Abstract
Members of the phylum Bacteroidetes are abundant in many marine ecosystems and are known to have a pivotal role in the mineralization of complex organic substrates such as polysaccharides and proteins. We studied the decomposition of the algal glycans laminarin and alginate by ‘Gramella forsetii’ KT0803, a bacteroidetal isolate from North Sea surface waters. A combined application of isotope labeling, subcellular protein fractionation and quantitative proteomics revealed two large polysaccharide utilization loci (PULs) that were specifically induced, one by alginate and the other by laminarin. These regulons comprised genes of surface-exposed proteins such as oligomer transporters, substrate-binding proteins, carbohydrate-active enzymes and hypothetical proteins. Besides, several glycan-specific TonB-dependent receptors and SusD-like substrate-binding proteins were expressed also in the absence of polysaccharide substrates, suggesting an anticipatory sensing function. Genes for the utilization of the beta-1,3-glucan laminarin were found to be co-regulated with genes for glucose and alpha-1,4-glucan utilization, which was not the case for the non-glucan alginate. Strong syntenies of the PULs of ‘G. forsetii’ with similar loci in other Bacteroidetes indicate that the specific response mechanisms of ‘G. forsetii’ to changes in polysaccharide availability likely apply to other Bacteroidetes. Our results can thus contribute to an improved understanding of the ecological niches of marine Bacteroidetes and their roles in the polysaccharide decomposition part of carbon cycling in marine ecosystems.
Similar content being viewed by others
Introduction
Bacteroidetes represent important contributors to the mineralization of complex organic substrates such as proteins and polysaccharides in the marine realm (Kirchman 2002; Bauer et al., 2006; González et al., 2008; Teeling et al., 2012; Fernández-Gómez et al., 2013). An important source of such organic matter is the marine phytoplankton that has been estimated to be responsible for about 50% of the global net primary production (Field et al., 1998). Polysaccharides constitute a substantial fraction of this primary production, and algae can be composed of up to 50% or more of polysaccharides (Kraan, 2012). Marine Bacteroidetes are assumed to have a key role in the utilization of these algal polysaccharides (Kirchman, 2002; Pinhassi et al., 2004; Bauer et al., 2006; González et al., 2008; Teeling et al., 2012; Fernández-Gómez et al., 2013), but only for few cultivable members this has been investigated in functional studies so far (Thomas et al., 2012). Recently, it could be shown that during a North Sea diatom bloom specialized populations of Bacteroidetes coacted in the successive decomposition of algal-derived organic matter (Teeling et al., 2012). This supports the assumption that the wealth of naturally occurring algal polysaccharide structures and compositions provides distinct ecological niches for heterotrophic marine bacteria (Grossart et al., 2005; Hehemann et al., 2010; Gomez-Pereira et al., 2010; Gomez-Pereira et al., 2012). However, so far respective niche specializations of different members of the Bacteroidetes are only poorly resolved. In order to gain deeper insights into the relationship between abundant algal polysaccharides and the gene repertoires of adapted Bacteroidetes, we studied the isolate ‘Gramella forsetii’ KT0803 (Eilers et al., 2001). This strain belongs to the class Flavobacteria of the phylum Bacteroidetes and has been isolated from eutrophic coastal surface waters near the island Helgoland in the German Bight (54°11′03′′N, 7°54′00′′E). A previous whole-genome analysis (Bauer et al., 2006) of ‘G. forsetii’ has revealed various genetic modules encoding carbohydrate-active enzymes (CAZymes) and transporters, frequently arranged in so-called polysaccharide utilization loci (PULs; Sonnenburg et al., 2010). Such PULs have also been reported for commensal Bacteroidetes from the human intestine, and are specifically induced by oligosaccharides (Martens et al., 2011), including such of algal origin (Hehemann et al., 2012). In this proteogenomic study, ‘G. forsetii’ was studied during growth on two typical algal glycans, laminarin and alginate, in comparison to a glucose-supplemented control. Beta-1,3-glucans of the laminarin type act as a storage polysaccharide in brown algae, Chrysophyceae, diatoms and haptophytes such as Phaeocystis spp. where they can make up about 30–50% of the cell dry weight (Beattie et al., 1961; Janse et al., 1996; Davis et al., 2003). Alginate on the other hand is a cell wall polysaccharide as well as an intracellular component in brown algae that can account for up to 40% of algal dry weight (Haug et al., 1966; Davis et al., 2003). It is an unbranched copolymer composed of 1,4-linked beta-D-mannuronate (M) and alpha-L-guluronate (G) that are arranged in alternating M-blocks, G-blocks and mixed M/G-blocks. Relevant proteins for the decomposition of these polysaccharides were determined by stable isotope labeling, followed by subcellular fractionation and high-accuracy mass spectrometry. This combinatory approach allowed for an enhanced precision in protein quantification and revealed two major PULs of ‘G. forsetii’ that were specifically controlled by either alginate or laminarin. Data of this study furthermore indicate a cross-regulation of genes acting on similar polysaccharides such as laminarin and 1,4-glucans and revealed that some proteins so far designated as ‘hypothetical’ might contribute to polysaccharide decomposition. Particular progress was achieved with respect to possible substrate specificities within the highly variable group of TonB-dependent receptors (TBDRs), which are known to be of crucial importance for uptake of glycans by marine Gram-negative bacteria (Tang et al., 2012).
