Abstract
Cyanobacterial organic matter excretion is crucial to carbon cycling in many microbial communities, but the nature and bioavailability of this C depend on unknown physiological functions. Cyanobacteria-dominated hypersaline laminated mats are a useful model ecosystem for the study of C flow in complex communities, as they use photosynthesis to sustain a more or less closed system. Although such mats have a large C reservoir in the extracellular polymeric substances (EPSs), the production and degradation of organic carbon is not well defined. To identify extracellular processes in cyanobacterial mats, we examined mats collected from Elkhorn Slough (ES) at Monterey Bay, California, for glycosyl and protein composition of the EPS. We found a prevalence of simple glucose polysaccharides containing either α or β (1,4) linkages, indicating distinct sources of glucose with differing enzymatic accessibility. Using proteomics, we identified cyanobacterial extracellular enzymes, and also detected activities that indicate a capacity for EPS degradation. In a less complex system, we characterized the EPS of a cyanobacterial isolate from ES, ESFC-1, and found the extracellular composition of biofilms produced by this unicyanobacterial culture were similar to that of natural mats. By tracing isotopically labeled EPS into single cells of ESFC-1, we demonstrated rapid incorporation of extracellular-derived carbon. Taken together, these results indicate cyanobacteria reuse excess organic carbon, constituting a dynamic pool of extracellular resources in these mats.
Similar content being viewed by others
Introduction
Cyanobacteria represent a 0.05% of global carbon biomass (Garcia-Pichel et al., 2003), and organic matter excretion by these primary producers, which can be as high as 24% of their net primary productivity (Bertlisson et al., 2005), drives C cycling in many microbial communities (Fogg, 1983). Understanding the metabolic pathways that lead to C excretion and quantifying C cycling rates in cyanobacterial-dominated systems is complicated by the high molecular diversity of the excreted C and the diverse physiological functions of excretion. For example, cyanobacteria can excrete fermentation products at night (Stal and Moezelaar, 1997), excrete osmolytes following osmotic shock (Hagemann, 2011; Reed et al., 1986) and release extracellular polymeric substances (EPSs; Decho, 1990). It is thought that EPS excretion serves multiple functions, including nutrient storage (organic compounds containing C, N or P and trace metals), structural organization and buffering against environmental stressors (Flemming and Wingender, 2010). In cyanobacterial communities, extracellular chemistry may have additional functions related to lithification (Decho et al., 2005; Dupraz et al., 2009), or protection from desiccation (Tamaru et al., 2005; Wright et al., 2005) and radiation (Garcia-Pichel and Castenholz, 1991; Garcia-Pichel and Castenholz, 1993). In addition, a number of factors such as inorganic nutrient C or N availability (Otero and Vincenzini, 2004) and light (De Brouwer et al., 2002) can affect EPS biosynthesis in photosynthetic organisms.
EPS may include polysaccharides, proteins and nucleic acids, as well as inorganic molecules and organic metabolites. Cyanobacterial exopolysaccharides, in particular, vary in molecular structure depending on the producing species (Pereira et al., 2009). Some cyanobacteria have encasing sheaths of ‘capsular polysaccharides’, often tightly bound to the cells, which are distinct from ‘released polysaccharides’ (De Philippis and Vincenzini, 1998; Pereira et al., 2009). It is not known whether these distinct exopolysaccharides have different functions, or how available they are to other community members.
Hypersaline laminated microbial mats represent excellent model systems to investigate C cycling because they are more or less closed (relatively little biomass is exported) and they are sustained by a single carbon source (photosynthesis) (Canfield and Des Marais, 1993). The thick, gelatinous extracellular matrix is an inherently recognizable component of such spatially organized microbial communities. However, the production, consumption and basic structure of hypersaline mat EPS is understudied, in part because of the complex chemical composition within a highly diverse community. There is, however, detailed information on the interlinked biogeochemical cycling of carbon, nitrogen and sulfur (Jørgensen and Des Marais, 1986; Canfield and Des Marais, 1993; Hoehler et al., 2001). These studies show that cyanobacteria accumulate photosynthate during the day, which is fermented to organic acids and H2 at night (Bebout et al., 2002; Hoehler et al., 2002; Burow et al 2012). Other community members such as Chloroflexi further metabolize the organic acids (Burow et al., 2013; Lee et al., 2014). In spite of the abundance of detailed studies linking C cycling to key microbial mat taxa involved in these transformations, the makeup and dynamics of the extracellular C pool are not known.
To elucidate the function of cyanobacterial C excretion in microbial mats, we characterized the basic composition of mat EPS and identified its key producers and consumers. We collected naturally complex microbial mats from the Moss Landing Wildlife Area at Elkhorn Slough (ES), Monterey Bay, California and compared these with unicyanobacterial biofilms containing the cyanobacterium ESFC-1 (referred to as a ‘cultured biofilm’) isolated from the same site (Woebken et al., 2012; Everroad et al., 2013). This sheathless cyanobacterium can comprise up to 10% of the mat cyanobacterial community (Woebken et al., 2012) and readily forms biofilms in culture. We postulated that the EPS contains cyanobacterial exopolysaccharides, proteins and nucleic acids, which are consumed predominantly by other dominant bacterial phyla in the photosynthetic layer of the mats, such as Chloroflexi and Gammaproteobacteria (Burow et al., 2013; Lee et al., 2014). To test this, we examined the glycosyl and protein composition of the EPS using glycosyl analysis and shotgun metaproteomics. We then tracked carbon usage in our model culture using isotope tracing and single-cell analysis. Our results lead us to propose that through degradation and uptake, cyanobacteria reuse a significant fraction of extracellular organic carbon.
Materials and methods
Field sampling
Triplicate cores (10 mm diameter) were collected on 13 November 2012 at 1445 hours from ES, Monterey Bay (36.812986°N; −121.784815°W). Cores (0–10 mm depth) were collected in sterile 50 ml Falcon tubes (BD Biosciences, San Jose, CA, USA) and placed on dry ice until storage at −20 °C.
