Abstract
Ammonia-oxidizing Archaea (AOA) are ubiquitous throughout the oceanic water column; however, our knowledge on their physiological and ecological diversity in different oceanic regions is rather limited. Here, we report the cultivation and characterization of two novel Nitrosopumilus strains, originating from coastal surface waters of the Northern Adriatic Sea. The combined physiological and genomic information revealed that each strain exhibits different metabolic and functional traits, potentially reflecting contrasting life modes. Strain NF5 contains many chemotaxis-related genes and is able to express archaella, suggesting that it can sense and actively seek favorable microenvironments such as nutrient-rich particles. In contrast, strain D3C is non-motile and shows higher versatility in substrate utilization, being able to use urea as an alternative substrate in addition to ammonia. Furthermore, it encodes a divergent, second copy of the AmoB subunit of the key enzyme ammonia monooxygenase, which might have an additional catalytic function and suggests further metabolic versatility. However, the role of this gene requires further investigation. Our results provide evidence for functional diversity and metabolic versatility among phylogenetically closely related thaumarchaeal strains, and point toward adaptations to free-living versus particle-associated life styles and possible niche differentiation among AOA in marine ecosystems.
Similar content being viewed by others
Introduction
Ammonia oxidation represents the first and rate-limiting step in nitrification and the only known oxidative biological process in nature that converts reduced inorganic nitrogen species into their oxidized form (Prosser, 1989). For more than a century, ammonia oxidation was thought to be mainly performed by certain lineages of Beta- and Gammaproteobacteria (Purkhold et al., 2000); however, the discovery of ammonia-oxidizing archaea (AOA) (Könneke et al., 2005; Treusch et al., 2005), which now comprise the class Nitrososphaeria (Stieglmeier et al., 2014), within the phylum Thaumarchaeota (Brochier-Armanet et al., 2008; Spang et al., 2010) has dramatically changed our perception of microbial nitrification and nitrogen cycling.
During the past decade, several studies based on 16 S rRNA and ammonia monooxygenase subunit A (amoA) gene sequences of AOA have shown that this archaeal group appears to be extremely diverse and abundant in various marine and terrestrial environments (Francis et al., 2005; Leininger et al., 2006; Zhang et al., 2008), constituting up to 20% of the ocean’s picoplankton (Karner et al., 2001; Teira et al., 2006). Cultivated thaumarchaeal ammonia oxidizers have been shown to be adapted to oligotrophic conditions, specifically to low ammonium concentrations (nm–μm range) (Martens-Habbena et al., 2009; Prosser and Nicol, 2012), as generally found in surface waters of the global oceans.
The majority of AOA present in marine ecosystems belongs to a specific subgroup referred to as group 1.1a or Marine Group I, recently defined as the candidate order Nitrosopumilales (Stieglmeier et al., 2014) which includes the first cultured representative, 'Candidatus Nitrosopumilus maritimus' SCM1, isolated from a tropical seawater aquarium (Könneke et al., 2005). Organisms closely related to 'Ca. N. maritimus' SCM1 have now been successfully enriched from arctic marine and estuarine sediments (Mosier et al., 2012b; Park et al., 2014) and were isolated from coastal surface waters (Qin et al., 2014). So far, all cultured representatives of group 1.1a couple ammonia oxidation to autotrophic carbon fixation, but vary in other metabolic traits such as the ability to utilize urea or the dependence on small amounts of organic compounds to sustain growth (Park et al., 2014; Qin et al., 2014). Such distinctive differences can support niche partitioning of AOA populations, potentially influencing nitrification rates in various marine habitats.
Despite the global distribution and ecological importance of members of group 1.1a, studies on their metabolic diversity and detailed genomic analyses are relatively sparse. In this study, two novel thaumarchaeal ammonia oxidizers were obtained from Northern Adriatic coastal waters. The two enriched organisms were designated 'Candidatus Nitrosopumilus sp.' strains NF5 and D3C, and their complete genomes were assembled. Physiological and genomic characterization revealed that both strains differ in their metabolic capabilities as well as in their functional traits, potentially reflecting contrasting life modes.
Materials and methods
Sampling and establishment of enrichment cultures
Seawater was collected in the Northern Adriatic Sea from approx. 0.5 m depth off Piran, Slovenia (45.518 N, 13.568 E) in two consecutive months (November and December 2011). At the time of sampling, seawater temperatures ranged between 13.1 and 15.5 °C and ammonium concentrations varied between 0.12 and 0.48 μm.
Enrichment cultures were initiated by inoculating 10 ml of surface seawater to 90 ml of modified Synthetic Crenarchaeota Medium (SCM) supplemented with KH2PO4, NaHCO3, NH4Cl, Fe-NaEDTA and non-chelated trace element solution, as described by Könneke et al. (2005). The medium also contained 5–10% of filter-sterilized (0.2 μm mesh) and autoclaved Adriatic seawater to reduce isolation stress. The pH of the medium was adjusted to 7.6 by adding 10 ml l−1 of HEPES buffer (1 m HEPES, 0.6 m NaOH). Cultures were incubated aerobically at 17 and 25 °C in the dark without shaking.
Ammonia-oxidizing activity in the enrichment cultures was monitored by measuring NH4+ consumption and NO2− production, applying fluorescence and absorbance spectroscopy methods using the OPA reagent (Holmes et al., 1999) and Griess reagent (Griess, 1879), respectively. Additionally, archaeal growth was monitored by flow cytometry (as described in Supplementary material).
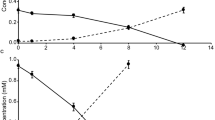
Subsequent sub-culturing was performed by inoculating fresh medium (5–10% vol/vol inoculum) with cells in mid-exponential phase, which corresponds to a residual NH4+ concentration of approx. 300 μm (Figure 3). As faster growth was observed at 25 °C than at 17 °C, sub-cultures were grown at 25 °C. In the latest stages of the enrichment process, addition of Adriatic Sea water was omitted from the medium. Different antibiotics were alternately added to the medium over several cell passages to further purify the enrichments. The following antibiotics were used: kanamycin (100 μg ml−1), streptomycin (100 μg ml−1), carbenicillin (100 μg ml−1), spectinomycin (100 μg ml−1), ofloxacin (25 μg ml−1) and chloramphenicol (15 μg ml−1).
This procedure resulted in the enrichment of two AOA strains thereafter referred to as NF5 and D3C (strain NF5 derived from samples taken in November 2011 whereas strain D3C derived from samples taken in December 2011). Temperature and pH optima were determined for both strains (see Supplementary material).
Sequencing, genome assembly and analyses
Genomic DNA was extracted from 1 l batch cultures in early stationary phase, and sequenced using a 454/FLX-Titanium sequencer (Roche, MWG Eurofins, Ebersberg, Germany). Both cultures were >90% enriched at the time of sampling for genomic sequencing as indicated by catalyzed reporter deposition fluorescence In Situ hybridization analyses. The total number of obtained reads was 114 359 and 80 867 and the average read length was 622 and 623 bp for the D3C and NF5 enrichment culture, respectively. The obtained reads were length- and quality-filtered (min. length: 150 bp, min. average quality: 25) using Prinseq (Schmieder and Edwards, 2011), and draft genomes were de novo assembled with Newbler 2.6 (Margulies et al., 2005). The draft genome assembly of the NF5 enrichment culture resulted in 23 contigs (N50=239 081bp) containing 63 618 reads. Ten contigs (500–1500 bp) of the NF5 genome could not be linked to the rest of the genome, suggesting that these are from bacterial contaminants, which was supported by the fact that they only gave bacterial hits in nucleotide homology (Blastn) searches. For the D3C enrichment, 89 704 reads were assembled into 3 contigs (N50=1 617 100bp). The remaining gaps in both genomes were closed via PCR, using specific primers targeting the start and end positions of each contig. The complete genomes were deposited under the accession numbers CP010868 and CP011070.