Materials and methods
Culture conditions and metabolic labeling
‘G. forsetii’ KT0803 was grown at 25 °C and 170 r.p.m. in an artificial seawater medium, as described by Schut et al. (1993), in batches of 100 ml in 500 ml shaking flasks until the mid-exponential phase (OD600≍0.5). The medium was supplemented with either 14N naturally labeled or 15N-labeled ammonium chloride (‘isotopic tracer’, 99%; Euriso-Top, Saint-Aubin Cedex, France). As algae-specific carbon sources, laminarin (Sigma-Aldrich, Munich, Germany; source: brown alga Laminaria digitata; composition: about 25 glucose units) and sodium alginate (Sigma-Aldrich; source: brown algae; composition: 61% mannuronic acid, 39% guluronic acid with 60–400 uronic acid units) were added to a final concentration of 0.2%. The control cultures were supplemented with glucose (Carl Roth GmbH & Co. KG, Karlsruhe, Germany) to a final concentration of 0.2%. All of these experiments were carried out in triplicates (Supplementary Figure S1A).
Proteome fractionation
Cellular protein extracts were separated into three different fractions: the intracellular soluble, the membrane-bound and the extracellular soluble proteome (Supplementary Figure S1B). For purification of extracellular proteins, cells were centrifuged (6600 g at 4 °C for 10 min) and the resulting supernatant was precipitated at 4 °C using 10% trichloroacetic acid, according to the protocol published by Antelmann et al. (2001). The precipitated proteins were pelleted via centrifugation (40 000 g at 4 °C for 60 min), washed with ethanol and solubilized in 8 M urea and 2 M thiourea. To extract the membrane-bound and intracellular protein fractions, the cells were disrupted in a French press (Simaminco SLM, Rochester, NY, USA; three cycles) followed by removal of cell debris by centrifugation (40 000 g at 4 °C for 20 min). Subsequent ultracentrifugation (100 000 g at 4 °C for 60 min) allowed for separation of intracellular soluble and membrane-bound proteins. Purification of the membrane fraction was performed, as described by Eymann et al. (2004), omitting the n-dodecyl-β-D-maltoside treatment. Accordingly, pelleted bacterial membranes were homogenized, washed for 30 min with a highly concentrated saline solution (20 mM Tris-HCl pH 7.5, 10 mM EDTA and 1 M NaCl) and ultracentrifuged (100 000 g at 4 °C for 60 min). Following this, the same steps were carried out using a carbonate buffer (100 mM Na2CO3-HCl pH 11, 10 mM EDTA and 100 mM NaCl). Finally, purified membrane and intracellular proteins were solubilized in 50 mM triethylammonium bicarbonate buffer (Sigma-Aldrich) supplemented with the Complete Protease Inhibitor (Roche, Berlin, Germany). After determination of the protein concentration, triplicates of 15N-labeled protein extracts from all three different subcellular fractions were combined and then mixed with equal amounts of individual 14N-labeled protein samples in a 1:1 ratio for all substrates before proteome analysis.
1D-PAGE and protein measurements
From each 14N-labeled triplicate combined with the 15N-labeled pooled protein extracts, 15 μg of each fraction were separated by 1D-polyacrylamide gel electrophoresis (10% acrylamide, protein ladder by Thermo Fisher Scientific Inc., Waltham, MA, USA), stained with Coomassie G-250 ‘blue silver’ (Candiano et al., 2004), eluted from the gel and digested using trypsin. Peptides were subsequently desalted by ZipTip columns (Millipore, Billerica, MA, USA) according to the manufacturer’s guidelines. For liquid chromatography-MS/MS measurements, peptides were subjected to a reversed-phase C18 column chromatography operated on a nanoACQUITY-UPLC (Waters Corporation, Milford, MA, USA) and separated as described by Otto et al. (2010). Mass spectrometry (MS) and MS/MS data were determined using an online-coupled LTQ-Orbitrap mass spectrometer (Thermo Fisher Scientific Inc.).