Cell culture and 13C-labeling
Unicyanobacterial cultures of cyanobacterium strain ESFC-1 were grown in modified artificial seawater media (ASN), which is a modified version of ASN-III (Rippka, 1988) described in Woebken et al. (2012). Cultures were grown at 23 °C with 20 μmol m-2 s-1 (4.16 W m-2) light with a 12 h:12 h light:dark cycle. Batch cultures were inoculated into 300 ml of ASN in acid-washed 1 liter glass flasks and grown as biofilms without shaking for 4 weeks. After 2 weeks, spent media were removed from the cultures and 300 ml of fresh ASN was added. To generate 13C-labeled material, liquid cultures were grown in ASN containing 3 mM 13C sodium bicarbonate (13C, 99% Cambridge Isotopes, Tewksbury, MA, USA) and transferred three times in sealed flasks (with no headspace). Although some unlabeled 12C bicarbonate may have equilibrated from the atmosphere during media preparation, we expect that most of the bicarbonate (>98%) in the media were 13C labeled. A supplement of 2 mM 13C bicarbonate was added every 5 days to ensure an adequate isotopically labeled inorganic carbon source. After three transfers, 13C-labeled cultures were inoculated into 500 ml 13C-labeled ASN and grown for an additional month, after which biofilms were harvested for EPS separation. Bulk isotope ratios for 13C/12C of this 13C-labeled material was determined by isotope-ratio mass spectrometry (ANCA-IRMS, PDZE Europa Limited, Crewe, England) at the University of California-Berkeley.
EPS separations
EPS extraction methods were based on previously described techniques, (Klock et al., 2007; Jiao et al., 2010), and are described in detail here. Biological triplicates of natural mat and culture samples were separated into three operationally defined fractions (two extracellular and one total): ‘EPS-L’ (loose), containing all soluble material recovered in the supernatant of a homogenized sample; ‘EPS-B’ (bound) from a secondary wash designed to strip loosely bound or outer membrane-associated material; and ‘Total’ intended to capture both intracellular and extracellular material from lysed cells. The upper phototrophic layer (0–2 mm) of frozen ES mat samples was excised using a sterile razor blade. Natural mats and cultured biofilms were weighed and homogenized on ice in sterile 10% NaCl (Wheaton Dounce homogenizer, pestle clearance 0.114±0.025 mm). One-tenth volume of the homogenate was kept for the Total fraction, sonicated to lyse cells (Misonix, Farmingdale, NY, USA; 50% intensity, six cycles of 30 s on and 2 min off on ice), followed by centrifugation at 12 000 × g at 4 °C for 10 min. The resulting supernatant defines the Total fraction. The remaining homogenate was incubated at 40 °C for 15 min and centrifuged at 15 000 × g at 4 °C for 20 min to separate cells and loosely bound EPS. Supernatant was filtered through a 0.2 μm polycarbonate filter, forming the EPS-L fraction. The pellet was resuspended in a buffer containing 50 mM Tris-HCl, pH 8, 50 mM EDTA and 15% sucrose (developed previously to isolate marine Synechococcus outer membranes (McCarren and Brahamsha, 2007)), incubated for 30 min on ice and centrifuged at 12 000 × g at 4 °C for 10 min to strip off tightly bound EPS. Supernatant was then separated from cells and debris by centrifugation (as above), defining the EPS-B fraction.
To test for cellular lysis during harvest, activity of glucose-6-phosphate dehydrogenase, an intracellular enzyme marker, was compared between Total and EPS fractions (Wingender et al., 2001) and frozen and thawed natural mat samples. Glucose-6-phosphate dehydrogenase activity was not detected in EPS-B. In EPS-L, activity was <10% of that in the Total fraction. Light microscopy confirmed there was no visible cell lysis during the fractionation procedure. Although it is difficult to discount cellular lysis in any global analysis targeting putative exoproteins, our method includes a paired Total fraction, allowing us to calculate significant enrichment in the extracellular fraction relative to the Total (see Methods in Supplementary Information). Localization predictions and common exoproteins published for other cyanobacteria substantiate our predictions.
Carbohydrate concentration was determined using the traditional phenol sulfuric acid method (Dubois et al., 1951). Nucleic acid concentrations were measured using PicoGreen and RiboGreen dyes for DNA and RNA, respectively, according to the manufacturer’s instructions (Life Technologies, Carlsbad CA, USA). Protein concentrations were determined using a Bradford assay (Bio-Rad, Hercules CA, USA) and absorbance at 280 nm. All concentrations were normalized to biomass wet weight.
Glycosyl analysis
Glycosyl composition was performed on four samples, extracted as described above, a Total and EPS-L fraction from each an ES mat sample (top 2 mm) and a cultured biofilm. Linkage analysis and NMR analysis was conducted on the material from the two EPS-L samples analyzed for glycosyl composition (one from an ES mat and one from a cultured biofilm). Samples were precipitated with cold 70% ethanol for 2 h, centrifuged at 15 000 × g, pellet dried and frozen at –80 °C until analysis. Glycosyl composition (Santander et al., 2013) and linkage (‘methylation’) (Heiss et al., 2009) analysis were performed as described previously. For NMR, the samples (~20 mg each) were deuterium-exchanged twice from D2O (99.9% D, Sigma-Aldrich, St Louis, MO, USA) and dissolved in 0.5 ml D2O (99.96% D, Cambridge Isotopes) containing 0.06% acetone. The 1D proton and 2D gCOSY NMR spectra were acquired on a Varian Inova-600 instrument (Varian Medical Systems, Palo Alto, CA, USA) at 25 °C. Chemical shifts were referenced to internal acetone (2.218 p.p.m.). The 1D proton spectra were signal averaged from 512 scans each. The 2D gCOSY spectra were acquired in 32 scans per increment and 128 increments. The anomeric peaks were identified by their chemical shifts and by their coupling patterns, as well as by the chemical shifts of 2H, which were determined using 2D-1H-1H COSY spectra (data not shown).
Cyanobacterial uptake of extracellular organic carbon
To test whether mat cyanobacteria can utilize extracellular organic material as a C source, 30 replicate cultured biofilms were grown for 1 month in six-well plates. Fluorescence (595/640 nm excitation/emission) was read on each plate every other day to estimate biofilm growth. ASN medium was replaced once a week (10 ml media per well). After 20 days of growth, 4% formaldehyde was added to half of the cultures and incubated for 1 h to generate killed controls. At the start of the light phase of the 12 h:12 h light–dark cycle, the medium was removed from all cultures and 3 ml of 13C-labeled EPS-L diluted 1:10 in sterile medium was added to each well. After 6 and 12 h (in the light), three replicate cultures and killed control wells were washed with sterile unlabeled ASN medium, incubated with 4% formaldehyde for 1–2 h, rinsed with 1X phosphate-buffered saline and frozen at −20 °C in 50% ethanol. For the second control experiment, a second batch of 13C-labeled EPS-L was diluted 1:30 in sterile media, and added to eight wells at the start of the light cycle. For this experiment, there were three treatments with two biological replicates including: dark incubated, light incubated and killed controls. All were incubated for 6 h and then fixed as described above. To prepare these samples for NanoSIMS analysis, subsamples were removed, homogenized gently to separate trichomes, rinsed with sterile milliQ water three times, and then 0.5 μl was spotted onto a silicon wafer, air dried and stored in an argon dry box.