Annotations of the draft genomes were performed using the MicroScope genome platform (Vallenet et al., 2013) and manually curated as previously described (Spang et al., 2012). Synteny statistics were generated by the respective tools of MaGe (Vallenet et al., 2013) (blastP alignment threshold=min. 35% sequence identity on min. 80% of the length of the smallest protein, gap parameter=5 genes). The circular genome plot (Supplementary Figure S4) was generated using the Circos software (version 0.54) (Krzywinski et al., 2009). Average nucleotide identity (ANI) values were calculated based on whole-genome sequences using nucleotide homology (Blastn) searches (Altschul et al., 1990).
Detailed descriptions of all methodological procedures used in this study can be found in the Supplementary material.
Results and Discussion
Enrichment of two novel marine AOA and their morphology
The two novel AOA strains described in this study were enriched from Northern Adriatic coastal surface waters (approx. 0.5 m depth) sampled off Piran, Slovenia. Ammonia consumption in the initial enrichment cultures was first observed after 8 weeks of incubation at 25 °C. Consecutive cell passages in medium containing 1 mm NH4Cl over a period of 2 years, and the addition of various antibiotics resulted in two stable, highly enriched cultures (>99%) (Supplementary Figure S1A and B). A total of 30 archaeal ammonia monooxygenase subunit A (amoA) gene fragments were cloned from each of the two cultures, and the corresponding sequences were nearly identical (>99.5% nucleotide identity) within each culture but differed slightly (94% nucleotide identity) between the two enrichments, suggesting that two different, but closely related AOA strains had been enriched, thereafter referred to as D3C and NF5.
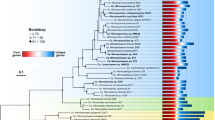
Strains D3C and NF5 are affiliated with the genus Nitrosopumilus, within the provisional order Nitrosopumilales (also known as Thaumarchaeota group I.1a). Based on the concatenated 16 S–23 S rRNA gene phylogeny of all cultivated Thaumarchaeota with sequenced genomes, strain D3C forms a sub-cluster with 'Ca. N. maritimus' SCM1, isolated from a tropical marine aquarium (Könneke et al., 2005), and 'Ca. N. koreensis' AR1, enriched from Arctic marine sediment (Park et al., 2014), whereas strain NF5 branches separately from all other cultivated Nitrosopumilus species (Figure 1). This topology is consistent with that of an amoA gene phylogeny including 'Ca. Nitrosopumilus sp.' strains PS0 and HCA1 (Qin et al., 2014), for which full rRNA gene sequences are not available (Supplementary Figure S2). The amoA sequence of strain NF5 is almost identical (>99% id) to an amoA sequence (KR072695) that was retrieved from a marine snow particle sampled at the same location off the coast of Piran (Vojvoda et al., unpublished).
Phylogenetic tree of concatenated 16S–23S rRNA genes from cultivated Thaumarchaeota with sequenced genomes (16 organisms) showing the affiliation of strains D3C and NF5. Full-length gene sequences were aligned with MAFFT (L-INS-I method) based on Archaea-specific structurally accurate seed alignments (1474 and 3026 aligned positions for 16S and 23S rRNA genes, respectively), and unreliable positions were filtered from the resulting alignments with TCS before concatenation (4443 aligned positions in the concatenated alignment). The tree was calculated by maximum likelihood with IQ-Tree based on the GTR+I+Γ4 model, with ultrafast bootstrap (UFBoot) and SH-aLRT support values inferred from 1000 replicates each (see Supplementary materials and methods for details). Support values ⩾85% are represented on the respective branches by semi-circles color-coded as indicated on the figure.
In electron microscopy, cells of strain NF5 appeared as small, regular rods with a diameter of 0.20–0.25 μm (mean±s.d.: 0.23±0.015 μm; n=50) and a length of 0.59–1.74 μm (mean±s.d.: 0.93±0.26 μm; n=50) and cells of strain D3C ranged in their diameter from 0.20 to 0.25 μm (mean±s.d.: 0.22±0.008; n=50) and in their length from 0.49 to 2.00 μm (mean±s.d.: 0.85±0.28 μm; n=50) (Figure 2). In agreement with previous observations of all other described Nitrosopumilus strains, we did not observe flagella on cells of strain D3C. In contrast, strain NF5 showed, at least occasionally, flagella-like appendages (21 out of 300 cells) with a diameter of 11–14 nm and hence, within the typical size range reported for archaeal flagella (archaella) (Jarrell et al., 2013). 'Ca. Nitrosopumilus sp.' NF5 is the first representative of the Nitrosopumilus genus shown to possess archaella. Thus far, within marine representatives of the Nitrosopumilales, archaella have only been observed for 'Ca. Nitrosoarchaeum limnia' strains SFB1 and BG20 (Blainey et al., 2011, Mosier et al., 2012b).
In Northern Adriatic coastal waters, marine snow is common and represents a nutrient-rich environment in the euphotic layer (Kaltenböck and Herndl, 1992), characterized by a patchy distribution of nutrients. Hence, the potential ability of flagellated microbes to sense and actively move toward favorable microhabitats might be beneficial for microorganisms in Northern Adriatic coastal waters, suggesting that strain NF5 could possibly be associated with such particles. Under culture conditions, nutrients are likely to be homogenously distributed, suggesting that the expression of archaella might be less important, potentially also explaining the low number of flagellated cells observed by TEM.
Physiological characterization
Both enrichment cultures converted ammonia into nitrite stoichiometrically, which was accompanied by growth of AOA as measured by flow cytometry (Figure 3). Both cultures showed very similar growth patterns. Typically after 7–12 days of incubation (starting from a 10% inoculum), 1 mm ammonium was consumed and cultures reached cell abundances of 5 × 107–108 cells ml−1. Ammonium uptake was also observed for very low ammonium concentrations ranging between 50 and 100 nm (limit of detection, data not shown), consistent with previous reports of low kinetic constants (Km 132 nm) of ammonium uptake of 'Ca. Nitrosopumilus maritimus' SCM1 (Martens-Habbena et al., 2009).
Generally, cells grew at temperatures between 14 and 34 °C (Supplementary Figure S3A), which is similar to the temperature range reported for 'Ca. N. maritimus' SCM1, and strains HCA1 and PS0 (Qin et al., 2014). The temperature optimum of strain NF5 was between 30 and 32 °C, whereas cells of strain D3C grew faster at slightly higher temperatures of 32–34 °C. When cultures were exposed to 37 °C, strain D3C maintained both ammonia oxidation and growth whereas strain NF5 maintained ammonia oxidation in the first days of exposure but rapidly decreased in cell abundance over time. In contrast to terrestrial thaumarchaeal enrichment and pure cultures (Tourna et al., 2011; Lebedeva et al., 2013), marine representatives of group I.1a have so far not been shown to be able to sustain growth at a temperature above 35 °C (Qin et al., 2014; Santoro et al., 2015). Unlike for strains HCA1 and PS0 (Qin et al., 2014), growth at 10 °C could not be observed for strains D3C and NF5, indicating a potential adaptation to Adriatic surface waters, where temperatures are typically well above 10 °C for most of the year.
Both strains grew well between pH 6.8 and 8.0, although their highest specific growth rates were observed at approx. pH 7.1 (Supplementary Figure S3B). Despite the average pH range in surface waters of 8.0–8.2, all cultured marine AOA described so far have typical growth optima at around pH 7 (Könneke et al., 2005; Qin et al., 2014). However, it should also be considered that the process of ammonia oxidation (uptake of NH3/NH4+ and generation of NO2−) possibly reduces the pH outside the cell, potentially creating microenvironments with lower pH that could facilitate growth in natural environments. Additionally, it has recently been hypothesized that some AOA populations have the capacity to adapt to lower pH and that ocean acidification might promote growth of AOA (Qin et al., 2014).