Data processing
The *.dta files were extracted from *.raw files with the BioworksBrowser 3.3.1 SP1 (Thermo Fisher Scientific Inc.) with no charge state deconvolution and deisotoping. The data were searched by SEQUEST version v28 (rev.12, Thermo Fisher Scientific Inc.) against a ‘G. forsetii’ target-decoy protein sequence database. A set of common laboratory contaminants was included and all data were compiled using the BioworksBrowser. The following search parameters were used: enzyme type, trypsin (KR); peptide tolerance, 10 p.p.m.; tolerance for fragment ions, 1 a.m.u.; b- and y-ion series; variable modification, methionine (15.99 Da). A maximum of three modifications per peptide was allowed. In the second iteration, the mass shift of all amino acids completely labeled with 15N-nitrogen was taken into account in the search parameters. Resulting *.dta and *.out files were assembled and filtered using DTASelect (version 2.0.25; parameters: -y 2 -c 2 -C 4 —here —decoy Reverse_ -p 2 -t 2 -u —MC 2 -i 0.3 —fp 0.005).
Relative protein quantification and analysis of variance
Relative quantification of the shotgun proteomics data was carried out according to the principle of pooled reference samples published by MacCoss et al. (2003) using the Census software (Park et al., 2008) as described in Otto et al. (2010). Basically, according to the filtered database search peptide identification data provided by DtaSelect, both intensities for 14N- (sample) and 15N- (pooled reference) mass traces were extracted for each peptide identification in the respective mass spectrometric data and were compared within Census to yield a ratio between the sample mass trace (14N) and the pooled reference mass trace (15N). These peptide ratios were then combined to yield sample/reference ratio values on the protein level. Finally, peptide ratios and combined protein ratios were exported (R2-values >0.7 and only unique peptides; proteins failing to be relatively quantified were checked manually in the graphical user interface for on/off proteins). Only proteins identified and quantified with at least two peptides were taken into account for further analyses. For correcting any potential systematic bias that might have been introduced by pooling of the 15N-labeled samples and mixing with unlabeled protein samples, the quantitative data of a complete GeLC-MS run were median-centered. For all ratios, log2-values and averages of the biological replicates were determined. TMEV (version 4.4; Saeed et al., 2003) was applied to carry out the analysis of variance with the following settings: number of groups 5; P-values based on permutation (1000) resulting in P-values <0.01 as significance threshold. Quantification and analysis of variance data are provided in Supplementary Table S1.
Gene functions and protein localization
Pairwise identities of homologous genes were determined by means of the Geneious alignment software (Drummond et al., 2010). Details on protein functions were obtained from precomputed data available on the UniProt Knowledgebase (Magrane, UniProt consortium 2011), the ENZYME database (Bairoch, 2000) and by using the JCoast software (Richter et al., 2008). Information on the classification of CAZymes was derived from the CAZy database (Cantarel et al., 2009). Subcellular localizations of all proteins were predicted according to Romine (2011); the data are provided in Supplementary Table S2.
Results and Discussion
Proteome analysis
In the present proteome analysis, 1691 proteins were identified, which corresponds to 47% of all protein-coding genes in ‘G. forsetii’. A considerable proportion (12%) of identified proteins could be assigned to the outer membrane, with one-third showing significant upregulation in alginate- or laminarin-grown cultures. Proteins enriched in the membrane fraction most frequently consisted of transporters (12%, mainly TBDRs), CAZymes (5%, primarily glycosyltransferases, GTs), signal-transducing components (3%), SusD-like proteins (2%) and proteins with unknown functions (35%).
A genome-wide in silico localization prediction indicated that the ‘G. forsetii’ extracellular proteins are in most cases attached to its outer membrane (Supplementary Table S2). This was supported by broad overlaps of proteins detected in the extracellular and the membrane proteome (Supplementary Figures S2A and S3). The exposure of proteins to the extracellular space is essential for bacteria in order to efficiently bind macromolecular substrates and decompose them into smaller components that are suitable for uptake into the cell. Conceivably, it would be an economical advantage for marine microorganisms to keep the majority of substrate-binding proteins and hydrolytic enzymes to themselves by attaching them to the cell envelope rather than secreting them into the surrounding water. Such a strategy would prevent a loss of these proteins by diffusion and increase the efficiency of breakdown/uptake coupling (see also Azam and Malfatti, 2007). Intriguingly, at the same time, such membrane-anchored enzyme systems would be a means of cell attachment to nutrient-rich particles like marine snow. Indeed, about 74% of all secreted proteins found in the extracellular fraction of ‘G. forsetii’ were anchored to the outer membrane and thus associated with the cell (Supplementary Figures S2 and S3, Supplementary Table S3).
The proteome analysis revealed that both laminarin and alginate notably affected the protein expression patterns of the membrane and extracellular fractions, whereas distinct quantitative changes of proteins from the intracellular fraction were observed in glucose-grown cultures (Supplementary Figure S2B). In total, a differential expression of 703 proteins (20% of all annotated genes) was observed. Seven outer membrane proteins showed a particular strong increase in abundance. These proteins are encoded in two large alginate- and laminarin-dependent PULs (Supplementary Figures S4A and B). The membrane fraction from glucose-grown cells was mainly dominated by inner membrane enzymes with functions in signal transduction, protein translocation, electron transport and ATP synthesis (Supplementary Figure S4C). Moreover, certain TBDRs were found to be highly abundant in all investigated cell samples.
