spliced leader (DinoSL) is a useful hook for fishing dinoflagellate transcripts from mixed microbial samples: Symbiodinium kawagutii as a case study. Protist 164: 510–527.
Zhuang Y, Zhang H, Hannick L, Lin S . (2015). Metatranscriptome profiling reveals versatile N-nutrient utilization, CO2 limitation, oxidative stress, and active toxin production in an Alexandrium fundyense bloom. Harmful Algae 42: 60–70.
Acknowledgements
We thank J Braddy and other members of the ModMon Team who assisted with sample collection and analysis. We are grateful to H Masters for assistance with RNA extractions and C Stackhouse and J Roach for assistance with sequence analysis. UNC Research Computing generously provided cluster time for sequence analysis. ModMon is funded by NC-Department of Environment and Natural Resources, Division of Water Resources, the Lower Neuse Basin Association and Neuse River Compliance Association. This research was supported by NC Sea Grants R/12-HCE-3—NA10OAR4170080 and R/14-HCE-2—NA14OAR4170073 to AM and HP as well as start-up funds to AM.
Author contributions
AM and HP designed research; WG, AM and JB performed research, WG and DMS contributed new analytical tools; WG, AM and NH analyzed data; and WG and AM wrote the paper.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies this paper on The ISME Journal website
Supplementary information
Rights and permissions
About this article
Cite this article
Gong, W., Browne, J., Hall, N. et al. Molecular insights into a dinoflagellate bloom. ISME J 11, 439–452 (2017). https://doi.org/10.1038/ismej.2016.129
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ismej.2016.129
This article is cited by
-
Quantitative proteomics provides insight into the response of the marine dinoflagellate Prorocentrum donghaiense to changes in ambient phosphorus
Journal of Oceanology and Limnology (2022)
-
Distribution patterns and seasonal variations in phytoplankton communities of the hypersaline Pulicat lagoon, India
Environmental Science and Pollution Research (2021)
-
Picophytoplankton dynamics in a large temperate estuary and impacts of extreme storm events
Scientific Reports (2020)
-
Strategies among phytoplankton in response to alleviation of nutrient stress in a subtropical gyre
The ISME Journal (2019)
-
Transcriptomic and microRNAomic profiling reveals multi-faceted mechanisms to cope with phosphate stress in a dinoflagellate
The ISME Journal (2017)