Neither growth nor nitrite production was observed when cells of strain NF5 were incubated in ammonium-free medium supplemented with 0.5 mm urea, whereas growth of strain D3C was comparable to that with an equivalent amount of ammonium. Growth of strain D3C on 0.5 mm urea was accompanied by the generation of approx. 1 mm nitrite, indicating that this strain can efficiently utilize urea as an alternative substrate for ammonia oxidation (Figure 4).
Archaeal urease genes appear to be predominantly present in marine polar waters (Alonso-Saez et al., 2012; Connelly et al., 2014) and were also identified in 'Ca. Nitrosopumilus sediminis' AR2, enriched from Arctic sediments (Park et al., 2012), and in the psychrophilic sponge symbiont 'Ca. Cenarchaeum symbiosum' (Hallam et al., 2006). Together with the recently described 'Ca. Nitrosopumilus sp.' PS0 that was enriched from Puget Sound coastal surface waters and has been shown to grow on urea (Qin et al., 2014), our findings indicate that urea metabolism might also be important in coastal waters where urea concentrations are typically high (Remsen, 1971; Painter et al., 2008).
Mixotrophy and potential interaction with heterotrophic bacteria
Amendment of the growth medium with alpha-ketoglutaric acid, pyruvic acid and oxaloacetic acid did not have any effect on the growth of either strain, suggesting that strains D3C and NF5 do not depend on organic molecules to sustain growth. However, the dependence on small organic molecules cannot be completely excluded due to the presence of remaining bacterial contaminants that could potentially produce required metabolites.
The co-cultured bacteria belong to the Alphaproteobacteria and share 99% 16 S rRNA gene sequence with Oceanicaulis alexandrii C116-3 (AJ309863). Oceanicaulis spp. belong to the family Hyphomonadaceae, which consists of chemoorganotrophic prosthecate marine bacteria (Strompl et al., 2003) known to persist under extremely oligotrophic conditions (Abraham and Rohde, 2014), as typically found in most parts of the ocean.
Recently, co-occurrence patterns of Thaumarchaeota with chemoheterotrophic Proteobacteria have been described in the deep chlorophyll maximum of the Southern California Bight (Beman et al., 2011). It was suggested that in concert, these groups might be involved in the remineralization of organic material and hence, nutrient cycling.
Physiological studies on Nitrososphaera viennensis EN76T revealed that pure cultures require pyruvate to reach growth rates comparable to those of enrichment cultures containing co-cultured Hyphomicrobium vulgare and Mesorhizobium sp. of the alphaproteobacterial order Rhizobiales (Tourna et al., 2011). A similar observation was made for the marine strains PS0 and HCA1, which require organic carbon for growth (Qin et al., 2014), indicating possible symbiotic interactions with heterotrophic bacteria in natural environments. Beneficial trophic interactions between autotrophic AOA and heterotrophic bacteria could potentially also explain the apparent ecological success of Thaumarchaeota in oligotrophic environments such as the open ocean.
Genomic characterization
Genomic plasticity
The complete genomes of the two newly characterized AOA strains D3C and NF5 were assembled. When compared with each other, both genomes share an ANI of 84%. Compared with the genome of 'Ca. Nitrosopumilus maritimus' SCM1, the genomes of strains D3C and NF5 show 88% and 81% nucleotide identity, respectively. These ANI values are far below the previously defined threshold of ⩾95% for species definition (Goris et al., 2007; Mende et al., 2013; Kim et al., 2014). Therefore, we propose two new species names for strains NF5 and D3C: 'Candidatus Nitrosopumilus adriaticus' NF5 and 'Candidatus Nitrosopumilus piranensis' D3C.
Both genomes contain several unique genomic regions that are absent in 'Ca. N. maritimus' SCM1 (Walker et al., 2010). The genome of 'Ca. N. adriaticus' NF5 contains 375 and 395 coding sequences that are not present in 'Ca. N. piranensis' D3C and 'Ca. N. maritimus' SCM1, whereas 'Ca. N. piranensis' D3C contains 319 and 329 coding sequences that are absent in the genomes of 'Ca. N. adriaticus' NF5 and 'Ca. N. maritimus' SCM1, respectively (Supplementary Figure S4). When compared with each other and with 'Ca. N. maritimus' SCM1, ⩽75% of the genes are conserved in syntenic gene clusters (NF5 vs D3C: 73%, D3C vs NF5: 75%, NF5 vs SCM1: 72%, D3C vs SCM1: 74%), despite their highly similar 16 S rRNA gene sequences.
Energy metabolism
Both genomes encode a complete set of genes that have been proposed to be involved in ammonia oxidation and electron transfer (Walker et al., 2010; Spang et al., 2012; Stahl and de la Torre, 2012), including ammonia monooxygenase genes amoAXCB and genes for several blue-copper domain-containing proteins and multicopper oxidases. Additionally, two genes for putative ammonium transporters and genes encoding putative copper-containing nitrite reductases (NirK) have been identified in both genomes.
The genomes of 'Ca. Nitrosopumilus adriaticus' NF5 and 'Ca. Nitrosopumilus maritimus' SCM1 contain two highly similar nirK gene copies (95% and 91% identity, respectively), in contrast to all other sequenced genomes of members of the Nitrosopumilales order, which contain one nirK gene copy each. The exact function of NirK in nitrifiers is not yet clear, although nitric oxide (NO), one of the reduction products of NO2−, might possibly have a central role in the ammonia oxidation process of AOA (Klotz and Stein, 2008; Shen et al., 2013 Martens-Habbena et al., 2014). So far, there are no apparent physiological differences related to ammonia oxidation of 'Ca. N. adriaticus' NF5 and 'Ca. N. maritimus' SCM1 as compared with other strains, suggesting a potential additional role of a second NirK copy in those organisms. Another possible function of NirK in nitrifiers could be nitrite detoxification, as proposed for ammonia-oxidizing bacteria (Cho et al., 2006; Cantera and Stein, 2007). AOA are typically present in environments where nitrite concentrations are far below toxic levels. However, 'Ca. N. maritimus' SCM1 was isolated from an aquarium where nitrite concentrations can occasionally be high. 'Ca. N. adriaticus' NF5 was enriched from Northern Adriatic coastal waters, where marine snow formation occurs regularly during the summer months (Müller-Niklas et al., 1994) and nitrite concentrations in these aggregates can be up to 10 times higher than in the surrounding seawater (Kaltenböck and Herndl, 1992). The capability to detoxify nitrite could thus be beneficial in such environments.
'Ca. N. piranensis' D3C harbors a second, highly divergent amoB gene ortholog (amoB2), a unique feature among the genomes of all characterized archaeal and bacterial ammonia oxidizers. Although several ammonia-oxidizing Betaproteobacteria harbor multiple amo gene copies, these are invariably identical or highly similar (Norton et al., 2002) and only duplications of the amoC gene have been described among characterized AOA genomes (Spang et al., 2012). The amoB2 gene of 'Ca. N. piranensis' D3C is 567 bp long and encodes a predicted mature protein and a signal peptide of 157 and 31 amino acids, respectively. The full polypeptide chain of 'Ca. N. piranensis' D3C AmoB1 has a similar length (189 aa) and is also predicted to include a signal peptide (35 aa). The AmoB2 shares only 48% amino-acid identity with the AmoB1 of 'Ca. N. piranensis' D3C and between 38% and 50% with all other thaumarchaeal AmoB proteins over the full polypeptide chain (between 42% and 55% with thaumarchaeal AmoB proteins for the predicted mature protein). These overall low amino-acid identities reflect the unusual protein sequence of AmoB2, which comprises regions of high similarity with other AmoB proteins, interspaced by unique amino-acid motifs (Supplementary Figure S5). The amoB2 gene is distantly located from the typical amoAXCB gene cluster of marine AOA and is present within a region of low similarity to other Nitrosopumilus genomes. This cluster comprises several genes encoding secreted and/or membrane-bound proteins of unknown function, suggesting that amoB2 might be a component of an unknown membrane protein complex.