Laminarin and alginate utilization loci
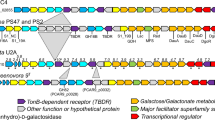
PUL-like systems are frequently found in members of the Bacteroidetes and typically comprise outer membrane Sus-like lipoproteins, such as SusC-like TonB-dependent transporters and SusD-like substrate-binding proteins, which function in concert with linked CAZymes, as described for members of the anaerobic genus Bacteroides of the human intestine (Martens et al., 2009, 2011). The proteome analysis of this study demonstrated that in ‘G. forsetii’ two PULs are involved in alginate and laminarin degradation. These PULs comprise common components (TBDR, SusD, PKD domain proteins and CAZymes) and were found to be present in other marine Flavobacteriaceae as well (Figures 1 and 2).
Expression profile (a) and gene organization of the laminarin utilization operon. (a) Protein ratios of the intracellular soluble and membrane protein fractions of cultures grown on laminarin compared with alginate are shown. Except for a signal transduction system (GFO_3462), all genes of the laminarin operon were overexpressed in the presence of laminarin. Consistency in protein levels is highlighted by the gray background that combines all data points of the corresponding substrate and indicates a uniform cluster regulation. Error bars refer to the s.d. of all biological replicates (n=3), median-centered protein ratios are given in Supplementary Table S4. (b) The gene organization of the laminarin-specific PUL in ‘G. forsetii’ is conserved among other marine Flavobacteriaceae as exemplified. Identity/similarity values are listed in Supplementary Table S9. htcs, hybrid two-component system; susC, SusC-like TBDR; susD, SusD-family protein; pkd, PKD-domain-containing protein; gh, glycoside hydrolase.
Differential expression (a) and genomic structure (b) of the alginate utilization cluster. (a) Shown are protein ratios of the intracellular soluble and membrane protein fractions of cultures grown on alginate compared with laminarin. The gray background marks the clustering of protein patterns in corresponding cultures. According to the CAZy database, GFO_1150 and 1151 are fragments of a single PL17 enzyme, which probably hindered its protein identification. Error bars indicate the s.d. of the biological replicates (n=3), protein ratio values can be found in Supplementary Table S9. (b) The four putative operons of the alginate-specific PUL are flanked by thermostable stem-loop structures of possible transcriptional terminators (T, terminator; changes in Gibbs free energy: delta G(T1) −6.9 kcal mol−1, delta G(T2) −20.0 kcal mol−1, delta G(T3) −21.3 kcal mol−1, delta G(T4) −8.1 kcal mol−1). Homologous gene equivalents are present in related marine Flavobacteriaceae (identity/similarity values are listed in Supplementary Table S5). Dotted lines mark genes that belong to a separate operon, but were considered in this overview, because of their orthology to corresponding genes in ‘G. forsetii’. susC, SusC-like TBDR; susD, SusD-family protein; pkd, PKD-domain-containing protein; pl, polysaccharide lyase; tim, TIM-barrel-domain-containing protein; kdgF, pectin degradation protein; gntR, GntR family transcriptional regulator; mfs, MFS permease; sdr, short-chain dehydrogenase/reductase family protein with a putative function as 2-dehydro-3-deoxy-gluconate-6-dehydrogenase; kdgK, 2-dehydro-3-deoxy-gluconokinase; kdgA, KDPG/KHG aldolase; fbp, fructose-1,6-bisphosphatase; lacI, LacI family transcriptional regulator; hyp, hypothetical protein.
The laminarin-dependent PUL of ‘G. forsetii’ encodes three 1,3-beta-glucanases, that is, two family-16 glycoside hydrolases (GH16 laminarinases) and one GH3 (Supplementary Table S4). On the basis of their localization at the cell surface and according to information in the CAZy database (Cantarel et al., 2009) both GH16 proteins (GFO_3466 and 3468) may constitute endo-acting enzymes that initiate polymer breakdown by randomly cleaving laminarin into smaller oligosaccharides. The latter can be taken up by a TonB-dependent transporter into the periplasmic space, where subsequent hydrolysis of terminal glucose residues by the presumably exo-acting GH3 (GFO_3467) may take place (Figure 3). Both, the hypothetical outer membrane protein (GFO_3465, Supplementary Table S7) encoded in this PUL, and one of the two GH16 laminarinases (GFO_3466) feature a PKD domain, known to participate in near-surface protein–protein interactions (Jing et al., 2002). This suggests the existence of a physical link between the proteins, possibly connecting the substrate-binding SusD and the glycoside hydrolases.
