Abstract
Community dynamics are often studied in subsets of pairwise interactions. Scaling pairwise interactions back to the community level is, however, problematic because one given interaction might not reflect ecological and evolutionary outcomes of other functionally similar species interactions or capture the emergent eco-evolutionary dynamics arising only in more complex communities. Here we studied this experimentally by exposing Pseudomonas fluorescens SBW25 prey bacterium to four different protist predators (Tetrahymena pyriformis, Tetrahymena vorax, Chilomonas paramecium and Acanthamoeba polyphaga) in all possible single-predator, two-predator and four-predator communities for hundreds of prey generations covering both ecological and evolutionary timescales. We found that only T. pyriformis selected for prey defence in single-predator communities. Although T. pyriformis selection was constrained in the presence of the intraguild predator, T. vorax, T. pyriformis selection led to evolution of specialised prey defence strategies in the presence of C. paramecium or A. polyphaga. At the ecological level, adapted prey populations were phenotypically more diverse, less stable and less productive compared with non-adapted prey populations. These results suggest that predator community composition affects the relative importance of ecological and evolutionary processes and can crucially determine when rapid evolution has the potential to change ecological properties of microbial communities.
Similar content being viewed by others
Introduction
One of the major goals of ecology is to try to understand the dynamics of complex communities. Traditionally, this question has been approached by decomposing food web complexity into more manageable subsets of interacting species, which are then studied in isolation from the rest of the community (Vandermeer, 1969; Billick and Case, 1994). This approach has shown that there are frequently emergent properties that arise only in the presence of multiple species (Sih et al., 1998; Strauss and Irwin, 2004) resulting in ecological and evolutionary outcomes that could not be predicted by on the basis of single- or even two-species dynamics (Iwao and Rausher, 1997; Strauss and Irwin, 2004; Thompson, 2005; Berenbaum and Zangerl, 2006; Parchman and Benkman, 2008; Friman and Buckling, 2013; Friman and Buckling, 2014). We were interested in whether part of the difficulty in predicting multi-species dynamics arises from the feedbacks between ecological and evolutionary processes that are dependent on the precise composition of the predator–prey community.
Recent results have shown that rapid evolution can significantly alter the ecological properties of predator–prey systems. Probably, the most convincing evidence comes from microbial predator–prey study systems, where rapid evolution of traits connected to prey defence and predator counter-defence has been observed to change the productivity, stability and diversity of predator–prey communities (Yoshida et al., 2003; Meyer and Kassen, 2007; Friman et al., 2008; Becks et al., 2010; Friman et al., 2014; Hiltunen and Becks, 2014). Even though most of this evidence comes from relatively simple two-species model communities, it has recently been shown that the presence of another predator can affect the temporal dynamics of one-prey-one-predator system (Hiltunen et al., 2013), while theory predicts that evolution is more likely to feedback to population dynamics when the prey evolves predator-specific defences (Ellner and Becks, 2011). However, we are not aware of the experiments that test the effects of predator community complexity on prey evolution.
Increasing the number of interacting species could affect predator–prey evolution via ecological and genetic constraints. First, competition for the shared prey is likely to affect the relative abundance of each competing predator species, which will then affect the strength of selection that every predator exerts on the given prey species (Friman and Buckling, 2013). If predator competition is asymmetrical, the most dominant predator species is expected to have strongest effect on prey evolution. If competition between different predators is more symmetrical, both predators are likely to exert selection on prey but these effects are likely to be weaker compared with the effects predators would be exerting on prey in the absence of competition. Second, trait correlations between defence mechanisms against different predators could affect the evolutionary dynamics in multi-predator communities (Iwao and Rausher, 1997; Strauss and Irwin, 2004; Strauss et al., 2005; Friman and Buckling, 2013). In the case of no correlation (independent predator effects), the combined effect of multiple predators may result in divergent selection for specialist defence strategies, where different sub-populations adapt to different interacting species (Futuyma and Moreno, 1988; Davies and Brooke, 1989; Nuismer and Thompson, 2006; Edeline et al., 2008). If defence correlations are negative, selection by one predator could reduce the selection imposed by another predator owing to trade-offs in morphology or physiology (Davies and Brooke, 1989; Stinchcombe and Rausher, 2001; Thompson and Cunningham, 2002; Nuismer and Thompson, 2006; Berenbaum and Zangerl, 2006; Friman and Buckling, 2013). It is also possible that defence against one predator correlates positively with the defence against other predator (for example, owing to functional similarity between different enemies). In this case, selection could be ‘diffuse’ where the prey species evolves in response to the predator community as a whole (Fox, 1988; Thompson, 2005) resulting in a generalist defence phenotype, which is resistant to all predators (Stinchcombe and Rausher, 2001; Thompson and Cunningham, 2002; Berenbaum and Zangerl, 2006; Craig et al., 2007; Gomez et al., 2009).