Phylogenetic analysis of available full-length archaeal amoB genes shows that the amoB2 gene of 'Ca. N. piranensis' D3C originates from within the order Nitrosopumilales, but inference of more detailed relationships was likely limited by the short length of thaumarchaeal amoB genes (between 567 and 576 bp, including predicted signal peptides) (Supplementary Figure S6). It is unclear whether amoB2 originates from a duplication event within the genome of 'Ca. N. piranensis' D3C, followed by rapid diversification, or results from horizontal gene transfer from another member of the Nitrosopumilales. Previous studies have indeed shown evidence for frequent horizontal gene transfers in marine Thaumarchaeota (Lopez-Garcia et al., 2004; Brochier-Armanet et al., 2011; Deschamps et al., 2014). The long branch of the amoB2 gene in the phylogenetic tree (Supplementary Figure S6) reflects the highly divergent codons encoding amino-acid motifs unique to this protein, despite the several conserved residues shared with other AmoB protein sequences (Supplementary Figure S5).
Extensive biochemical characterization of particulate methane monooxygenases, the only structurally characterized copper-dependent membrane-bound monooxygenase complexes (which include the archaeal and bacterial AMOs), has shown that the particulate methane monooxygenases active site is a dicopper center coordinated by three histidines in the PmoB subunit (Balasubramanian et al., 2010; Culpepper and Rosenzweig, 2012). Furthermore, recent crystallization of the AmoB of the thaumarchaeon 'Ca. Nitrosocaldus yellowstonii' HL72 indicated that these three conserved histidines coordinate copper in this protein, possibly constituting the catalytic center of the thaumarchaeal AMO (Lawton et al., 2014). The AmoB2 protein of 'Ca. N. piranensis' D3C also contains the three histidine residues conserved in all other AmoB and nearly all PmoB proteins (Supplementary Figure S5) (Lawton et al., 2014), suggesting that it might also bind copper and has a catalytic role. Nevertheless, given the lack of functional evidence, the location of the active site of AMO complexes remains uncertain, as earlier studies indirectly suggested that AmoA might be the catalytic subunit of the bacterial AMO (Hyman and Wood, 1985).
In addition to genes involved in ammonia oxidation, 'Ca. N. piranensis' D3C harbors a ure gene cluster consisting of three urease-encoding subunits (ureABC), four urease accessory proteins (ureDGFE) and two urea transporters. The presence of urease confirms our observations that 'Ca. N. piranensis' D3C is able to grow on urea as a sole energy source. Genes for urea metabolism have only been found in a few members of group I.1a (Hallam et al., 2006; Park et al., 2012), but appear to be rather conserved among different species (Supplementary Figure S7). Although urea might represent an important alternative substrate for ammonia oxidation, AOA could scavenge ammonia/nitrogen also through cleavage of peptides. Thus far, all described Nitrosopumilales members with sequenced genomes, including 'Ca. N. adriaticus' NF5 and 'Ca. N. piranensis' D3C, contain an ABC transport system for di-/oligopeptides and genes encoding membrane-bound aminopeptidases (alanyl-, methionyl-, and leucyl-) as well as various endopeptidases.
Central carbon metabolism
As for all Thaumarchaeota that have been characterized thus far, both genomes contain all the enzymes required for a modified 3-hydroxypropionate/4-hydroxybutyrate (3HP/4HB) pathway for the fixation of inorganic carbon as well as all gluconeogenic enzymes for the generation of glucose-6-phosphate during autotrophic growth. The 3HP/4HB cycle has recently been proposed to be the most energy-efficient aerobic pathway for CO2 fixation (Könneke et al., 2014). Both 'Ca. Nitrosopumilus adriaticus' NF5 and 'Ca. Nitrosopumilus piranensis' D3C contain one putative bicarbonate transporter (Offre et al., 2014), providing the ability to actively take up inorganic carbon in the form of bicarbonate, which is used to form malonyl-CoA/methylmalonyl-CoA from acetyl-/propionyl-CoA by the acetyl-CoA/propionyl-CoA carboxylase in the first and seventh steps of the 3HP/4HB cycle (Berg et al., 2007).
In addition to the genomic repertoire supporting autotrophy, genes encoding candidate enzymes for all steps of an oxidative tricarboxylic acid (TCA) cycle are present in 'Ca. N. adriaticus' NF5 and 'Ca. N. piranensis' D3C. However, as recently described in Spang et al. (2012), it remains unclear whether Thaumarchaeota are able to exploit classical TCA bypass reactions or use an incomplete TCA cycle exclusively for biosynthetic purposes. There is no conclusive evidence for the operation of a reverse TCA cycle, as Thaumarchaeota, including 'Ca. N. adriaticus' NF5 and 'Ca. N. piranensis' D3C, harbor a putative citrate/citryl-coA lyase but lack citryl-coA synthetase which together catalyze the ATP-dependent cleavage of citrate into acetyl-CoA and oxaloacetate, representing the key step of the rTCA cycle (Aoshima et al., 2004a, 2004b).
'Ca. N. adriaticus' NF5 contains a gene encoding a NAD(P)-dependent malic enzyme, which is present in the genomes of 'Ca. Nitrosopumilus salaria' BD31 (Mosier et al., 2012a), 'Ca. Nitrosoarchaeum limnia' SFB1 (Blainey et al., 2011) and members of the order Nitrososphaerales (Spang et al., 2012), but appears to be absent in all other Nitrosopumilus species. This alternative route for PEP (phosphoenolpyruvate) synthesis using a NAD(P)-dependent malic enzyme and pyruvate phosphate dikinase is energetically less favorable than the direct conversion from oxaloacetate via ATP-dependent PEP carboxykinase (Siebers and Schönheit, 2005). The presence of this alternative pathway, however, likely provides higher flexibility in the utilization of the central metabolite pyruvate.
Motility and chemotaxis
In contrast to all other characterized representatives of the Nitrosopumilus genus, 'Ca. Nitrosopumilus adriaticus' NF5 encodes a number of genes associated with archaeal flagella (archaella) assembly and chemotaxis. The thaumarchaeal flagella apparatus shares genetic features with those of Euryarchaeota and Crenarchaeota. As typically found in Crenarchaeota, Thaumarchaeota lack flaCDE genes (Blainey et al., 2011; Spang et al., 2012; Lebedeva et al., 2013), which are thought to be responsible for switching rotation of the archaella. In contrast, both Thaumarchaeota and Euryarchaeota contain the preflagellin peptidase FlaK, which has been shown to specifically process pre-archaellins, not being able to process pre-pilins like the more versatile prepilin-like peptidase PibD found in Crenarchaeota (Szabo et al., 2007; Jarrell et al., 2013).
The archaella-associated gene cluster in 'Ca. N. adriaticus' NF5 comprises five flagellin genes (flaB1–5), the putative flagellar accessory genes flaFGHIJ and one gene encoding the preflagellin peptidase FlaK. The identified chemotaxis-related genes include the chemotaxis proteins cheCABW, three methyltransferases (cheR), and several response regulators (cheB and cheY), most of which are located adjacent to the fla gene cluster.