To estimate the maximum amount of 13CO2 that could be generated by other bacteria in the cultured biofilm, we calculated biovolume and net fixation of carbon. For bacterial counts and biovolume measurements, portions of fixed biofilms were mounted on a slide with 4,6-diamidino-2-phenylindole DNA stain (Porter and Feig, 1980). Biofilms from the 12-h time point and killed controls were imaged, with 10 fields of view at × 100 magnification, with 9 μm thick z-stacks. Z-stacks were combined into a maximum intensity projection image, using MetaMorph Imaging software (Molecular Devices, Sunnyvale, CA, USA). For cyanobacteria, biovolumes were calculated as a cylinder volume, using heights and radii from images (Garcia-Pichel et al., 1994). For bacteria, an average length (L) and width (W) was calculated from 40 cell measurements from images (1.4 and 0.35 μm, respectively). Volume (V) calculated according to Equation (1), which is applicable to both rods and spheres (Bratbak, 1985).

C content was estimated using biovolume-to-carbon conversion factors from literature (2.2 × 10-13 g C μm–3 for bacteria and 1.8 × 10-13 g C μm–3 filamentous diazotrophic cyanobacteria) (Bratbak, 1985; Goebel et al., 2008). C content was extrapolated to μg C per well for the six-well plates used. Net fixation of 13C and percentage of C incorporated relative to initial C content (Popa et al., 2007) was calculated for the bacteria and extrapolated to calculate μg C fixation per well. We assumed the lowest possible bacterial growth efficiency (amount of new biomass produced per unit organic C assimilation) of 0.01 (Del Giorgio and Cole, 1998) in order to estimate maximum 13CO2 production by bacteria and calculated μg 13C produced as 13CO2. We assumed complete assimilation of 13CO2 to calculate the resulting 13C/12C ratio in the cyanobacterium. This provides an estimate of an upper bound for estimates of cyanobacterial 13CO2 incorporation.
Results
Bulk and carbohydrate composition analyses
In the ES natural mat, the dominant organic constituent of EPS-L (loose soluble extracellular fraction) was carbohydrate, and the next most abundant constituent was protein (Figure 1). The majority of the carbohydrate was polysaccharide containing both starch- and cellulose-like linkages (Figure 2). Gas chromatography-mass spectrometry-based glycosyl analysis revealed that glucose was by far the most abundant residue in EPS-L (Figure 2a). In contrast, seven additional residues were detected in the Total fraction, which included lysed cells (Figure 2a). Free glucose was below the detection limit in EPS-L (data not shown), indicating a predominance of glucose polymers. In EPS-L, gas chromatography-mass spectrometry-based linkage analysis indicated that (1,4) glucose linkages were more common than terminal glucose, (4,6) or (1,6) glucose linkages (Figure 2b). This analysis also determined a 2:1 ratio of (1,4) linkages to terminal glucose units, inferring a prevalence of short glucan chains (oligosaccharides). 1D proton and 2D homonuclear gCOSY spectroscopy indicated the presence of both α- (starch-like) and β- (cellulose-like) glucans (Figure 2c).
Carbohydrate composition analyses for ES mat EPS-L fraction. (a) Glycosyl composition analysis of one replicate for each EPS-L and Total fractions. ‘GlcNAc’ is N-acetylglucosamine. (b) Linkage analysis of EPS-L from one ES mat sample, showing the types of pyranosides detected. ‘Glc’ is glucose and ‘t-Glc’ is a terminal glucose linkage. (c) Anomeric portion of the 1D-1H NMR spectra of EPS-L from one ES mat sample showing the regions used for line fitting. Numbers under peaks are integrated area under curve.
The composition of EPS-L from the cultured biofilm was similar to that extracted from ES mats (Figure 1, Supplementary Figure S1). However, the culture sample also contained a minor amount of mannose residues in α-(1,2) linkages (peak area of 0.57; Supplementary Figure S1C). Based on the integrated peak area, the number of α-glucose linkages was 6.2-fold higher than β-glucose linkages in the culture, (Supplementary Figure S1C), which was not the case in the natural mats (Figure 2c).
Exoproteome of natural microbial mats
We performed shotgun proteomics to compare the the two extracellular fractions, EPS-L (loose) and EPS-B (bound), and the Total (intracellular and extracellular proteins) fraction. We collectively refer to proteins significantly enriched in each of the extracellular fractions relative to the Total as the ‘exoproteome’. In the ES mat samples, we identified 2042 unique proteins based on the following criteria: 95% peptide identity, 99% protein identity, 1.1% false discovery rate and at least two peptides/protein. To distinguish putative extracellular proteins from those in the EPS as a result of lysis, we searched for proteins that were significantly overrepresented (t-test, P<0.05) in the EPS fractions over the Total fraction. We identified 151 proteins significantly overrepresented in EPS-L and 33 in EPS-B. There were 10 proteins overrepresented in both fractions, yielding a total of 174 proteins in the ES mat exoproteome (Supplementary Table S1). Protein localization predictions, using the nearest genome full protein sequence, indicated that 34% of the EPS-L exoproteins and 42% of the EPS-B exoproteins were likely non-cytoplasmic (Supplementary Table S1).
The majority of proteins in the metaproteome and the metaexoproteome were assigned to cyanobacteria, 56% and 79%, respectively (Figure 3). Another large proportion of the identified proteins originated from proteobacteria, particularly from the class Gammaproteobacteria (Figure 3).
Taxonomic classification of proteins from ES mat metaproteomics. (a) Total fraction phylum level association of identified proteins (cyanobacteria and Proteobacteria also subclassified at the order and class level, respectively), includes 1682 proteins in at least one of three biological replicates. (b) Exoproteome phylum level association of proteins, 176 proteins that were significantly enriched (P<0.05) in one or both of the two extracellular fractions (EPS-L and EPS-B) over the Total fraction across three biological replicates.