We used laboratory microbial communities to ask how predator community composition affects the prey evolution and eco-evolutionary dynamics of predator–prey communities. Specifically, Pseudomonas fluorescens SBW25, a prey bacterium, was exposed to four different bacterivorous protists (Tetrahymena pyriformis, Tetrahymena vorax, Chilomonas paramecium and Acanthamoeba polyphaga) in all single-predator, two-predator and four-predator communities for hundreds of prey generations (for ~4 weeks, 24 days); a sufficient timescale to observe changes both in ecological and evolutionary dynamics (Friman and Buckling, 2013; Friman et al., 2014). All selected protist species consumed bacteria and potentially imposed selection for prey defence. Furthermore, T. vorax is polymorphic having small microstome and large macrostome morphs (Gronlien et al., 2002). Macrostome morphs are able to feed on other protists (Gronlien et al., 2002) and T. vorax could thus potentially affect eco-evolutionary dynamics via intraguild predation.
We concentrated on both the population and evolutionary dynamics and investigated (i) how prey evolutionary responses depend on the predator species identity in single-predator communities, (ii) whether pairwise predator–prey interactions predict prey evolutionary responses in multi-predator communities and (iii) whether prey evolution in single- vs multi-predator communities altered the ecological properties of the study system in terms of prey diversity, stability and productivity.
Materials and methods
Study species, culture conditions and selection experiment
We used SBW25 P. fluorescens as a prey for four protist species (T. pyriformis ciliate; CCAP #1630/1W, T. vorax ciliate; CCAP #1630/3C, C. paramecium flagellate; CCAP #977/2A and A. polyphaga amoebae; CCAP #1501/18). The strain SBW25 was originally isolated from a sugar beet leaf (Rainey and Bailey, 1996), and protist cultures were ordered from the Culture Collection for Algae and Protozoa (CCAP). All selected protist species were originally isolated from aquatic environments (Elliott, 1959; Patterson, 1996), were able to feed on the study bacterium and, hence, potentially exerted selection for prey defence (Friman and Buckling, 2013; Friman and Buckling, 2014).
All protists species were cultured axenically in the absence of bacteria before starting the experiment (both Tetrahymena ciliates on PPY medium: 20 g l−1 peptone and 2.5 g l−1 of yeast extract; C. paramecium on CHM medium: 1 g l−1 sodium acetate trihydrate and 1 g l−1 ‘Lab-Lemco’ powder (Oxoid L29); and A. polyphaga on PPG medium: 15 g l−1 peptone, 18 g l−1 d-glucose in Page’s Amoeba Saline solution (CCAP)). Bacterial stocks were prepared by growing bacteria overnight on Luria Broth (LB) medium (Sigma-Aldrich, St Louis, MO, USA; 10 g l−1 of tryptone, 5 g l−1 of yeast extract and 5 g l−1 of NaCl) resulting in final densities of approximately 9 × 107 bacterial cells per ml.
We used 24-well cell culture plates, each containing 2 ml of 0.5% LB (described above) as microcosms during the selection experiment. The SBW25 bacterium was grown alone and in the presence of all protists in one-, two-, and four-protist species combinations at 22 °C in non-shaken conditions. All treatments (12 in total) were replicated five times (N=5) resulting in a total of 60 experimental populations. When initiating the experiment, approximately 2 × 105 bacterial cells ml−1 were first added to all populations. All single-predator treatments were subsequently inoculated with ~400 protist cells. All two-protist treatments were inoculated with ~200 cells per protist species, and four-protist treatment was inoculated with ~100 cells per protist species. Microcosms were renewed every fourth day for a total of six times (24 days) by first mixing the contents thoroughly with pipette and then replacing 1 ml of sample with 1 ml of fresh media. Subsamples of all populations were frozen at −80 °C in 20% glycerol at every sampled time point. Rest of the sample was used to define bacterial and protist population densities. Bacterial densities were estimated with Accuri C6 flow cytometer (Becton Dickinson, Oxford, UK; fast flow rate, 25 μl of sample, a minimum forward scatter threshold of 8000 based on negative controls containing only media). Protist densities were directly counted under the microscope (Motic AE2000, Motic Deutschland GmbH (Germany), Wetzlar, Germany; inverted light microscope).
Measuring bacterial defence against protists
Evolutionary changes in bacterial defence against protists were measured at the end of 24-day long selection experiment. Defence was measured at the level of colony types in order to link bacterial phenotype to a specific defence strategy and to increase measurement accuracy compared with population-level measurements. To this end, we randomly isolated eight independent bacterial colonies per replicate population (50 colonies per treatment; a total of 600 colonies), inoculated selected colonies into liquid 0.5% LB medium and incubated overnight at 22 °C and, finally, froze the colonies in 20% glycerol. Even though isolating eight colonies per replicate population might not capture rare colony types, it has been shown to effectively separate defending and non-defending bacterial genotypes at the population level (Friman et al., 2014). Before the defence measurements, all colonies were thawed and grown to similar densities in 96-well plates (24 h, 22 °C and in 200 μl of 0.5% LB medium; Biotek UK (Swindon, UK), optical density (OD) 600 nm; mean OD of 0.093±0.001; treatment: F11,48=0.572, P=0.842). By equilibrating the initial bacterial densities, subsequent protist growth was only affected by differences in the strength of bacterial antipredatory defence (Friman and Buckling, 2013). Bacterial defence was estimated as the relative fitness in terms of comparing the growth of with-predator-evolved and alone-evolved bacterial selection lines in the presence of ancestral stock predators. To this end, all bacterial selection lines (eight clones from each) were grown individually with every predator species they had been exposed to during the selection experiment. Briefly, all protist measurement plates were inoculated with 20 μl of ancestral stock protist (approximately 100 cells ml−1), and after 48 h of co-cultivation at 22 °C, bacterial defence was determined as the amount of bacterial biofilm biomass; previous studies have shown that bacteria use biofilm aggregation as a size-dependent defence mechanism against protist predators (Matz et al., 2004; Friman and Laakso, 2011; Friman et al., 2013). Bacterial biofilm growth was measured by adding 50 μl of 1% crystal violet solution to microplate wells and rinsed off with distilled water after 10 min. Crystal violet-stained bacteria were dissolved in 96% ethanol and the amount of biofilm measured as OD at 600 nm (O'Toole and Kolter, 1998).