'Ca. N. adriaticus' NF5 lacks a gene encoding the deamidase CheD, which is located between cheC and cheR in 'Ca. Nitrosoarchaeum limnia' SFB1 (Blainey et al., 2011) and 'Ca. Nitrososphaera gargensis' Ga9.2 (Spang et al., 2012) (Supplementary Figure S7). Deamidation was proposed to be necessary for receptor activation, and without CheD, receptors are undermethylated resulting in poor kinase activation (Rosario et al., 1995). Although Archaea usually harbor cheD genes, the function of CheD in some Bacteria, such as Escherichia coli, is carried out by CheB (Szurmant and Ordal, 2004). Additionally, the 'Ca. N. limnia' SFB1 genome only contains a fragment of cheD (Blainey et al., 2011), suggesting that some Thaumarchaeota may not require CheD for chemotaxis, and possess alternative methylation pathways, or have different requirements for methylation.
The gene organization of archaella-associated genes is rather conserved among Thaumarchaeota (Supplementary Figure S7), with the only exception being the number of flagellins/archaellins: one in 'Ca. N. gargensis' Ga9.2 (Spang et al., 2012), three in N. viennensis EN76 (Kerou et al., in preparation), four in 'Ca. N. limnia' SFB1 (Blainey et al., 2011) and five in 'Ca. N. adriaticus' NF5 (this study). The number, organization and amino-acid sequences of chemotaxis-related genes are more diverse than archaella-associated genes (Supplementary Figure S7), possibly indicating adaptations in signal sensing in different environments (soil, hot spring, estuarine and marine).
Flagella appear to be favorable attachment sites for bacteriophages (Rakhuba et al., 2010), and it has recently been shown that the Campylobacter jejuni bacteriophage NCTC 12673 is able to specifically recognize flagellin modified with acetamidino-modified pseudaminic acid (Javed et al., 2014). In Archaea, post-translational modification of archaellins is widespread and the amount of N-linked glycosylation sites appears to be variable (up to 16 in flaB2 of Methanothermococcus thermolithotrophicus) (Jarrell et al., 2013).
A higher diversity of flagellins, as present in 'Ca. N. adriaticus' NF5 which contains five different archaellins, may be associated with a potentially higher flexibility of flagellin modifications that could possibly be beneficial for reducing virus infections.
Protection against UV-induced DNA damage
The genomes of 'Ca. Nitrosopumilus adriaticus' NF5 and 'Ca. Nitrosopumilus piranensis' D3C encode putative cyclobutane pyrimidine dimer photolyases (CPD), which may confer greater resistance to ultraviolet radiation (UVR). Photolyases are known to directly repair UVR-induced DNA damage, and, in some cases, are also able to stimulate excision repair nucleases (Ozer et al., 1995). In Northern Adriatic surface waters, biologically effective UV radiation reaches down to 10 m depth (Herndl et al., 1993) and UVR may therefore represent a potential stressor for AOA inhabiting surface waters. This observation is supported by single cell genomic data showing that epipelagic Thaumarchaeota, in contrast to mesopelagic populations, harbor putative photolyases (Luo et al., 2014).
Photolyases belong to the broad cryptochrome/photolyase family, widespread in Bacteria, Archaea and Eukarya, although in Thaumarchaeota they have only been found in 'Ca. Nitrosoarchaeum limnia' strains SFB1 and BG20 (Blainey et al., 2011; Mosier et al., 2012b), 'Ca. Nitrosopelagicus brevis' CN25 (Santoro et al., 2015) and 'Ca. Nitrosotenuis uzonensis' N4 (Lebedeva et al., 2013). Comprehensive phylogenetic analyses show that the putative photolyases of 'Ca. N. piranensis' D3C and 'Ca. N. adriaticus' NF5 cluster within a distinct Thaumarchaeota-specific clade, which, together with Crenarchaeota (Sulfolobales), Euryarchaeota (Thermoplasmatales and Methanosarcinales), Mimivirus and diverse bacterial phyla, represent a previously unrecognized lineage within the cryptochrome/photolyase family (Supplementary Figure S8). The only biochemically and structurally characterized photolyase within this lineage is the CPD class I of Sulfolobus tokodaii (Fujihashi et al., 2007), which shares 43% and 41% amino-acid identity with that of 'Ca. N. adriaticus' NF5 and 'Ca. N. piranensis' D3C, respectively. Additionally, the related photolyase of Sulfolobus acidocaldarius functions in vivo in a light-dependent manner and not as part of a default DNA repair mechanism as, at least, some photolyases (Sakofsky et al., 2011). This further indicates that the related photolyases of 'Ca. N. adriaticus' NF5 and 'Ca. N. piranensis' D3C are likely involved in the adaptation of the organisms to epipelagic waters. Considering their low similarity with characterized photolyases, it is nevertheless possible that these enzymes might have additional light-dependent regulatory functions, as shown for other CPD I involved in both UVR-damage repair and development regulation (Bayram et al., 2008).
Provisional classification
We propose the following candidate status:
Nitrososphaeria class, Nitrosopumilales order, Nitrosopumilaceae fam., 'Nitrosopumilus adriaticus' sp. nov. Etymology: L. adj. m. nitrosus: nitrous (nitrite producer); L. adj. m. pumilus: dwarf; L. adriaticus, obtained from the Adriatic Sea.
Nitrososphaeria class, Nitrosopumilales order, Nitrosopumilaceae fam., 'Nitrosopumilus piranensis' sp. nov. Etymology: L. adj. m. nitrosus: nitrous (nitrite producer); L. adj. m. pumilus: dwarf; L. piranensis, obtained off the Piran coast.
Locality: Northern Adriatic coastal surface waters (Piran, Slovenia).
Diagnosis: Chemolithoautotrophic nitrifiers of the domain Archaea, closely related to 'Ca. Nitrosopumilus maritimus' SCM1. 'Ca. Nitrosopumilus piranensis' D3C can alternatively utilize urea as a substrate. Cells appear as straight rods with a diameter of 0.20–0.25 μm and a length of 0.49–2.00 μm ('Ca. Nitrosopumilus piranensis' D3C) and 0.59–1.74 μm ('Ca. Nitrosopumilus adriaticus' NF5). 'Ca. Nitrosopumilus adriaticus' NF5 cells occasionally appear to be flagellated. The species names allude to the habitat of the organisms.
Conclusions
Our results provide new insight into the functional diversity and metabolic versatility of marine Thaumarchaeota. Although 'Ca. Nitrosopumilus adriaticus' NF5 and 'Ca. Nitrosopumilus piranensis' D3C are phylogenetically closely related and were obtained from the same location, both strains appear to exhibit different metabolic traits and functional adaptations (Table 1), potentially reflecting contrasting life strategies.
The presence of motility and chemotaxis genes in the genome of 'Ca. N. adriaticus' NF5 indicates that it might actively sense and move toward more favorable microenvironments in marine surface waters such as marine snow. Northern Adriatic coastal waters are known for large marine snow formation (Müller-Niklas et al., 1994). These aggregates represent a heterogeneous, nutrient-rich environment (Kaltenböck and Herndl, 1992) and potentially, a favorable niche for flagellated microbes. In addition, the amoA sequence of 'Ca. N. adriaticus' NF5 is identical to an amoA sequence retrieved from marine snow sampled at the same location (Vojvoda et al., unpublished), supporting the hypothesis that this organism might be associated with marine snow-type particles.