Tentative model of the alginate (a) and laminarin (b) utilization pathways in ‘G. forsetii’. Protein localizations were predicted in silico according to (Romine, 2011); Supplementary Table S2). (a) Four outer membrane alginate lyases of polysaccharide families 6 and 7 (PL6 and PL7, endo-acting according to ExPASy) probably catalyze the depolymerization of alginate into hetero- or homo-oligomeric alpha-L-guluronic acid (GA) and beta-D-mannuronic acid (MA). The PKD-domain-containing protein (PKD) and a SusD-related oligosaccharide-binding protein (SusD) may function as bridges delivering the oligomeric substrate to the TBDR. Transfer through the outer membrane is mediated by a linkage of the TBDR to the energizing TonB–ExbBD complex. According to their suggested localization, periplasmic alginate lyases (putative oligoalginate lyase PL17 and guluronate/ mannuronate lyase PL7) might exhibit oligoalginate specificity. This allows for breakdown into 4-deoxy-5-keto-mannuronic or guluronic acid (KMA and KGA). The monomers can be transported into the cytoplasm by means of an MFS permease and step by step converted into 2-keto-3-deoxygluconate (KDG; by a putative 2-dehydro-3-deoxy-gluconate-6-dehydrogenase, Sdr), keto-deoxy-phospho-gluconate (KDPG; by a 2-dehydro-3-deoxy-gluconokinase, KdgK) and pyruvate (Pyr) as well as glyceraldehyde-3-phosphate (GAP; by KDPG aldolase, KdgA). (b) The polysaccharide laminarin is metabolized into monomeric glucose by two outer membrane endo-1,3-beta-glucanases (GH16) and a periplasmic exo-1,3-beta-glucanase (GH3). Nutrient-binding and outer membrane transport are carried out by the same protein families as present in the alginate utilization pathway (PKD, SusD and TBDR). Uptake of glucose into the cytoplasm is conducted by a constitutively expressed MFS permease (MFS) or ABC-type transporter (ABC), followed by glycolysis resulting in the generation of pyruvate.
The much larger alginate-dependent PUL encompasses four operons (Figure 2, Supplementary Tables S5). Upstream of the typical activity unit (operon 3 with TBDR, SusD, PKD and CAZymes) three polysaccharide lyase (PL) genes are located (operon 1 and 2 with PL6, PL7 and PL17). Downstream, an inner membrane transporter and genes for cytoplasmic processing of carbohydrates are encoded (operon 4 with genes for MFS permease, Sdr, KdgK and KdgA). On the basis of our expression analysis and consistent with transcriptome studies on the Flavobacterium species Zobellia galactanivorans (Thomas et al., 2012), ‘G. forsetii’ gradually catabolizes alginate into 2-keto-3-deoxy-6-phosphogluconate (KDPG), which is likely directly funneled into the Entner–Doudoroff pathway (Preiss and Ashwell, 1962; Figure 3). A highly coordinated action of all alginate lyases (in operon 1, 2 and 3) and the cytoplasmic proteins 2-dehydro-3-deoxy-D-gluconate 6-dehydrogenase (Sdr), 2-dehydro-3-deoxygluconokinase (KdgK) and KDPG aldolase (KdgA; operon 4) is strongly supported by the proteome analysis, which revealed a consistent regulation pattern of these proteins within the alginate PUL (Supplementary Table S5).
The PKD-domain-containing proteins present in both, the alginate (GFO_1155) and the laminarin cluster (GFO_3465), most likely perform different functions as is indicated by their low similarity (51.4%) and striking differences in molecular weight (34.0 kDa for the alginate- and 96.6 kDa for the laminarin-induced proteins). PKD-domain-containing proteins and alginate lyases or laminarinases, respectively, were only upregulated in the two PULs described.
Carbohydrate transporters and associated SusD-related proteins
Efficient nutrient uptake is a critical step in the competition of marine bacterioplankton. In heterotrophic Gram-negative bacteria, TBDRs/transporters have a pivotal role in the uptake of complex organic matter such as from algal polysaccharides. This is especially reflected by a high number of TBDR-encoding genes in the genomes of marine Flavobacteria, compared with monomer specialists within the class Alphaproteobacteria (Fernández-Gómez et al., 2013). The importance of TBDR in marine bacterioplankton is corroborated by in situ proteome studies of samples from the South Atlantic (in particular at coastal upwelling zones; Morris et al., 2010), the Antarctic Southern Ocean (Williams et al., 2012) and the coastal North Sea (Teeling et al., 2012) where TBDRs dominated the set of expressed transporters. These transporters consist of an outer membrane TBDR that is linked to the cytoplasmic membrane via a TonB–ExbBD complex. The latter allows usage of the proton motive force across the cytoplasmic membrane for transport through the outer membrane by the selective opening of an outer membrane pore (Krewulak and Vogel 2011). Owing to their substrate-binding function and high sequence diversity, the TBDR component is suggested to mediate substrate specificity (Schauer et al., 2008; Tang et al., 2012).