Measuring eco-evolutionary changes in prey communities
Changes in bacterial community diversity were estimated on the basis of colony morphology. SBW25 bacterium can rapidly diversify into different colony types by growing in the air–liquid interface (wrinkly spreader colony types), liquid media (smooth colony types) or by sinking to the bottom of the culture vessels (fuzzy spreader colony type) (Rainey and Travisano, 1998). All these colony types have a fitness advantage when rare and can be maintained in the population via negative frequency-dependent selection (Rainey and Travisano, 1998). In addition to spatial heterogeneity, protist predation can drive SBW25 diversification by favouring wrinkly spreader types (Meyer and Kassen, 2007), which differ genetically from ancestral smooth colony type (Spiers, 2014). We quantified bacterial diversification at the end of the experiment (last sampling point) by counting the number of different colony types from each treatment (plates containing at least 100 individual bacterial colonies). Prey population diversities were estimated with Shannon diversity index (Friman et al., 2008). Prey population stability was determined by calculating the coefficient of variation for each replicate population by using whole time series: higher coefficient values denote higher variability (Friman et al., 2008). Prey population productivity was measured as maximum densities in the absence of predators after 48 h growth at 22 °C (200 μl of 0.5% LB medium).
Statistical analyses
A general linear mixed model (Gaussian family) was used to analyse all data. In all models, the dependent variable was explained with experimental treatment, focal protist species, measurement environment, sampling time and their interactions. For repeated-measures analyses, populations were set as subjects and time as a repeated factor. Replicates were nested under treatments and fitted as a random factor. Additional general linear mixed models were carried out when significant interactions were found. Log-transformed values were used for analysing protist densities owing to unequal variances between the treatments. Arcsin-transformed values were used to analyse differences in colony-type frequencies. Bonferroni-adjusted P-values were used for multiple pairwise comparisons.
Results
Predator effects on bacterial population dynamics
Only T. pyriformis and T. vorax reduced bacterial densities in single-predator treatments (treatment: F4,19.53=13.9, P<0.001, Figures 1a and b), while A. polyphaga or C. paramecium had no effect on bacterial densities (P=0.365 and P=0.183, respectively, Figures 1c and d). The T. pyriformis-driven decrease in bacterial densities was attenuated only in the presence of T. vorax in both two- and four-predator communities (treatment: F5,23.78=81.2, P<0.001; A. polyphaga or C. paramecium had no effect: P=0.559 and P=0.456, respectively, Figure 1a). Similarly, the T. vorax-driven decrease in bacterial densities was attenuated in the presence of T. pyriformis but only in the two-predator communities (treatment: F5,21.99=23, P<0.001; A. polyphaga or C. paramecium had no effect: P=0.906 and P=0.881, respectively, Figure 1b). Finally, the presence of A. polyphaga had no effect on C. paramecium and vice versa (P=0.158 and P=0.600, respectively, Figures 1c and d).
Together these results show that only the two Tetrahymena species decreased bacterial densities, while this effect was constrained only by the presence of the other Tetrahymena species.
Predator effects on protist population dynamics
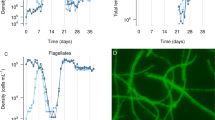
The dynamics of the predator communities are summarised in Figure 2. T. pyriformis reached highest, A. polypahaga second highest and T. vorax and C. paramecium reached lowest densities in single-predator treatments (F3,13.86=21.97, P<0.001, Figures 2a and d). We observed several types of interaction among the protists, including negative, positive and neutral interactions (focal protist density difference between single- and multi-protist treatments). Overall, T. pyriformis was little affected by the presence of the other species and grew well in all combinations except those in which T. vorax was present, where it was strongly depressed (F4,18=197.86, P<0.001). Similarly, T. pyriformis had a negative effect on T. vorax (F4,16.47=5.9, P=0.004). C. paramecium experienced a strong positive response to T. pyriformis (treatment × time: F20,14.59=6.25, P<0.001, Figure 2c). Finally, A. polyphaga grew well on its own or in the presence of C. paramecium, but its growth was depressed by the two ciliates (F4,20.18=349.6, P<0.007).