In contrast, 'Ca. N. piranensis' D3C is non-motile and shows more versatility than 'Ca. N. adriaticus' NF5 in substrate utilization, as shown by its ability to grow on urea as an alternative substrate. Although easily accessible ammonia seems to be preferentially taken up, the potential to utilize urea may be beneficial when ammonium is in short supply. We suggest that urea uptake by marine AOA might not solely be predominant in polar waters as recently proposed (Alonso-Saez et al., 2012; Connelly et al., 2014), but might also represent an important alternative energy source for AOA in temperate coastal waters. Moreover, the presence of an additional distinct amoB subunit gene (amoB2) and respective genomic context suggests a potential for the use of additional alternative substrates and/or additional regulatory mechanisms of ammonia oxidation.
It has recently been shown that extremely closely related (>99% ANI) bacterioplankton populations have different behavioral strategies to interact with microscale nutrient patches in the ocean (Yawata et al., 2014). Whereas one population colonizes particles, the other population rapidly migrates to new nutrient sources, indicating that spatiotemporal structuring contributes to fine-scale ecological differentiation. Our results indicate that 'Ca. N. adriaticus' NF5 and 'Ca. N. piranensis' D3C occupy different niches in marine coastal waters, corresponding to a more particle-associated life mode versus a free-living life mode, respectively. Niche complementation leads to a reduction of competition and improves resource utilization (Cordero and Polz, 2014), which might be beneficial for survival under changing environmental conditions, potentially explaining the remarkable diversity and ecological success of AOA in the global oceans.
Cultivation of marine Thaumarchaeota often follows the culturing conditions established for 'Ca. Nitrosopumilus maritimus' SCM1 (for example, in this study) (Könneke et al., 2005). The relatively high ammonium concentrations (0.25–1 mm) and/or high temperatures (25–30 °C) may thus be responsible for the selection of AOA strains from the genus Nitrosopumilus in cultivation studies, despite the much greater diversity identified in environmental surveys (Francis et al., 2005; Sintes et al., 2013). This is further illustrated by the fact that the recently cultivated 'Ca. Nitrosopelagicus brevis' CN25, which represents a distinct lineage within the order Nitrosopumilales widespread in oceanic waters, grows optimally at ~22 °C (Santoro et al., 2015).
Given the functional and genomic diversity observed among closely related AOA and the possible bias introduced by current culturing approaches, it is likely that the current range of cultivated AOA greatly underestimates their actual metabolic and functional diversity in the ocean. Thaumarchaeota are ubiquitous and constitute a large fraction of the microbial biomass in the oceans. Therefore, a broader knowledge about the functional adaptations of their populations on different taxonomic levels will be essential to better understand their role in the nutrient dynamics of these ecosystems.
References
Abraham W-R, Rohde M . (2014). The family Hyphomonadacaea. In: The Prokaryotes. Rosenberg E, DeLong EF, Lory S, Stackebrandt E, Thompson F (eds). Springer-Verlag: Berlin, Heidelberg, 4: 283–297.
Alonso-Saez L, Waller AS, Mende DR, Bakker K, Farnelid H, Yager PL et al. (2012). Role for urea in nitrification by polar marine Archaea. Proc Natl Acad Sci USA 109: 17989–17994.
Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ . (1990). Basic local alignment search tool. J Mol Biol 215: 403–410.
Aoshima M, Ishii M, Igarashi Y . (2004a). A novel enzyme, citryl-CoA synthetase, catalysing the first step of the citrate cleavage reaction in Hydrogenobacter thermophilus TK-6. Mol Microbiol 52: 751–761.
Aoshima M, Ishii M, Igarashi Y . (2004b). A novel enzyme, citryl-CoA lyase, catalysing the second step of the citrate cleavage reaction in Hydrogenobacter thermophilus TK-6. Mol Microbiol 52: 763–770.
Balasubramanian R, Smith SM, Rawat S, Yatsunyk LA, Stemmler TL, Rosenzweig AC . (2010). Oxidation of methane by a biological dicopper centre. Nature 465: 115–119.
Bayram O, Biesemann C, Krappmann S, Galland P, Braus GH . (2008). More than a repair enzyme: Aspergillus nidulans photolyase-like CryA is a regulator of sexual development. Mol Biol Cell 19: 3254–3262.
Beman JM, Steele JA, Fuhrman JA . (2011). Co-occurrence patterns for abundant marine archaeal and bacterial lineages in the deep chlorophyll maximum of coastal California. ISME J 5: 1077–1085.
Berg IA, Kockelkorn D, Buckel W, Fuchs G . (2007). A 3-hydroxypropionate/4-hydroxybutyrate autotrophic carbon dioxide assimilation pathway in Archaea. Science 318: 1782–1786.
Blainey PC, Mosier AC, Potanina A, Francis CA, Quake SR . (2011). Genome of a low-salinity ammonia-oxidizing archaeon determined by single-cell and metagenomic analysis. PLoS One 6: e16626.
Brochier-Armanet C, Boussau B, Gribaldo S, Forterre P . (2008). Mesophilic Crenarchaeota: proposal for a third archaeal phylum, the Thaumarchaeota. Nat Rev Microbiol 6: 245–252.
Brochier-Armanet C, Deschamps P, Lopez-Garcia P, Zivanovic Y, Rodriguez-Valera F, Moreira D . (2011). Complete-fosmid and fosmid-end sequences reveal frequent horizontal gene transfers in marine uncultured planktonic archaea. ISME J 5: 1291–1302.
Cantera JJ, Stein LY . (2007). Molecular diversity of nitrite reductase genes (nirK) in nitrifying bacteria. Environ Microbiol 9: 765–776.
Cho CM, Yan T, Liu X, Wu L, Zhou J, Stein LY . (2006). Transcriptome of a Nitrosomonas europaea mutant with a disrupted nitrite reductase gene (nirK). Appl Environ Microbiol 72: 4450–4454.
Connelly TL, Baer SE, Cooper JT, Bronk DA, Wawrik B . (2014). Urea uptake and carbon fixation by marine pelagic bacteria and archaea during the Arctic summer and winter seasons. Appl Environ Microbiol 80: 6013–6022.
Cordero OX, Polz MF . (2014). Explaining microbial genomic diversity in light of evolutionary ecology. Nat Rev Microbiol 12: 263–273.
Culpepper MA, Rosenzweig AC . (2012). Architecture and active site of particulate methane monooxygenase. Crit Rev Biochem Mol Biol 47: 483–492.
Deschamps P, Zivanovic Y, Moreira D, Rodriguez-Valera F, Lopez-Garcia P . (2014). Pangenome evidence for extensive interdomain horizontal transfer affecting lineage core and shell genes in uncultured planktonic thaumarchaeota and euryarchaeota. Genome Biol Evol 6: 1549–1563.
Francis CA, Roberts KJ, Beman JM, Santoro AE, Oakley BB . (2005). Ubiquity and diversity of ammonia-oxidizing archaea in water columns and sediments of the ocean. Proc Natl Acad Sci USA 102: 14683–14688.
Fujihashi M, Numoto N, Kobayashi Y, Mizushima A, Tsujimura M, Nakamura A et al. (2007). Crystal structure of archaeal photolyase from Sulfolobus tokodaii with two FAD molecules: implication of a novel light-harvesting cofactor. J Mol Biol 365: 903–910.
Goris J, Konstantinidis KT, Klappenbach JA, Coenye T, Vandamme P, Tiedje JM . (2007). DNA-DNA hybridization values and their relationship to whole-genome sequence similarities. Int J Syst Evol Microbiol 57: 81–91.
Griess P . (1879). Bemerkungen zu der Abhandlung der H.H. Weselsky und Benedikt “Ueber einige Azoverbindungen”. Chem Ber 12: 426–428.
Hallam SJ, Konstantinidis KT, Putnam N, Schleper C, Watanabe Y, Sugahara J et al. (2006). Genomic analysis of the uncultivated marine crenarchaeote Cenarchaeum symbiosum. Proc Natl Acad Sci USA 103: 18296–18301.