When grouped into broad functional categories, many of the cyanobacterial proteins in the ES mat exoproteome had predicted functions related to degradation or modification of amino acids, carbohydrates, proteins and nucleic acids (Table 1). These include several classes of peptidases, a glycosyl hydrolase, aminotransferases and a ribonuclease (Supplementary Table S1). These functional categories were also prevalent in the exoproteins of bacteria other than cyanobacteria (Table 1), although there were more transport binding proteins such as sugar and amino-acid transport and extracellular solute binding proteins assigned to non-cyanobacterial groups (Supplementary Table S1). We found putative phosphate acquisition exoproteins, such as phosphate- and phosphonate transporters, and nucleic acid degradation enzymes, from several groups including Cyanobacteria, Alphaproteobacteria and Gammaproteobacteria (Supplementary Table S1). We also identified proteins assigned to expected categories including cell wall, capsule and secreted proteins (Table 1). Oxidative stress response proteins assigned to phyla Cyanobacteria and Planctomycetes were also detected (Table 1).
Comparison of natural mat and cultured biofilm exoproteomes
We performed shotgun proteomics in unicyanobacterial cultured biofilms, and identified 849 proteins that match genes of the cyanobacterium ESFC-1 genome as a search database, detected 849 proteins, of which 212 were enriched in the extracellular fraction (148 in the EPS-L and 87 in the EPS-B). Protein localization predictions indicated 24% and 47% of the EPL-L and EPS-B exoproteins, respectively, were of non-cytoplasmic origin (Supplementary Table S1). After grouping exoproteins into functional categories, we observed similar patterns as in the natural ES mats, including amino acid, carbohydrate and protein metabolism in the EPS-L fraction (Table 1). This included a 4-α-glucanotransferase that may be involved in glycogen or starch breakdown and a large number of peptidases and proteases (Supplementary Table S1). As the cultured biofilm was not axenic, we also mapped the peptide spectra to the metagenomic database used for the ES mat samples. Using the same thresholds as above, we identified 24 non-cyanobacterial proteins present in at least two biological replicates (Supplementary Table S1). The concentration of these proteins was too low to obtain a significant enrichment value in the EPS-B or EPS-L fractions when compared with the Total fraction. The most abundant of these proteins originated from Gammaproteobacteria and included an amino-acid transporter.
Protein homologs were identified in the exoproteins of both the ES mat and cultured biofilm that are predicted to degrade organic compounds such as those in the EPS (Figure 4). Of the 22 homologs identified, 21 in ES mats originate from an abundant filamentous cyanobacterium, Coleofasciculus chthonoplastes (formerly known as Microcoleus chthonoplastes). Not closely related at the genome level, Cyanobacterium ESFC-1 and C. chtonoplastes share 1700 homologous genes that account for only 35% of the protein coding genes of ESFC-1 and 20% of those from C. chthonoplastes. Most of these 21 proteins were above the median abundance in the exoproteome (normalized spectral count abundances of 9.7 and 9.5 in the ES mat and culture, respectively) (Figure 4; Supplementary Table S2). They include two homologous Zn-dependent peptidases (one predicted as extracellular in the cultured biofilm), a C-terminal peptidase, an enzyme involved in lysine metabolism, and a carboxymethylenebutenolidase (predicted as extracellular in C. chthonoplastes). Among the more abundant exoprotein homologs in the two sample types were an inorganic pyrophosphatase, nucleic acid degradation enzymes and a sulfate adenyltransferase. There were also some differences between the natural mats and cultured biofilm, for example, there were more peptidases, proteases and proteins related to RNA metabolism in the cultured biofilm exoproteome than in that of the natural ES mat.
Homologous and abundant exoproteins identified in each (a) ES mat and (b) cultured biofilm exoproteomes. Protein abundance values are average normalized spectral counts over triplicate biological replicates (error bars represent 1 s.d.). All homologs enriched in ES mat and cultured biofilm exoproteomes with predicted functions are included and indicated in bold. Also included are the top seven most abundant proteins in the EPS-L fraction and top three in the EPS-B fraction. Exoproteins are enriched in EPS-L unless indicated by (EPS-B) following the name and are of cyanobacterial origin unless indicated. Dotted line indicates median protein abundance in each set of samples.
Extracellular enzyme degradation capabilities
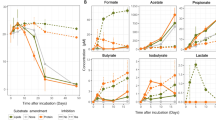
As the annotation of putative function does not always correlate with activity, we assayed for general types of degradative enzymes in the EPS-L fraction of ES mats and cultured biofilms (Figure 5). Based on the most abundant proteins identified in the exoproteome, we tested for activities on substrates that fluoresce upon hydrolysis of peptide bonds (peptidases and proteases), cellobiose (cellulases, cellobiohydrolases), and both α- and β-linked glucose residues (glucosidases) (Figure 5). In the mat EPS samples, we detected significant hydrolysis of substrates including proteins, cellulose, and α- and β-linked glycans (Figure 5). Assays of the culture confirmed high peptidase activity (Figure 5, Table 1). Glucosidase activity (α and β) was also detectable in the culture samples, confirming the potential for cyanobacterial degradation of carbohydrates, but cellulase activity was below detection (Figure 5).
Extracellular enzyme activity for four types of degradative enzymes in EPS-L fractions from ES mat and cultured biofilms. Activity was calculated by rate of degradation of fluorescent substrate over 3 h in the EPS-L fractions of triplicate biological replicates of natural mats and cultured biofilms. Error bars represent 1 s.d. for three biological replicates.
Cyanobacterial uptake of EPS-L-derived organic carbon
The glycosyl composition, exoproteome and enzyme activity collectively indicate the potential for cyanobacteria-mediated organic carbon, amino acid and protein degradation in the extracellular environment. To directly address whether cyanobacteria re-assimilate extracellular organic matter, we tested whether cyanobacterium ESFC-1 could incorporate its own extracellular carbon. We grew cultured biofilms in 13C-labeled media, extracted EPS-L and incubated biofilm cultures with this 13C-labeled EPS-L (59.6 13C atom percent excess (APE) diluted 1:10 in media) for 12 h in the light phase of a 12:12 diel cycle. Individual cyanobacterial trichomes were harvested and analyzed at two time points (24 at 6 h and 29 at 12 h) with high-resolution secondary ion mass spectrometry (NanoSIMS) in order to assess 13C incorporation. Growth rate of the cultured biofilm under these experimental conditions was 0.114 d-1 (a doubling time of ~6 days, Supplementary Figure S2).