Bacterial defence evolution in single- and multi-predator communities
In single-predator communities, bacteria evolved defence to protist predation only in the presence of T. pyriformis (F1,8=15.9, P=0.004; none of the other protists increased bacterial defence in any single-predator treatments: all P>0.05, Figure 3). The T. pyriformis-driven increase in bacterial defence was affected by the presence of other protists (F5,24=5.65, P=0.001, Figure 3a): concurrent selection by T. vorax repressed defence evolution in both two- and four-predator communities (P<0.001 and P=0.007, respectively), while bacterial defence against T. pyriformis also evolved less strongly in the presence of C. paramecium (P=0.039; A. polyphaga had no effect: P=0.497). Bacteria did not evolve defence against T. vorax or C. paramecium in any of the treatments (treatment for T. vorax: F5,24=2.7, P=0.09; treatment for C. paramecium: F5,24=1.96, P=0.12; Figures 3b and c). However, bacteria evolved defence against A. polyphaga in the A. polyphaga+T. pyriformis, A. polyphaga+T. vorax and four-protist treatments (F5,24=11.56, P<0.001; P<0.03 in all pairwise comparisons).
Bacterial defence measured against T. pyriformis (a), T. vorax (b), C. paramecium (c) and A. polyphaga (d) protists for bacteria originating from different experimental treatments after the selection experiment. Bacterial defence is calculated as the relative growth of protist-evolved vs alone-evolved bacterial populations. B, SBW25 P. fluorescens bacterium; protists: TP, T. pyriformis; TV, T. vorax; CP, C. paramecium; AP, A. polyphaga. White bars denote single-predator, light grey bars two-predator, and dark grey bars four-predator communities. All data points show mean of five replicate populations and ±1 s.e.m.
Together these results suggest that only T. pyriformis impose detectable selection for bacterial defence evolution in single-predator communities. In multi-protist communities, selection by T. pyriformis was attenuated in the presence of some other protists (T. vorax and C. paramecium), while in some cases bacteria evolved defence only in the presence of several protist species (for example, A. polyphaga-ciliate treatments).
Eco-evolutionary dynamics in single- and multi-predator communities
Predator-driven bacterial phenotypic diversification
Only T. pyriformis predation led to bacterial phenotypic diversification within single-predator treatments (Shannon index; F4,20=61.36, P<0.001, Figure 4a). Diversification was due to an increase in the frequency of wrinkly spreader (WS; F4,16=35.96, P<0.001; 36% of all colonies), and petite colony types (PT; P=0.37; 5% of all colonies; non-significant owing to variation between replicates), resulting in a decrease of ancestral, smooth colony type (SM; F4,20=97.26, P<0.001; 59% of all colonies vs 100% of all colonies in bacterium-only treatment).
Protist-driven bacterial phenotypic diversification (a) and the evolution of different defence strategies in phenotypically diverse experimental communities (b–d). B, SBW25 P. fluorescens bacterium; protists: TP, T. pyriformis; TV, T. vorax; CP, C. paramecium; AP, A. polyphaga. In panel (a), left and right y axes show colony-type frequencies and Shannon diversity index, respectively. Panels (b–d) show WS, PT and TT colony types’ defence relative to SM colony types within T. pyriformis-only (b), T. pyriformis+C. paramecium (c) and T. pyriformis+A. polyphaga (d) experimental treatments. Colony types’ defence was measured in the presence of T. pyriformis (TP), C. paramecium (CP) and A. polyphaga (AP) protists. All data points show mean of five replicate populations and ±1 s.e.m.
Bacterial diversification was further shaped by the presence of other enemies (F4,16=35.96, P<0.001, Figure 4a). Although T. vorax repressed diversification in the presence of T. pyriformis (Shannon index; F5,24=66.38, P<0.001; 100% of colonies SM type), both C. paramecium and A. polyphaga altered T. pyriformis-driven bacterial diversification by selecting for transparent colony types (TT) that were not observed in the T. pyriformis bacterium treatment (0% vs 17% and 23% of all colonies, respectively). Similar to the T. pyriformis-only treatment, PT colony types (10% of all colonies) also emerged in the presence of C. paramecium, whereas no PT colony types were observed in the presence of A. polyphaga.
Together these results suggest that T. pyriformis was the main driver of bacterial phenotypic diversification, while this process was further promoted by both C. paramecium and A. polyphaga and completely repressed by T. vorax.
Phenotypic diversification and evolution of different defence strategies
To assess whether bacterial phenotypic diversification was connected to evolution of different defensive strategies, we measured the defence of different bacterial colony types separately against all protist species they had been exposed to during the selection experiment. WS colony types isolated from the T. pyriformis monocultures were clearly more defensive compared with SM colony types (F2,16.48=30.52, P<0.001, Figure 4b). However, SM or PT colony types originating from the T. pyriformis monoculture treatment were equally poor at defending as SM colony types originating from bacterium-only treatment (F1,8.6=0.529, P>0.05 in both cases, Figure 4b).
We next compared the defence of evolved bacteria originating from the T. pyriformis+C. paramecium treatment (Figure 4c). We found that WS colony types evolved equal levels of defence in the T. pyriformis monoculture and the T. pyriformis+C. paramecium treatments (F2,56.54=1.41, P=0.252, Figure 4c). WS colony types originating from T. pyriformis+C. paramecium treatment were only slightly better at defending against C. paramecium compared with SM colony types. This suggests that defence against T. pyriformis was traded-off with defence against C. paramecium (colony type × predator species: F12,42.07=6.87, P<0.001, Figure 4c). The PT colony types were equally defensive against C. paramecium as the WS types (PT vs SM: P=0.017; PT vs WS: P=0.952, Figure 4c). However, PT colony types were equally susceptible to T. pyriformis as SM colony types (PT vs SM: P=0.912, Figure 4c), which suggests that PT types specialised to defend against C. paramecium. The TT colony types that emerged in small frequency were not particularly good defenders against any predator.