Our proteome study identified 33 of the 40 ‘G. forsetii’ TBDRs (83%) in the membrane fraction, 21 (53%) of which were differentially regulated under the three investigated substrate conditions (Figure 4). These findings suggest that some TBDRs are constitutively expressed and act as substrate sensors, whereas other TBDRs are dynamically upregulated only when these substrates become available. Laminarin exerted the most significant influence on TBDR expression (28% of TBDR genes responded to laminarin, 13% to alginate; Supplementary Figure S5). Among the laminarin-induced TBDRs we found (I) one encoded in the laminarin-specific PUL (GFO_3463), (II) one encoded next to a gene for a highly induced hypothetical inner membrane protein (Supplementary Table S7, GFO_2191) and (III) three TBDRs that were also identified in glucose-grown cultures (GFO_0361, 2138 and 1,4-alpha-glucanase associated; GFO_1029 next to two large outer membrane lipoproteins with adhesion domains), indicating regulatory overlaps between laminarin and its monomeric component glucose.
Regulation patterns of (a) TBDRs and (b) TBDR-SusD PULs frequently clustered with CAZymes. Alginate- and laminarin-dependent relative quantitative changes in protein amounts of SusD-independent and SusD-associated TBDRs as enriched in the membrane fraction. Protein profiles illustrate similarities in regulation patterns of TBDRs and corresponding SusD proteins. Colocalization of CAZymes was almost exclusively determined for operons containing SusD-related proteins with domain PF07980. Except for those associated with the major alginate or laminarin-specific PUL (in bold), all CAZyme genes in these operons remained unexpressed (in italics). Locus tags of ‘G. forsetii’ (GFO) proteins are indicated.
Half of all TBDR genes (20 of 40) were located next to genes encoding outer membrane SusD-like proteins. These proteins are unique to Bacteroidetes (Thomas et al., 2011), and have been suggested to be involved in oligosaccharide binding as well as to interact with TBDRs (Shipmann et al., 2000; Cho and Salyers, 2001). In addition to the 15 known SusD family genes (PF07980; Figure 4b upper section; Bauer et al., 2006), this study revealed five additional genes with novel SusD-like domains (PF12771 and PF12741; Figure 4b, lower section) in the genome of ‘G. forsetii’. Fourteen (70%) of these in total 20 susD-related genes were expressed and exhibited similar expression patterns as their colocalized TBDRs, but in comparison with the TBDRs a lower fraction of the susD-like genes were regulated (only 25% of susD genes were regulated, mainly laminarin-dependent with glucose overlaps). Alginate- or laminarin-induced upregulation of TBDR genes occurred irrespective of whether these were colocated with susD genes (50%, Figure 4b) or not (45%, Figure 4a).
Tandems of susC and susD genes in ‘G. forsetii’ are frequently associated with CAZyme genes (45%; Bauer et al., 2006). However, only few SusD-like proteins from GH-associated PULs were expressed under the tested conditions (44%, in contrast to 82% in non-GH-containing operons). Such GH-linked susD-like genes belonged to (I) the alginate and laminarin PUL (GFO_1154 and 3464) and were also associated with genes for (II) an alpha-amylase (GH13; SusD-like GFO_2139) and (III) a fructan-specific hydrolase (GH32; SusD-like GFO_0011). The GH32-associated susD-like gene belongs to an operon (GFO_0008-11, Supplementary Figure S6, Supplementary Table S9) that comprises a glucose-specific TBDR gene (GFO_0010). Surprisingly, the GH13-associated susD-like gene is linked to an aforementioned laminarin-induced susC-like gene (GFO_2138), which is part of a 1,4-alpha-glucan utilizing PUL (GFO_2129-41; Figure 5, Supplementary Table S9).