Six hours after the labeled EPS-L was introduced, cyanobacterial cells were significantly 13C enriched relative to killed controls (t-test, P<0.05, mean 13C APE of 1.40) and average enrichment levels increased significantly by 12 h (P<0.05, mean 13C APE 2.70, Figure 6), under light, oxic conditions. We observed high variability in the 13C enrichment of individual analyzed cells, likely due to variability in single-cell activity within the culture (Popa et al., 2007; Finzi-Hart et al., 2009).
NanoSIMS analysis of cyanobacterial trichomes incubated with 13C EPS. (a) Plot of 13C enrichment (APE) of trichomes at 6 h (n=24) and 12 h (n=29) following the addition of 13C label, with points representing values from three biological replicates (1, 2 or 3). ‘Killed’ represents control cells that were fixed before incubation. (b) Scanning electron microscopy image of representative trichomes after 12- h incubation with 13C EPS, taken after NanoSIMS analysis. (c) 12C 14N NanoSIMS image. (d) 13C APE NanoSIMS image. White outlines in bottom two images correspond to areas analyzed in regions of interest.
Relative quantitation of bacterial and cyanobacterial organic carbon incorporation
As the unicyanobacterial culture was not fully axenic, we conducted a second experiment to test whether the cyanobacterium directly incorporates organic carbon, or incorporates 13CO2 liberated because of extracellular organic matter degradation by the other bacteria present in the culture. In this experiment, we compared extracellular organic carbon incorporation at 6 h in dark conditions and compared this with light incorporation. For this second experiment, 13C-labeled EPS-L was harvested from a separate cultured biofilm batch than in the first experiment (67.6 13C APE, diluted 1:30 in media), therefore enrichment levels cannot be directly compared between these two experiments. Dark and light treatments showed significant enrichment over the killed controls (t-test, P<0.05), indicating organic C uptake occurred in the light and dark (Supplementary Figure S3). We calculated maximum bacterial C respiration to account for potential C remineralization. On average, bacteria other than cyanobacteria comprised 1.71%±1.05 of the biofilm biovolume (Supplementary Figure S4,Table S3). When converted from biovolume to μg C per well, the bacteria represented 1.95% (±0.52%) of the total C in biofilms (1.53 and 76.75 μg C per well for bacteria and cyanobacteria, respectively). The bacteria had a mean enrichment at 12 h of 5.44±2.25 13C APE (n=11) and a net carbon fixation of 10.44% (percentage of C incorporated relative to initial C content) (Popa et al., 2007). Given this net carbon fixation, bacteria incorporated approximately 0.16 μg C per well/12 h-1. Assuming a minimum bacterial growth efficiency (amount of new biomass produced per unit organic C assimilation) of 0.01 (Del Giorgio and Cole, 1998), 0.14 μg C could have been released per well in 12 h. Given the 13C enrichment of the EPS-L substrate (0.59 APE), this is approximately 0.08 μg 13C released in 12 h. If the cyanobacterium assimilated all of this respired 13CO2, this would have resulted in a 13C APE of 0.0011. This is only 2.7% of the average observed enrichment in the cyanobacterium at 12 h in the light, indicating that the observed enrichment is mostly a result of assimilation of organic carbon.
Discussion
In microbial mats, EPSs are a large and mostly uncharacterized reservoir of organic C. We set out to determine the basic composition of hypersaline microbial mat EPS and to identify its producers and consumers. Our work suggests that the extracellular milieu of microbial mats is a rich pool of organic compounds, mostly of cyanobacterial origin, and that several community members, including cyanobacteria, degrade and assimilate it.
The glycosyl composition of soluble material recovered in the supernatant of ES mats and the cultured biofilms (loose EPS or EPS-L) was primarily composed of starch- and cellulose-like glucan oligosaccharides, which can only be hydrolyzed to sugars by distinct enzymes. Other studies have reported more complex cyanobacterial exopolysaccharides with approximately 75% of the 140 cultures examined utilizing six or more different monosaccharides (reviewed in Pereira et al., 2009). We detected α-mannose linkages in the EPS-L from our cultured biofilms, indicating more complex structures were secreted than in the EPS from natural mats. We found a large reservoir of loosely bound α- and β-linked (1,4) glucans in EPS fractions from both natural mats and cultured biofilm. This may be common to mat-forming cyanobacteria, many of which secrete glucan exopolysaccharides, but these specific configurations are often not determined. The presence of β-linked (1,4) glucans is not unprecedented, however, as some cyanobacteria produce cellulose (de Winder et al., 1990) and it is often localized in the sheath (Nobles et al., 2001). Within the cyanobacterial mat community, individual members such as those with smaller genomes and specialized lifestyles may not possess the arsenal of degradative enzymes specific for α- and β-linked (1,4) glucans, as well as other less abundant types of oligosaccharides (Medie et al., 2012). The presence of the two linkage types could provide distinct pools of glucose for different organisms in the mat community.
Although there are few published studies describing cyanobacterial exoproteomes, some of the exoproteins we identified have also been detected in other individual cyanobacteria. Investigations in Nostoc commune, Nostoc punctiforme and Anabaena PCC 7120 provide evidence of oxidative stress response proteins in extracellular fractions similar to those found in this study (for example, superoxide dismutase) (Shirkey et al., 2000; Vilhauer et al., 2014; Oliveira et al., 2015). Adhesion, cell wall and secreted proteins were also detected in N. punctiforme (Vilhauer et al., 2014) grown in liquid culture and in an exoproteome from Synechocystis PCC6803 grown in mixotrophic conditions (Gao et al., 2014). Furthermore, active extracellular proteases have been described in N. punctiforme and Synechocystis PCC6803 (Gao et al., 2014; Vilhauer et al., 2014). In contrast to these reports, our work in the ES mat and the cultured biofilm shows carbohydrate and amino-acid metabolism categories contain the largest number of exoproteins (not including unknown proteins).
Contrary to our expectations, exoproteomic data and extracellular enzyme activity measurements in the ES mat and cultured biofilm suggested that the dominant primary producers, cyanobacteria, were degrading oligosaccharides, proteins and nucleic acids. Although it is likely that other bacteria also degrade extracellular organic matter, we were not able to detect as many of their exoenzymes relative to cyanobacterial exoenzymes. We were able to detect exoproteins assigned to Gammaproteobacteria, including several proteins involved in binding and uptake of sugars and amino acids, indicating a role for scavenging by these community members. Enzyme activities confirm that ES mats and unicyanobacterial cultured biofilms are capable of extracellular protein degradation, and α- and β-glucan hydrolysis. Moreover, in both ES mat and cultured biofilm samples we identified 21 homologous cyanobacterial exoproteins, indicating that there are commonly secreted proteins among mat cyanobacteria, and several of these are involved in protein and organic C metabolism. Although EPS biosynthetic and secretion pathways have been proposed in cyanobacteria (Pereira et al., 2009), there are no proposed pathways for EPS degradation or utilization, and regulatory controls of secretion are not well understood. With further study of their regulation, our identified exoproteins may be suitable as cyanobacterial biomarkers for extracellular degradation processes.