Finally, we assessed the defence of evolved bacteria originating from the T. pyriformis+A. polyphaga treatment (Figure 4d). We found that WS colony types evolved equally defensive in T. pyriformis monoculture and T. pyriformis+A. polyphaga treatments (F2,56.54=1.41, P=0.252, Figure 4d). WS colony types originating from the T. pyriformis+A. polyphaga treatment were also clearly better at defending against A. polyphaga compared with ancestral SM colony types. This suggests that defence against T. pyriformis correlated positively with defence against A. polyphaga (colony type × predator species: F12,43.5=4.45, P<0.001, Figure 4d). Moreover, TT colony types evolved higher levels of defence against A. polyphaga (TT vs SM: P=0.046, Figure 4d). However, this specialist defence strategy correlated negatively with defence against T. pyriformis: TT colony types were as susceptible to T. pyriformis as ancestral SM colony types (TT vs SM: P=0.517).
These results suggest that T. pyriformis selected for generalist defenders in two-predator communities (WS colony types) that were highly defended against both enemies they had been exposed to during the selection experiment. Furthermore, C. paramecium and A. polyphaga selected for specialist defenders in two-predator communities (PT and TT colony types, respectively) that were poor at defending against T. pyriformis but good at defending against C. paramecium and A. polyphaga, respectively.
Changes in stability and productivity of prey populations
Prey selection lines that evolved defence against protists (T. pyriformis monoculture, T. pyriformis+A. polyphaga and T. pyriformis+C. paramecium) became temporally more variable compared with the control selection line (bacterium alone) or selection lines that did not evolve defence against any protists (F1,50=14.6, P<0.001; P<0.001 in all pairwise comparisons) in both single- and two-predator communities (F1,50=0.004, P=0.95; Figure 5a). Non-evolved and control selection lines were equally variable (P=0.2). Similarly, prey selection lines that evolved defence against protists became less productive compared with the control selection line or selection lines that did not evolve defence against any protists (F1,50=7.7, P<0.001; P<0.001 in all pairwise comparisons) in both single- and two-predator communities (F1,50=0, P=0.98; Figure 5b). Non-evolved and control selection lines were equally productive (P=0.8). At the colony-type level, reduced productivity was due to poorer growth of WS, PT and TT colony types relative to ancestral-like SM colony types (F3,28=4.41, P=0.012; P<0.05 in all pairwise comparisons; Figure 5c). Of specialist defenders, TT colony type suffered highest reduction in growth (WS vs TT: P=0.018), while PT colony types suffered intermediate reduction in growth (WS vs PT: P=0.216 and TT vs PT: P=0.27; Figure 5c).
Comparison of prey population stability (a) and productivity (b–c) after selection experiment. In panels (a and b), grey bars show means for evolved treatments (T. pyriformis only, T. pyriformis+A. polyphaga and T. pyriformis+C. paramecium) and white bars show means for non-evolved treatments (all other protist communities). x Axis in panels (a and b) denotes the number of protists prey selection lines evolved during the selection experiment; white bar with 0 protists denotes control selection line (bacterium-only). Panel (c) shows productivity at the colony-type level within phenotypically most diverse experimental communities. B, SBW25 P. fluorescens bacterium; protists: TP, T. pyriformis; TV, T. vorax; CP, C. paramecium; AP, A. polyphaga. In all panels, error estimate is ±1 s.e.m.
Discussion
Here we studied experimentally the role of predator species identity and community complexity for the prey population dynamics, prey defence evolution and potential ecological feedbacks (results summarised in Figure 6). We found that T. pyriformis was a key driver of defence evolution in both single- and two-protist communities. Although other protists did not select for prey defence in single-protist treatment, concurrent selection by T. pyriformis and C. paramecium and T. pyriformis and A. polyphaga led to evolution of specialised defence strategies. Prey defence evolution was repressed in the presence of the intraguild predator, T. vorax, because of indirect competition for shared bacterial prey or because T. vorax was able to efficiently feed on T. pyriformis cells. At the ecological level, adapted prey populations became phenotypically more diverse, less stable and less productive compared with non-adapted prey populations. Together these results suggest that increasing predator community richness can increase prey diversity via selection for specialist defence strategies. However, introduction of intraguild top predator tipped the balance from evolutionary to purely ecological community dynamics. Predator–prey interactions are thus more likely to evolve in communities with weak predator–predator interactions.
Schematic description of the eco-evolutionary dynamics observed during the selection experiment in pairwise predator–prey communities (a), two predator–one prey communities (b) and four predator–one prey communities (c). In all panels, blue and red solid lines denote for negative and positive effects on species population dynamics, respectively, black dashed lines depict for bacterial defence evolution against given protist predators and pie charts depict relative protist abundances. Pairwise predator–prey and two predator–one prey communities were characterised by both ecological and evolutionary dynamics, while four predator–one prey communities were dominated by ecological dynamics. B, SBW25 P. fluorescens bacterium; protists: TP, T. pyriformis; TV, T. vorax; CP, C. paramecium; AP, A. polyphaga.