Herndl GJ, Müller-Niklas G, Frick J . (1993). Major role of ultraviolet-B in controlling bacterioplankton growth in the surface layer of the ocean. Nature 361: 717–719.
Holmes RM, Aminot A, Kérouel R, Hooker BA, Peterson BJ . (1999). A simple and precise method for measuring ammonium in marine and freshwater ecosystems. CanJ Fish Aquat Sci 56: 1801–1808.
Hyman MR, Wood PM . (1985). Suicidal inactivation and labelling of ammonia mono-oxygenase by acetylene. Biochem J 227: 719–725.
Jarrell KF, Ding Y, Nair DB, Siu S . (2013). Surface appendages of archaea: structure, function, genetics and assembly. Life (Basel) 3: 86–117.
Javed MA, van Alphen LB, Sacher J, Ding W, Kelly J, Nargang C et al. (2014). A receptor-binding protein of Campylobacter jejuni bacteriophage NCTC 12673 recognizes flagellin glycosylated with acetamidino-modified pseudaminic acid. Mol Microbiol 95: 101–115.
Kaltenböck E, Herndl GJ . (1992). Ecology of amorphous aggregations (marine snow) in the Northern Adriatic Sea. IV. Dissolved nutrients and the autotrophic community associated with marine snow. Mar Ecol Prog Ser 87: 147–159.
Karner MB, DeLong EF, Karl DM . (2001). Archaeal dominance in the mesopelagic zone of the Pacific Ocean. Nature 409: 507–510.
Klotz MG, Stein LY . (2008). Nitrifier genomics and evolution of the nitrogen cycle. FEMS Microbiol Lett 278: 146–156.
Kim M, Oh HS, Park SC, Chun J . (2014). Towards a taxonomic coherence between average nucleotide identity and 16S rRNA gene sequence similarity for species demarcation of prokaryotes. Int J Syst Evol Microbiol 64: 346–351.
Könneke M, Bernhard AE, de la Torre JR, Walker CB, Waterbury JB, Stahl DA . (2005). Isolation of an autotrophic ammonia-oxidizing marine archaeon. Nature 437: 543–546.
Könneke M, Schubert DM, Brown PC, Hugler M, Standfest S, Schwander T et al. (2014). Ammonia-oxidizing archaea use the most energy-efficient aerobic pathway for CO2 fixation. Proc Natl Acad Sci USA 111: 8239–8244.
Krzywinski M, Schein J, Birol I, Connors J, Gascoyne R, Horsman D et al. (2009). Circos: an information aesthetic for comparative genomics. Genome Res 19: 1639–1645.
Lawton TJ, Ham J, Sun T, Rosenzweig AC . (2014). Structural conservation of the B subunit in the ammonia monooxygenase/particulate methane monooxygenase superfamily. Proteins 82: 2263–2267.
Lebedeva EV, Hatzenpichler R, Pelletier E, Schuster N, Hauzmayer S, Bulaev A et al. (2013). Enrichment and genome sequence of the group I.1a ammonia-oxidizing Archaeon 'Ca. Nitrosotenuis uzonensis' representing a clade globally distributed in thermal habitats. PLoS One 8: e80835.
Leininger S, Urich T, Schloter M, Schwark L, Qi J, Nicol GW et al. (2006). Archaea predominate among ammonia-oxidizing prokaryotes in soils. Nature 442: 806–809.
Lopez-Garcia P, Brochier C, Moreira D, Rodriguez-Valera F . (2004). Comparative analysis of a genome fragment of an uncultivated mesopelagic crenarchaeote reveals multiple horizontal gene transfers. Environ Microbiol 6: 19–34.
Luo H, Tolar BB, Swan BK, Zhang CL, Stepanauskas R, Ann Moran M et al. (2014). Single-cell genomics shedding light on marine Thaumarchaeota diversification. ISME J 8: 732–736.
Margulies M, Egholm M, Altman WE, Attiya S, Bader JS, Bemben LA et al. (2005). Genome sequencing in microfabricated high-density picolitre reactors. Nature 437: 376–380.
Martens-Habbena W, Berube PM, Urakawa H, de la Torre JR, Stahl DA . (2009). Ammonia oxidation kinetics determine niche separation of nitrifying Archaea and Bacteria. Nature 461: 976–979.
Martens-Habbena W, Qin W, Horak RE, Urakawa H, Schauer AJ, Moffett JW et al. (2014). The production of nitric oxide by marine ammonia-oxidizing archaea and inhibition of archaeal ammonia oxidation by a nitric oxide scavenger. Environ Microbiol 7: 2261–2274.
Mende DR, Sunagawa S, Zeller G, Bork P . (2013). Accurate and universal delineation of prokaryotic species. Nat Methods 10: 881–884.
Mosier AC, Allen EE, Kim M, Ferriera S, Francis CA . (2012a). Genome sequence of "Candidatus Nitrosopumilus salaria" BD31, an ammonia-oxidizing archaeon from the San Francisco Bay estuary. J Bacteriol 194: 2121–2122.
Mosier AC, Lund MB, Francis CA . (2012b). Ecophysiology of an ammonia-oxidizing archaeon adapted to low-salinity habitats. Microb Ecol 64: 955–963.
Müller-Niklas G, Schuster S, Kaltenböck E, Herndl GJ . (1994). Organic content and bacterial metabolism in amorphous aggregations of the northern Adriatic Sea. Limnol Oceanogr 39: 58–68.
Norton JM, Alzerreca JJ, Suwa Y, Klotz MG . (2002). Diversity of ammonia monooxygenase operon in autotrophic ammonia-oxidizing bacteria. Arch Microbiol 177: 139–149.
Offre P, Kerou M, Spang A, Schleper C . (2014). Variability of the transporter gene complement in ammonia-oxidizing archaea. Trends Microbiol 22: 665–675.
Ozer Z, Reardon JT, Hsu DS, Malhotra K, Sancar A . (1995). The other function of DNA photolyase: stimulation of excision repair of chemical damage to DNA. Biochemistry 34: 15886–15889.
Painter SC, Sanders R, Waldron HN, Lucas MI, Torres-Valdes S . (2008). Urea distribution and uptake in the Atlantic Ocean between 50° N and 50° S. Mar Ecol Prog Ser 368: 53–63.
Park SJ, Kim JG, Jung MY, Kim SJ, Cha IT, Ghai R et al. (2012). Draft genome sequence of an ammonia-oxidizing archaeon, "Candidatus Nitrosopumilus sediminis" AR2, from Svalbard in the Arctic Circle. J Bacteriol 194: 6948–6949.
Park SJ, Ghai R, Martin-Cuadrado AB, Rodriguez-Valera F, Chung WH, Kwon K et al. (2014). Genomes of two new ammonia-oxidizing archaea enriched from deep marine sediments. PLoS One 9: e96449.
Prosser JI . (1989). Autotrophic nitrification in bacteria. Adv Microb Physiol 30: 125–181.
Prosser JI, Nicol GW . (2012). Archaeal and bacterial ammonia-oxidisers in soil: the quest for niche specialisation and differentiation. Trends Microbiol 20: 523–531.
Purkhold U, Pommerening-Roser A, Juretschko S, Schmid MC, Koops HP, Wagner M . (2000). Phylogeny of all recognized species of ammonia oxidizers based on comparative 16 S rRNA and amoA sequence analysis: implications for molecular diversity surveys. Appl Environ Microbiol 66: 5368–5382.
Qin W, Amin SA, Martens-Habbena W, Walker CB, Urakawa H, Devol AH et al. (2014). Marine ammonia-oxidizing archaeal isolates display obligate mixotrophy and wide ecotypic variation. Proc Natl Acad Sci USA 111: 12504–12509.