Our isotope tracing results show a potential for rapid cyanobacterial turnover of the extracellular C pool, which we define here as ‘reuse’. Along with the degradation capabilities detected, we found that our cultured cyanobacterium rapidly (within 6 h) acquired C from its own extracellular material. Assimilation of extracellular-derived carbon occurred in the light and the dark. Furthermore, the rate of carbon uptake by the cyanobacterium was rapid relative to the slow growth rate of this organism in culture. The biovolume of non-cyanobacteria at the time of the experiment was low relative to the total biovolume, and we calculated that at most 2.7% of the cyanobacterial C uptake could have been a result of bacterial remineralization in the light. This degradation and re-incorporation of organic matter, especially under light conditions, is surprising, given the photoautotrophic role assigned to cyanobacteria in hypersaline laminated microbial mats. Previous work in microbial mats has shown some uptake of glucose and mixed amino acids by mat cyanobacteria during the day (Paerl et al., 1993), and there is evidence of uptake of simple organic carbon compounds by cyanobacteria in culture, including amino acids and phosphonate and even re-uptake of various metabolites (Dyhrman et al., 2006; Gomez-Baena et al., 2008; Zubkov, 2009; Baran et al., 2011). In the case of amino-acid uptake in the marine picocyanobacterium, Prochlorococcus, a day–night uptake rhythm has been observed (Mary et al., 2008). Our results add a new dimension to the understanding of primary producer physiology by demonstrating that a mat cyanobacterium assimilates its excreted organic C, and moreover, excretes enzymes involved in the degradation of more complex organic matter. In further studies, the activity of these exoenzymes can be measured to quantify degradation and could be used to estimate incorporation.
Cyanobacteria likely dictate the bioavailability of extracellular C in natural mats through a balance of degradation and uptake. The turnover of EPS in some mats is 5–8% in 20–30 min (Decho et al., 2005; Dupraz et al., 2009), but the contribution of cyanobacteria to this turnover has not been assessed. Our NanoSIMS results indicate that other bacteria in the cultured biofilm can access a fraction of extracellular organic carbon, and that their average enrichment is higher than that observed in the cyanobacterium. The diversity of proteobacterial transport proteins further supports the bioavailability of EPS carbon to these groups. However, the overall quantitative contribution from other bacteria to this process is likely to be much smaller than that of the cyanobacteria as they comprise a relatively minor component of the total microbial biomass of the cultures (1.71%±1.05%), and in the natural ES mats. In the upper phototrophic layers of natural mats, molecular and microscopic-based data demonstrate that cyanobacteria represent a majority (45–90%) of the active community (Kruschel and Castenholz, 1998; Burow et al., 2013). Given their dominance in terms of overall microbial biomass, even a low level of organic carbon incorporation by cyanobacteria could change the nature of the extracellular pool available to other mat members.
Our observation of degradation and uptake of extracellular organic carbon by cyanobacteria in mats changes our understanding of extracellular organic carbon availability and, consequently, C cycling. Our data suggest that the extracellular space serves as a C store for mat cyanobacteria. Mats are known to be dynamic, with fluctuations of light, pH and temperature on millimeter and hour scales (for example, Bebout et al., 1987), hence being able to store C and take it up quickly under C limitation or osmotic stress could be an advantage. For example, at midday, inorganic C in microbial mats may be virtually unavailable given that the pH is in the range of 9.5–10 (Revsbech et al., 1983). Although it is less likely that these pH changes are occurring in the thin cultured biofilms, this adaptation to mat life may have been retained. Our results provide evidence that the relationship of primary producers with C in dynamic microbial communities is more complicated than fluxes of inorganic carbon alone would indicate. The degradative exoproteins and links between degradation and assimilation that we have identified confirm the important role of cyanobacterial organic C excretion and utilization in complex microbial communities.
References
Baran R, Bowen BP, Northen TR . (2011). Untargeted metabolic footprinting reveals a surprising breadth of metabolite uptake and release by Synechococcus sp. PCC 7002. Mol BioSyst 7: 3200–3206.
Bebout BM, Paerl HW, Crocker KM, Prufert LE . (1987). Diel interactions of oxygenic photosynthesis and N2 fixation (acetylene reduction) in a marine microbial mat community. Appl Environ Microbiol 53: 2353–2362.
Bebout BM, Carpenter SP, Des Marais DJ, Discipulo M, Embaye T, Garcia-Pichel F et al. (2002). Long-term manipulations of intact microbial mat communities in a greenhouse collaboratory: simulating Earth's present and past field environments. Astrobiology 2: 383–402.
Bertlisson S, Berglund O, Pullin MJ, Chisholm SW . (2005). Release of dissolved organic matter by Prochlorococcus. Vie Milieu 55: 225–232.
Bratbak G . (1985). Bacterial biovolume and biomass estimations. Appl Environ Microbiol 49: 1488–1493.
Burow LC, Woebken D, Bebout BM, McMurdie PJ, Singer SW, Pett-Ridge J et al. (2012). Hydrogen production in photosynthetic microbial mats in the Elkhorn Slough estuary, Monterey Bay. ISME J 6: 863–874.
Burow LC, Woebken D, Marshall IP, Lindquist EA, Bebout BM, Prufert-Bebout L et al. (2013). Anoxic carbon flux in photosynthetic microbial mats as revealed by metatranscriptomics. ISME J 7: 817–829.
Canfield DE, Des Marais DJ . (1993). Biogeochemical cycles of carbon, sulfur, and free oxygen in a microbial mat. Geochimica et Cosmochimica Acta 57: 3971–3984.
De Brouwer JFC, Wolfstein K, Stal LJ . (2002). Physical characterization and diel dynamics of different fractions of extracellular polysaccharides in an axenic culture of a benthic diatom. Eur J Phycol 37: 37–44.