T. pyriformis was the only predator species that significantly reduced prey populations and was the only predator consistently associated with the evolution of prey defence and diversification. These results are broadly consistent with previous studies (Meyer and Kassen, 2007; Friman and Buckling, 2013; Friman et al., 2014). C. paramecium and A. polyphaga were more weakly linked with prey bacteria and did not significantly decrease bacterial densities in single-protist cultures, which could also explain relatively weak selection for prey defence. Bacteria did not evolve detectable defence against T. vorax either in single-protist cultures, despite the clear reduction in bacterial densities. One explanation for this could be that large T. vorax (maximum cell length of ~200 μm) were able to effectively consume bacterial biofilm aggregates owing to their larger orifice, whereas the relatively smaller T. pyriformis (~60 μm in cell length) could not.
Even though C. paramecium and A. polyphaga did not select for detectable changes in prey defence in single-protist cultures, they affected the diversification of bacterial defensive strategies in T. pyriformis co-cultures. First, the frequency of wrinkly colony types (WS) increased T. pyriformis, T. pyriformis+C. paramecium and T. pyriformis+A. polyphaga treatments. This is in line with previous studies where predation by T. pyriformis and T. thermophila, a closely related species (Brunk et al., 2003), has been shown to drive bacterial diversification in defensive phenotypes (Meyer and Kassen, 2007; Mikonranta et al., 2012; Friman and Buckling, 2014). WS colony types were equally defensive against T. pyriformis regardless if they had evolved in the presence of C. paramecium or A. polyphaga. Similarly, WS colony types that emerged in two-protist treatments were able to defend against C. paramecium and A. polyphaga compared with non-defending SM colony types. This suggests that WS colony types exerted generalist defence strategy probably because small protists were unable to feed on large bacterial biofilm aggregates. Moreover, bacteria diversified into petite (PT) and transparent (TT) colony types in T. pyriformis+C. paramecium and T. pyriformis+A. polyphaga treatments. These colony types were specialised to defend against C. paramecium and A. polyphaga but were at the same time susceptible to predation by T. pyriformis. As a result, concurrent selection by two different protists led to coexistence of generalist and specialist defenders (Berenbaum and Zangerl, 2006; Parchman and Benkman, 2008; Friman and Buckling, 2013), resulting in increased intra-bacterial diversity. Even though these specialist defenders (PT and TT) had a fitness advantage over the non-defending SM colony types at least in the presence of one predator, they always had lower or equally high fitness with a generalist defender (WS). Why were not these specialists driven into extinction? One possibility is that, by testing each colony type in isolation, we have not accounted for interactions with the other colony types. Alternatively, slow-growing PT and TT colony types could have been organised in the bottom of mixed biofilms resulting in enhanced protection against protist predation (Kim et al., 2014) or could have hitchhiked along with SM and WS colony types in the mixed biofilms (Popat et al., 2012; Friman et al., 2013). Although further experiments are needed to test these hypotheses, our results suggest that concurrent selection by two protists potentially changes the topology of bacterial fitness landscape in ways that allow bacterial adaptation against multiple enemies (Flynn et al., 2013).
We also found that protists had negative, positive and neutral effects on each other in multi-protist cultures. Although both T. pyriformis and T. vorax ciliates reduced bacterial densities efficiently in the absence of other predators, their independent effects were attenuated in the presence of each other. This can be explained by indirect and direct interference. First, T. pyriformis likely reduced the T. vorax effect on bacterial prey by indirectly competing for the same bacterial resource. Second, macrostome morphs of T. vorax can directly consume T. pyriformis (Banerji and Morin, 2009), which could have reduced T. pyriformis densities leading to weakened selection for bacterial defence. Defence evolution against T. pyriformis was also weakened in the presence of C. paramecium. As C. paramecium did not affect T. pyriformis densities in co-cultures, this result is more likely explained by the evolution of specialist defenders that were vulnerable to C. paramecium (PT and TT colony types). Unexpectedly, T. pyriformis enhanced C. paramecium growth. Even though the mechanism for this is unknown, one explanation could be that C. paramecium was able to cross feed on T. pyriformis waste metabolites—a common process often observed between different bacteria (Lawrence et al., 2012). We also found that concurrent selection by A. polyphaga and T. pyriformis, or A. polyphaga and T. vorax, led to increased bacterial defence against A. polyphaga. Together these results suggest that protist predators can exert conflicting or diffuse selection (Janzen, 1980; Strauss and Irwin, 2004) leading to specialist or generalist defensive strategies in multi-predator communities.