Proposed pathway for the decomposition of alpha-1,4-glucans in ‘G. forsetii’ (a), organization of the cluster and its protein profile (b). (a) An alpha-1,4-linked glucose-polymer (glycogen, starch and amylose) is converted into oligomeric dextrin (by GH13 alpha-amylase, GFO_2132, GFO_2141 or GFO_2133), dimeric maltose (by dextrinase function of GFO_2133), glucose 1-phosphate (Glc-1P; by GH65 maltose phosphorylase, GFO_2134) and finally glucose-6-phosphate (Glc-6P; by PgmB, GFO_2135). The latter metabolite can be directly used by glycolysis and metabolized into pyruvate (Pyr). (b) Intracellular components contained in the operon located upstream were upregulated in both glucose- and laminarin-supplied cultures (†, highest protein quantity changes in glucose-grown cultures), whereas membrane proteins of the downstream located operon exhibited upregulation particularly in laminarin-grown cultures (‡P<0.01). Except for the two alpha-amylases (GFO_2132 and GFO_2141) and the LacI family regulator, all genes were found to be expressed in this proteome study. According to the predicted subcellular localization, log2 ratio values are shown for the respective subcellular fraction of each identified protein (error bars: s.d.; protein ratios: see Supplementary Table S9). Locus tags are given as numbers below the genes. hyp, hypothetical protein; gapA, glyceraldehyde-3-phosphate dehydrogenase A; pfkA, 6-phosphofructokinase; gh, glycoside hydrolase family; pgmB, beta-phosphoglucomutase; mfs, MFS permease—possibly alpha-glucoside transporter; lacI, LacI family transcriptional regulator; susC, SusC-like TBDR; susD, SusD-family protein; susE, putative SusE-like protein.
Among the group of inner membrane transporters, differential expression was detected for three genes of the major facilitator superfamily (MFS) transporters (5 of 18 genes identified on protein level). On the basis of their annotation, the regulated secondary carriers are specific for hexuronate/hexarate (GFO_1159, upregulated in alginate), alpha-glucosides and galactose/glucose (GFO_2136 and 3326, both exclusively detected in glucose cultures). In contrast, high numbers of ATP-binding cassette (ABC) transporters (20 of 28 genes) were found in the proteome of all examined substrates, but exhibited relatively constant, and therefore substrate-independent, expression levels. Like other marine Bacteroidetes, ABC transporters of ‘G. forsetii’ almost entirely lack periplasmic solute-binding proteins that usually mediate substrate specificity (Bauer et al., 2006). This stands in stark contrast to the expression of such periplasmic-binding proteins in Alphaproteobacteria, which was recently found in a metaproteomic study of Antarctic surface waters (Williams et al., 2012), and might indicate a housekeeping function of ABC transporters in ‘G. forsetii’.
CAZymes
GTs participate in the biosynthesis of glycans and glycoconjugates that mediate various functions such as surface adhesion, biofilm formation, signaling or energy and carbon storage (Taniguchi et al., 2002). Genome analyses of several marine Bacteroidetes (Bauer et al., 2006; Fernández-Gómez et al., 2013) have shown high abundances of GT genes. Of the 60 genes predicted for ‘G. forsetii’ (Bauer et al., 2006), 39 were proteomically identified in this study, with a dominance of the families GT1 and GT2. Probably owing to their basic cellular function, expression of GTs was similar under all substrate conditions.
Many members of glycoside hydrolase families exhibited non-differential expression or were not identified (Supplementary Table S8). Interestingly, however, two colocated genes, one encoding a GH13 (amylase/cyclomaltodextrinase, GFO_2133) and one encoding a GH65 (maltose phosphorylase, GFO_2134) were significantly upregulated in glucose- and laminarin-grown cultures. Both the genes affiliated to a large alpha-1,4-D-glucan PUL (Supplementary Figure 5, Supplementary Table S9) together with adjacent glycolytic genes. The maltose phosphorylase likely obtains its substrate maltose from the adjacent amylase/cyclomaltodextrinase that, owing to its dual function, may decompose both maltodextrins (1,4-linked D-glucose oligomers) and its precursors glycogen, starch and amylose (long-chained glucose polymers). Absence of two further cluster-encoded amylases (GFO_2132 and 2141) on the protein level suggests focusing on dextrins as parent substrates. Two additional components of this cluster, an alpha-glucoside-specific MFS permease and a beta-phosphoglucomutase, were identified in a single replicate of glucose-grown cells that provide the cytoplasmic glycolysis machinery with glucose-6-phosphate. ‘G. forsetii’ notably regulated cell surface and intracellular utilization genes of the alpha-1,4-D-glucan PUL in a differential manner (Figure 5). The outer membrane proteins TBDR and SusD were selectively activated in the presence of polymeric laminarin, whereas intracellular tri-/di-/monosaccharide processing enzymes and a transporter were strongly induced by monomeric glucose.