De Philippis R, Vincenzini M . (1998). Exocellular polysaccharides from cyanobacteria and their possible applications. FEMS Microbiol Rev 22: 151–175.
de Winder B, Stal LJ, Mur LR . (1990). Crinalium epipsammum sp. nov.: a filamentous cyanobacterium with trichomes composed of elliptical cells and containing poly-β-(1, 4) glucar (cellulose). J Gen Microbiol 136: 1645–1653.
Decho AW . (1990). Microbial exopolymer secretions in ocean environments: their role (s) in food webs and marine processes. Oceanogr Mar Biol 28: 73–153.
Decho AW, Visscher PT, Reid RP . (2005). Production and cycling of natural microbial exopolymers (EPS) within a marine stromatolite. Palaeogeography Palaeoclimatology Palaeoecology 219: 71–86.
Del Giorgio PA, Cole JJ . (1998). Bacterial growth efficiency in natural aquatic systems. Annu Rev Ecol Syst 29: 503–541.
Dubois M, Gilles K, Hamilton JK, Rebers PA, Smith F . (1951). A colorimetric method for the determination of sugars. Nature 168: 167.
Dupraz C, Reid RP, Braissant O, Decho AW, Norman RS, Visscher PT . (2009). Processes of carbonate precipitation in modern microbial mats. Earth-Sci Rev 96: 141–162.
Dyhrman ST, Chappell PD, Haley ST, Moffett JW, Orchard ED, Waterbury JB et al. (2006). Phosphonate utilization by the globally important marine diazotroph Trichodesmium. Nature 439: 68–71.
Everroad RC, Woebken D, Singer SW, Burow LC, Kyrpides N, Woyke T et al. (2013). Draft genome sequence of an oscillatorian cyanobacterium, strain ESFC-1. Genome Announc 1: e00527.
Finzi-Hart JA, Pett-Ridge J, Weber PK, Popa R, Fallon SJ, Gunderson T et al. (2009). Fixation and fate of C and N in the cyanobacterium Trichodesmium using nanometer-scale secondary ion mass spectrometry. Proc Natl Acad Sci USA 106: 6345–6350.
Flemming HC, Wingender J . (2010). The biofilm matrix. Nat Rev Microbiol 8: 623–633.
Fogg G . (1983). The ecological significance of extracellular products of phytoplankton photosynthesis. Botanica Marina 26: 3–14.
Gao L, Huang X, Ge H, Zhang Y, Kang Y . (2014). Profiling and compositional analysis of the exoproteome of Synechocystis sp. PCC 6803. J Metabolomics Syst Biol 1: 8.
Garcia-Pichel F, Castenholz RW . (1991). Characterization and biological implications of scytonemin, a cyanobacterial sheath pigment. J Phycol 27: 395–409.
Garcia-Pichel F, Castenholz RW . (1993). Occurrence of UV-absorbing, mycosporine-like compounds among cyanobacterial isolates and an estimate of their screening capacity. Appl Environ Microbiol 59: 163–169.
Garcia-Pichel F, Mechling M, Castenholz RW . (1994). Diel migrations of microorganisms within a benthic, hypersaline mat community. Appl Environ Microbiol 60: 1500–1511.
Garcia-Pichel F, Belnap J, Neuer S, Schanz F . (2003). Estimates of global cyanobacterial biomass and its distribution. Algol Stud 109: 213–227.
Goebel NL, Edwards CA, Carter BJ, Achilles KM, Zehr JP . (2008). Growth and carbon content of three different-sized diazotrophic cyanobacteria observed in the subtropical North Pacific. J Phycol 44: 1212–1220.
Gomez-Baena G, Lopez-Lozano A, Gil-Martinez J, Lucena JM, Diez J, Candau P et al. (2008). Glucose uptake and its effect on gene expression in Prochlorococcus. PLoS One 3: e3416.
Hagemann M . (2011). Molecular biology of cyanobacterial salt acclimation. FEMS Microbiol Rev 35: 87–123.
Heiss C, Klutts JS, Wang Z, Doering TL, Azadi P . (2009). The structure of Cryptococcus neoformans galactoxylomannan contains beta-D-glucuronic acid. Carbohyd Res 344: 915–920.
Hoehler TM, Bebout BM, Des Marais DJ . (2001). The role of microbial mats in the production of reduced gases on the early Earth. Nature 412: 324–327.
Hoehler TM, Albert DB, Alperin MJ, Bebout BM, Martens CS, Des Marais DJ . (2002). Comparative ecology of H2 cycling in sedimentary and phototrophic ecosystems. Antonie van Leeuwenhoek 81: 575–585.
Jiao Y, Cody GD, Harding AK, Wilmes P, Schrenk M, Wheeler KE et al. (2010). Characterization of extracellular polymeric substances from acidophilic microbial biofilms. Appl Environ Microbiol 76: 2916–2922.
Jørgensen BB, Des Marais DJ . (1986). Competition for sulfide among colorless and purple sulfur bacteria in cyanobacterial mats. FEMS Microbiol Ecol 38: 179–186.
Klock J-H, Wieland A, Seifert R, Michaelis W . (2007). Extracellular polymeric substances (EPS) from cyanobacterial mats: characterisation and isolation method optimisation. Mar Biol 152: 1077–1085.
Kruschel C, Castenholz RW . (1998). The effect of solar UV and visible irradiance on the vertical movements of cyanobacteria in microbial mats of hypersaline waters. FEMS Microbiol Ecol 27: 53–72.
Lee JZ, Burow LC, Woebken D, Everroad RC, Kubo MD, Spormann AM et al. (2014). Fermentation couples Chloroflexi and sulfate-reducing bacteria to Cyanobacteria in hypersaline microbial mats. Front Microbiol 5: 61.
Mary I, Garczarek L, Tarran GA, Kolowrat C, Terry MJ, Scanlan DJ et al. (2008). Diel rhythmicity in amino acid uptake by Prochlorococcus. Environ Microbiol 10: 2124–2131.
McCarren J, Brahamsha B . (2007). SwmB, a 1.12-megadalton protein that is required for nonflagellar swimming motility in Synechococcus. J Bacteriol 189: 1158–1162.
Medie FM, Davies GJ, Drancourt M, Henrissat B . (2012). Genome analyses highlight the different biological roles of cellulases. Nat Rev Microbiol 10: 227–234.
Nobles DR, Romanovicz DK, Brown RM . (2001). Cellulose in Cyanobacteria. Origin of vascular plant cellulose synthase? Plant Physiol 127: 529–542.
Oliveira P, Martins N, Santos M, Couto N, Wright P, Tamagnini P . (2015). The Anabaena sp. PCC 7120 exoproteome: taking a peek outside the box. Life 5: 130–163.