In addition to increased bacterial phenotypic diversity, prey defence evolution changed other ecological aspects of predator–prey communities. First, evolved prey populations were more variable in time (higher coefficient of variation) compared with non-evolved or control populations, and there are at least two possible causes for this pattern. Prey defence evolution can destabilise predator–prey dynamics, for example, by changing the amplitude and phase of predator–prey cycles (Abrams, 2000; Yoshida et al., 2003; Becks et al., 2010). On the other hand, competitive interactions between different prey phenotypes could increase population instability via frequency-dependent selection (Yoshida et al., 2003; Meyer and Kassen, 2007). Unfortunately, we cannot separate these hypotheses with our data, as we quantified evolutionary changes only at the end of the experiment. However, we did find that evolved prey populations became less productive compared with non-evolved or control populations. At the colony-type level, reduced growth was linked with specialist and generalist defender prey phenotypes, suggesting that evolving prey defence was traded-off with prey competitive ability, a commonly found trade-off in microbial predator–prey systems (Yoshida et al., 2003; Meyer and Kassen, 2007; Friman and Laakso, 2011; Friman and Buckling, 2013; Friman et al., 2015). Such a trade-off could also have affected prey population instability (Abrams, 2000; Yoshida et al., 2003; Ellner and Becks, 2011). Our results also showed that evolved prey populations were equally variable in single- and two-predator communities even though some two-predator communities had higher phenotypic prey richness (T. pyriformis–C. paramecium). This suggests that relatively more abundant SM and WS colony types were associated with the largest effect on destabilisation of evolved prey populations. Together these results suggest that multiple predators can have emergent evolutionary effects on prey that cannot be predicted on the basis of pairwise interactions.
To conclude, our results show that predator community composition is important in defining the relative importance of ecological and evolutionary dynamics of microbial communities. In general, increasing protist community richness increased prey diversity by allowing the evolution of specialist defence strategies. However, ecological dynamics dominated in the presence of top predator owing to reduction in the densities of T. pyriformis—a key driver of bacterial adaptation. Intraguild predation could thus indirectly constrain evolution of predator–prey interactions.
References
Abrams PA . (2000). The evolution of predator-prey interactions: theory and evidence. Annu Rev Ecol Syst 31: 79–105.
Banerji A, Morin PJ . (2009). Phenotypic plasticity, intraguild predation and anti-cannibal defences in an enigmatic polymorphic ciliate. Funct Ecol 23: 427–434.
Becks L, Ellner SP, Jones LE, Hairston NG Jr . (2010). Reduction of adaptive genetic diversity radically alters eco-evolutionary community dynamics. Ecol Lett 13: 989–997.
Berenbaum MR, Zangerl AR . (2006). Parsnip webworms and host plants at home and abroad: trophic complexity in a geographic mosaic. Ecology 87: 3070–3081.
Billick I, Case TJ . (1994). Higher-order interactions in ecological communities - what are they and how can they be detected. Ecology 75: 1529–1543.
Brunk CF, Lee LC, Tran AB, Li JL . (2003). Complete sequence of the mitochondrial genome of Tetrahymena thermophila and comparative methods for identifying highly divergent genes. Nucleic Acids Res 31: 1673–1682.
Craig TP, Itami JK, Horner JD . (2007). Geographic variation in the evolution and coevolution of a tritrophic interaction. Evolution 61: 1137–1152.
Davies NB, Brooke MDL . (1989). An experimental study of co-evolution between the cuckoo, Cuculus canorus, and its hosts. I. Host egg discrimination. J Anim Ecol 58: 207–224.
Edeline E, Ben Ari T, Vollestad LA, Winfield IJ, Fletcher JM, Ben James J et al. (2008). Antagonistic selection from predators and pathogens alters food-web structure. PNAS 105: 19792–19796.
Elliott AM . (1959). Biology of Tetrahymena. Annu Rev Microbiol 13: 79–96.
Ellner SP, Becks L . (2011). Rapid prey evolution and the dynamics of two-predator food webs. Theor Ecol 4: 133–152.
Flynn KM, Cooper TF, Moore FB, Cooper VS . (2013). The environment affects epistatic interactions to alter the topology of an empirical fitness landscape. PLoS Genet 9: e1003426.
Fox LR . (1988). Diffuse coevolution within complex communities. Ecology 69: 906–907.
Friman V-P, Diggle SP, Buckling A . (2013). Protist predation can favour cooperation within bacterial species. Biol Lett 9: 20130548.
Friman V-P, Buckling A . (2014). Phages can constrain protist predation-driven attenuation of Pseudomonas aeruginosa virulence in multienemy communities. ISME J 8: 1820–1830.
Friman V-P, Guzman LM, Reuman DC, Bell T . (2015). Bacterial adaptation to sublethal antibiotic gradients can change the ecological properties of multitrophic microbial communities. Proc R Soc Lond B Biol Sci 282: 20142920.
Friman VP, Hiltunen T, Laakso J, Kaitala V . (2008). Availability of prey resources drives evolution of predator-prey interaction. Proc Biol Sci R Soc 275: 1625–1633.
Friman VP, Laakso J . (2011). Pulsed-resource dynamics constrain the evolution of predator-prey interactions. Am Nat 177: 334–345.
Friman VP, Buckling A . (2013). Effects of predation on real-time host-parasite coevolutionary dynamics. Ecol Lett 16: 39–46.
Friman VP, Jousset A, Buckling A . (2014). Rapid prey evolution can alter the structure of predator–prey communities. J Evol Biol 27: 374–380.
Futuyma DJ, Moreno G . (1988). The evolution of ecological specialization. Annu Rev Ecol Syst 19: 207–233.
Gomez JM, Perfectti F, Bosch J, Camacho JPM . (2009). A geographic selection mosaic in a generalized plant-pollinator-herbivore system. Ecol Monogr 79: 245–263.
Gronlien HK, Berg T, Lovlie AM . (2002). In the polymorphic ciliate Tetrahymena vorax, the non-selective phagocytosis seen in microstomes changes to a highly selective process in macrostomes. J Exp Biol 205: 2089–2097.