Rakhuba DV, Kolomiets EI, Dey ES, Novik GI . (2010). Bacteriophage receptors, mechanisms of phage adsorption and penetration into host cell. Pol J Microbiol 59: 145–155.
Remsen CC . (1971). The distribution of urea in coastal and oceanic waters. Limnol Oceanogr 16: 732–740.
Rosario MM, Kirby JR, Bochar DA, Ordal GW . (1995). Chemotactic methylation and behavior in Bacillus subtilis: role of two unique proteins, CheC and CheD. Biochemistry 34: 3823–3831.
Sakofsky CJ, Runck LA, Grogan DW . (2011). Sulfolobus mutants, generated via PCR products, which lack putative enzymes of UV photoproduct repair. Archaea 2011: 864015.
Santoro AE, Dupont CL, Richter RA, Craig MT, Carini P, McIlvin MR et al. (2015). Genomic and proteomic characterization of "Candidatus Nitrosopelagicus brevis": an ammonia-oxidizing archaeon from the open ocean. Proc Natl Acad Sci USA 112: 1173–1178.
Schmieder R, Edwards R . (2011). Quality control and preprocessing of metagenomic datasets. Bioinformatics 27: 863–864.
Shen T, Stieglmeier M, Dai J, Urich T, Schleper C . (2013). Responses of the terrestrial ammonia-oxidizing archaeon Ca. Nitrososphaera viennensis and the ammonia-oxidizing bacterium Nitrosospira multiformis to nitrification inhibitors. FEMS Microbiol Lett 344: 121–129.
Siebers B, Schönheit P . (2005). Unusual pathways and enzymes of central carbohydrate metabolism in Archaea. Curr Opin Microbiol 8: 695–705.
Sintes E, Bergauer K, De Corte D, Yokokawa T, Herndl GJ . (2013). Archaeal amoA gene diversity points to distinct biogeography of ammonia-oxidizing Crenarchaeota in the ocean. Environ Microbiol 15: 1647–1658.
Spang A, Hatzenpichler R, Brochier-Armanet C, Rattei T, Tischler P, Spieck E et al. (2010). Distinct gene set in two different lineages of ammonia-oxidizing archaea supports the phylum Thaumarchaeota. Trends Microbiol 18: 331–340.
Spang A, Poehlein A, Offre P, Zumbragel S, Haider S, Rychlik N et al. (2012). The genome of the ammonia-oxidizing Candidatus Nitrososphaera gargensis: insights into metabolic versatility and environmental adaptations. Environ Microbiol 14: 3122–3145.
Stahl DA, de la Torre JR . (2012). Physiology and diversity of ammonia-oxidizing archaea. Annu Rev Microbiol 66: 83–101.
Stieglmeier M, Klingl A, Alves RJ, Rittmann SK, Melcher M, Leisch N et al. (2014). Nitrososphaera viennensis gen. nov., sp. nov., an aerobic and mesophilic, ammonia-oxidizing archaeon from soil and a member of the archaeal phylum Thaumarchaeota. Int J Syst Evol Microbiol 64: 2738–2752.
Strompl C, Hold GL, Lunsdorf H, Graham J, Gallacher S, Abraham WR et al. (2003). Oceanicaulis alexandrii gen. nov., sp. nov., a novel stalked bacterium isolated from a culture of the dinoflagellate Alexandrium tamarense (Lebour) Balech. Int J Syst Evol Microbiol 53: 1901–1906.
Szabo Z, Stahl AO, Albers SV, Kissinger JC, Driessen AJ, Pohlschroder M . (2007). Identification of diverse archaeal proteins with class III signal peptides cleaved by distinct archaeal prepilin peptidases. J Bacteriol 189: 772–778.
Szurmant H, Ordal GW . (2004). Diversity in chemotaxis mechanisms among the bacteria and archaea. Microbiol Mol Biol Rev 68: 301–319.
Teira E, Lebaron P, van Aken H, Herndl GJ . (2006). Distribution and activity of Bacteria and Archaea in the deep water masses of the North Atlantic. Limnol Oceanogr 51: 2131–2144.
Tourna M, Stieglmeier M, Spang A, Könneke M, Schintlmeister A, Urich T et al. (2011). Nitrososphaera viennensis, an ammonia oxidizing archaeon from soil. Proc Natl Acad Sci USA 108: 8420–8425.
Treusch AH, Leininger S, Kletzin A, Schuster SC, Klenk HP, Schleper C . (2005). Novel genes for nitrite reductase and Amo-related proteins indicate a role of uncultivated mesophilic crenarchaeota in nitrogen cycling. Environ Microbiol 7: 1985–1995.
Vallenet D, Belda E, Calteau A, Cruveiller S, Engelen S, Lajus A et al. (2013). MicroScope—an integrated microbial resource for the curation and comparative analysis of genomic and metabolic data. Nucleic Acids Res 41: D636–D647.
Walker CB, de la Torre JR, Klotz MG, Urakawa H, Pinel N, Arp DJ et al. (2010). Nitrosopumilus maritimus genome reveals unique mechanisms for nitrification and autotrophy in globally distributed marine crenarchaea. Proc Natl Acad Sci USA 107: 8818–8823.
Yawata Y, Cordero OX, Menolascina F, Hehemann JH, Polz MF, Stocker R . (2014). Competition-dispersal tradeoff ecologically differentiates recently speciated marine bacterioplankton populations. Proc Natl Acad Sci USA 111: 5622–5627.
Zhang CL, Ye Q, Huang Z, Li W, Chen J, Song Z et al. (2008). Global occurrence of archaeal amoA genes in terrestrial hot springs. Appl Environ Microbiol 74: 6417–6426.
Acknowledgements
We thank Maria Pinto and Elisabeth Haberleitner for technical assistance with cultivations, Thomas Weinmaier for assistance with bioinformatic analyses, Thomas Reinthaler for assistance during 454 sequencing, Michaela Stieglmeier for helpful discussions and Claus Pelikan for critically reviewing the manuscript. We also thank the Core Facility Cell Imaging and Ultrastructure Research staff of the Faculty of Life Sciences of the University of Vienna. Furthermore, we are grateful to the LABGeM bioinformatics team of Genoscope for providing access to the MicroScope platform. The experimental work was supported by the European Research Council under the European Community’s Seventh Framework Program (FP7/2007–2013)/ERC Advanced Grant ‘MEDEA’ agreement No. 268595 and by the Austrian Science Fund (FWF) project I486-B09 both to GJH.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies this paper on The ISME Journal website
Supplementary information
Rights and permissions
This work is licensed under a Creative Commons Attribution-NonCommercial-ShareAlike 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-sa/4.0/
About this article
Cite this article
Bayer, B., Vojvoda, J., Offre, P. et al. Physiological and genomic characterization of two novel marine thaumarchaeal strains indicates niche differentiation. ISME J 10, 1051–1063 (2016). https://doi.org/10.1038/ismej.2015.200
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ismej.2015.200
This article is cited by
-
Increased prokaryotic diversity in the Red Sea deep scattering layer
Environmental Microbiome (2023)
-
Nitrification and beyond: metabolic versatility of ammonia oxidising archaea
The ISME Journal (2023)
-
Phylotype resolved spatial variation and association patterns of planktonic Thaumarchaeota in eastern Chinese marginal seas
Marine Life Science & Technology (2023)
-
Genomics, Phylogeny, and in Silico Phenotyping of Nitrosopumilus Genus
Current Microbiology (2023)
-
HT-SIP: a semi-automated stable isotope probing pipeline identifies cross-kingdom interactions in the hyphosphere of arbuscular mycorrhizal fungi
Microbiome (2022)