Otero A, Vincenzini M . (2004). Nostoc (Cyanophyceae) goes nude: extracellular polysaccharides serve as a sink for reducing power under unbalanced C/N metabolism. J Phycol 40: 74–81.
Paerl HW, Bebout BM, Joye SB, Des Marais DJ . (1993). Microscale characterization of dissolved organic-matter production and uptake in marine microbial mat communities. Limnol Oceanogr 38: 1150–1161.
Pereira S, Zille A, Micheletti E, Moradas-Ferreira P, De Philippis R, Tamagnini P . (2009). Complexity of cyanobacterial exopolysaccharides: composition, structures, inducing factors and putative genes involved in their biosynthesis and assembly. FEMS Microbiol Rev 33: 917–941.
Popa R, Weber PK, Pett-Ridge J, Finzi JA, Fallon SJ, Hutcheon ID et al. (2007). Carbon and nitrogen fixation and metabolite exchange in and between individual cells of Anabaena oscillarioides. ISME J 1: 354–360.
Porter KG, Feig YS . (1980). The use of DAPI for identifying and counting aquatic microflora. Limnol Oceanogr 25: 943–948.
Reed RH, Warr SRC, Kerby NW, Stewart WDP . (1986). Osmotic shock-induced release of low molecular weight metabolites from free-living and immobilized cyanobacteria. Enzyme Microbial Technol 8: 101–104.
Revsbech NP, Jorgensen B, Blackburn TH, Cohen Y . (1983). Microelectrode studies of the photosynthesis and O2, H2S, and pH profiles of a microbial mat. Limnol Oceanogr 28: 1062–1074.
Rippka R . (1988). Isolation and purification of cyanobacteria. Methods Enzymol 167: 3–27.
Santander J, Martin T, Loh A, Pohlenz C, Gatlin DM III, Curtiss R III . (2013). Mechanisms of intrinsic resistance to antimicrobial peptides of Edwardsiella ictaluri and its influence on fish gut inflammation and virulence. Microbiology (Reading, England) 159: 1471–1486.
Shirkey B, Kovarcik DP, Wright DJ, Wilmoth G, Prickett TF, Helm RF et al. (2000). Active Fe-containing superoxide dismutase and abundant sodF mRNA in Nostoc commune Cyanobacteria after years of desiccation. J Bacteriol 182: 189–197.
Stal LJ, Moezelaar R . (1997). Fermentation in cyanobacteria. FEMS Microbiol Rev 21: 179–211.
Tamaru Y, Takani Y, Yoshida T, Sakamoto T . (2005). Crucial role of extracellular polysaccharides in desiccation and freezing tolerance in the terrestrial cyanobacterium Nostoc commune. Appl Environ Microbiol 71: 7327–7333.
Vilhauer L, Jervis J, Ray WK, Helm RF . (2014). The exo-proteome and exo-metabolome of Nostoc punctiforme (Cyanobacteria) in the presence and absence of nitrate. Arch Microbiol 196: 357–367.
Wingender J, Strathmann M, Rode A, Leis A, Flemming HC . (2001). Isolation and biochemical characterization of extracellular polymeric substances from Pseudomonas aeruginosa. Methods Enzymol 336: 302–314.
Woebken D, Burow LC, Prufert-Bebout L, Bebout BM, Hoehler TM, Pett-Ridge J et al. (2012). Identification of a novel cyanobacterial group as active diazotrophs in a coastal microbial mat using NanoSIMS analysis. ISME J 6: 1427–1439.
Wright DJ, Smith SC, Joardar V, Scherer S, Jervis J, Warren A et al. (2005). UV irradiation and desiccation modulate the three-dimensional extracellular matrix of Nostoc commune (Cyanobacteria). J Biol Chem 280: 40271–40281.
Zubkov MV . (2009). Photoheterotrophy in marine prokaryotes. J Plankton Res 31: 933–938.
Acknowledgements
We thank Leslie Prufert-Bebout (NASA) for isolating and providing ESFC-1 and for comments on the manuscript; Whitney Stannard (LLNL) for technical assistance with cultures; Mark Boggs (LLNL) for initial NMR analyses of EPS samples; Heather Dang (UC Berkeley) for isotope-ratio mass spectrometry analysis; Michelle Salemi and Brett Phinney for analysis at the UC Davis Proteomics Core Facility; and Parastoo Azadi and Christian Heiss for analyses carried out at the Complex Carbohydrate Research Center at the University of Georgia, Athens, supported in part by DOE grant DE-FG02-93ER20097, ‘Center for Plant and Microbial Complex Carbohydrates’. We also thank Jeff Cann, Associate Wildlife Biologist, Central Region, California Department of Fish and Wildlife for coordinating access to the Moss Landing Wildlife Area; and Tijana Glavina del Rio and the DOE Joint Genome Institute (JGI) staff for sequencing and bioinformatics support (as part of JGI Community Sequencing Project #701). This work was performed under the auspices of the US Department of Energy by Lawrence Livermore National Laboratory under Contract DE-AC52-07NA27344, and was supported by the US Department of Energy, Office of Science, under contract number SCW1039 supported by the Genome Sciences Program of the Office of Biological and Environmental Research. Institution Paper Number LLNL-JRNL-667325.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies this paper on The ISME Journal website
Supplementary information
Rights and permissions
This work is licensed under a Creative Commons Attribution-NonCommercial-ShareAlike 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-sa/4.0/
About this article
Cite this article
Stuart, R., Mayali, X., Lee, J. et al. Cyanobacterial reuse of extracellular organic carbon in microbial mats. ISME J 10, 1240–1251 (2016). https://doi.org/10.1038/ismej.2015.180
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ismej.2015.180
This article is cited by
-
Impact of different nitrogen additions on microbes and exopolysaccharides excretion in cyanobacterial biocrusts
Plant and Soil (2023)
-
Biocidal H2O2 treatment emphasizes the crucial role of cyanobacterial extracellular polysaccharides against external strong oxidative stress
Environmental Science and Pollution Research (2023)
-
Metagenomic, (bio)chemical, and microscopic analyses reveal the potential for the cycling of sulfated EPS in Shark Bay pustular mats
ISME Communications (2022)
-
Novel functional insights into a modified sugar-binding protein from Synechococcus MITS9220
Scientific Reports (2022)
-
Reduction of the wind erosion potential in dried-up lakebeds using artificial biocrusts
Frontiers of Earth Science (2022)