Hiltunen T, Jones LE, Ellner SP, Hairston NG . (2013). Temporal dynamics of a simple community with intraguild predation: an experimental test. Ecology 94: 773–779.
Hiltunen T, Becks L . (2014). Consumer co-evolution as an important component of the eco-evolutionary feedback. Nat Commun 5: 5226.
Iwao K, Rausher MD . (1997). Evolution of plant resistance to multiple herbivores: quantifying diffuse coevolution. Am Nat 149: 316–335.
Janzen DH . (1980). When is it coevolution. Evolution 34: 611–612.
Kim W, Racimo F, Schluter J, Levy SB, Foster KR . (2014). Importance of positioning for microbial evolution. PNAS 111: E1639–E1647.
Lawrence D, Fiegna F, Behrends V, Bundy JG, Phillimore AB, Bell T et al. (2012). Species interactions alter evolutionary responses to a novel environment. PLoS Biol 10: e1001330.
Matz C, Bergfeld T, Rice SA, Kjelleberg S . (2004). Microcolonies, quorum sensing and cytotoxicity determine the survival of Pseudomonas aeruginosa biofilms exposed to protozoan grazing. Environ Microbiol 6: 218–226.
Meyer JR, Kassen R . (2007). The effects of competition and predation on diversification in a model adaptive radiation. Nature 446: 432–435.
Mikonranta L, Friman VP, Laakso J . (2012). Life history trade-offs and relaxed selection can decrease bacterial virulence in environmental reservoirs. PLoS One 7: e43801.
Nuismer SL, Thompson JN . (2006). Coevolutionary alternation in antagonistic interactions. Evolution 60: 2207–2217.
O'Toole GA, Kolter R . (1998). Initiation of biofilm formation in Pseudomonas fluorescens WCS365 proceeds via multiple, convergent signalling pathways: a genetic analysis. Mol Microbiol 28: 449–461.
Parchman TL, Benkman CW . (2008). The geographic selection mosaic for ponderosa pine and crossbills: a tale of two squirrels. Evolution 62: 348–360.
Patterson DJ . (1996) Free-lIving Freshwater Protozoa: A Colour Guide. John Wiley & Sons: New York, NY, USA.
Popat R, Crusz SA, Messina M, Williams P, West SA, Diggle SP . (2012). Quorum-sensing and cheating in bacterial biofilms. Proc Biol Sci R Soc 279: 4765–4771.
Rainey PB, Bailey MJ . (1996). Physical and genetic map of the Pseudomonas fluorescens SBW25 chromosome. Mol Microbiol 19: 521–533.
Rainey PB, Travisano M . (1998). Adaptive radiation in a heterogeneous environment. Nature 394: 69–72.
Sih A, Englund G, Wooster D . (1998). Emergent impacts of multiple predators on prey. Trends Ecol Evol 13: 350–355.
Spiers AJ . (2014). A mechanistic explanation linking adaptive mutation, niche change, and fitness advantage for the wrinkly spreader. Int J Evol Biol 2014: 10.
Stinchcombe JR, Rausher MD . (2001). Diffuse selection on resistance to deer herbivory in the ivyleaf morning glory, Ipomoea hederacea. Am Nat 158: 376–388.
Strauss SY, Irwin RE . (2004). Ecological and evolutionary consequences of multispecies plant-animal interactions. Annu Rev Ecol Evol Syst 35: 435–466.
Strauss SY, Sahli H, Conner JK . (2005). Toward a more trait-centered approach to diffuse (co)evolution. New Phytol 165: 81–89.
Thompson JN, Cunningham BM . (2002). Geographic structure and dynamics of coevolutionary selection. Nature 417: 735–738.
Thompson JN . (2005) The Geographic Mosaic of Coevolution. University of Chicago Press: Chicago, IL, USA.
Vandermeer JH . (1969). The competitive structure of communities: an experimental approach with protozoa. Ecology 50: 362–371.
Yoshida T, Jones LE, Ellner SP, Fussmann GF, Hairston NG . (2003). Rapid evolution drives ecological dynamics in a predator-prey system. Nature 424: 303–306.
Acknowledgements
This study was funded by Imperial College Junior Research Fellow scheme (to V-PF), Grant-in-Aid funding from The Natural History Museum (to DJB), NHM-UCL PhD studentship (to AD) and the Royal Society and the Natural Environment Research Council (to TB).
Author contributions
V-PF, AD, DB and TB designed the study; AD and V-PF performed the research; VP-F wrote the first draft of the manuscript and all authors contributed substantially to revisions.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Rights and permissions
About this article
Cite this article
Friman, VP., Dupont, A., Bass, D. et al. Relative importance of evolutionary dynamics depends on the composition of microbial predator–prey community. ISME J 10, 1352–1362 (2016). https://doi.org/10.1038/ismej.2015.217
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ismej.2015.217
This article is cited by
-
The genetic architecture underlying prey-dependent performance in a microbial predator
Nature Communications (2022)
-
Protist communities are more sensitive to nitrogen fertilization than other microorganisms in diverse agricultural soils
Microbiome (2019)
-
Phage combination therapies for bacterial wilt disease in tomato
Nature Biotechnology (2019)