Sensing of substrates
The majority of hybrid sensor/response regulator signal transduction systems was identified in the membrane or intracellular fraction (9 of 12), some of which responded specifically to laminarin (GFO_1684, 1697 and 2345), but this response was insignificant in comparison with glucose-grown cells (Supplementary Table S9). Also, high numbers of sensors and regulators of conventional non-hybrid two-component systems were determined in the proteome (28 of 62 predicted signal transduction elements, 45%). However, except for a few glucose-induced response regulators (GFO_1017, 1420 and 1778), most of these proteins showed no differential expression. These findings, together with the omnipresence of various two-component signaling proteins under all substrate conditions, suggest a constitutive function, in which rather specificities than high quantities of signal transduction systems mediate the distinct upregulation of polysaccharide-associated genes. We observed a limited expression of ECF-sigma/anti-sigma factors (6 of 21 genes; 29%), which indicates that sensing of alginate, laminarin and glucose was not predominantly carried out via TonB-dependent transducers (TBDRs that are linked to a cytoplasmic sigma factor by inner membrane-localized anti-sigma factors; Koebnik, 2005). Instead, sensing was likely initiated by TBDR-enabled uptake of oligosaccharides into the periplasm in conjunction with (hybrid) two-component signal transduction to the cytoplasm. Only one sigma/anti-sigma system (GFO_1018-19) and an ECF-type sigma factor (GFO_3248) had an elevated expression in laminarin-supplied cultures, however, these genes were not found adjacent to a TBDR gene.
Glycan-specificity and suggested functional assignments of proteins with unknown functions
Of all identified proteins, 27% were annotated as hypothetical ones. Of these hypothetical proteins, 70 displayed laminarin- or alginate-dependent expression (Supplementary Tables S6 and S7) and were predominantly detected in the membrane-bound fraction. Presence of the two polysaccharides resulted in increased expression levels of proteins that were likely involved in outer membrane assembly and maintenance of the cell envelope’s integrity (alginate-dependent GFO_0835 and 1564; laminarin-dependent GFO_0244). Moreover, laminarin induced a marked upregulation of an imelysin-like protein with putative laminarin-binding function (GFO_2514) and its neighboring hypothetical protein (GFO_2515; Supplementary Figure S7). A variety of proteins with unknown functions also seemed to participate in TonB-dependent transport in laminarin-grown cultures (GFO_0462, 2190 and 2728). In addition a TonB-like hypothetical protein (GFO_2900) showed increased expression in glucan-supplied cells. Its functional assignment is supported by a similar membrane expression pattern detected for the associated TBDR component ExbD (GFO_2902). Furthermore, a laminarin-dependent putative SprE-like gliding protein (GFO_3274) and an alginate-dependent hydrolase (GFO_0138) were upregulated in polysaccharide-supplied cultures.
Ecological implications
‘G. forsetii’ KT0803 is able to specifically respond to the presence of the algal polysaccharides laminarin and alginate. Its glycan utilization strategy comprises a distinct regulation of regulons with mostly cell surface proteins that allow for binding, uptake and degradation of complex sugars. The availability of either laminarin or alginate induced two PULs that share common characteristics (TBDR, SusD, PKD and CAZymes). Upregulation of the PKD-domain-containing proteins and laminarinases or alginate lyases, respectively, was restricted to these major PULs and thus highlights the importance of such clusters for polysaccharide decomposition. Moreover, the presence of homologs of PUL-encoded genes in related marine Flavobacteriaceae indicates a conserved glycan utilization process. To accommodate the varied composition and complexity of polysaccharides, PULs seem to be organized as matching modules, in which genes can be rearranged, modified, added or removed in order to allow an evolutionary adaptation to the breakdown of specific substrates. Accordingly, utilization of the low-complexity substrate laminarin involves a smaller PUL compared with that of the more complex polysaccharide alginate.
‘G. forsetii’ lives in coastal surface waters and occupies an ecological niche that is defined by polysaccharide resources from algae. A respective adaptation is evident from the high number of susC/susD-like gene pairs and CAZymes in ‘G. forsetii’ that notably exceed those found in the oligotrophic ocean-adapted Flavobacteria, Polaribacter spp. and Dokdonia spp. (Fernández-Gómez et al., 2013). One-third of all TBDRs in ‘G. forsetii’ were involved in the laminarin-specific response. In addition, partial co-regulation of TBDRs by both laminarin and its monomer glucose indicates that not only the glycosidic linkage type but also sugar composition controls PUL expression in Bacteroidetes. Likewise, crosstalk between the PULs specific for the beta-1,3-glucan laminarin and the alpha-1,4-glucans in ‘G. forsetii’ demonstrates how the catabolism of two polysaccharides that are composed of the same monomer, but differ in linkage type can be efficiently co-regulated. A selected set of TBDRs and SusD-like proteins was found to be constitutively expressed under all tested substrate conditions. This suggests that TBDRs do not only act together with SusD-like proteins in binding and uptake of specific substrates, but also have a general function in the early recognition of nutrient availability, probably via periplasmic signal transduction systems. Likewise, Thomas et al. (2012) suggested the presence of ‘sentry’ proteins waiting to recognize complex sugars and to trigger the alginate degradation pathway in the marine flavobacterium Zobellia galactanivorans. Moreover, a transcriptome study of the gut bacterium Bacteroides plebeius by Hehemann et al. (2012) showed a basal expression of selected CAZymes without an algal polysaccharide, but in the presence of its respective monomer. Such ‘sensing’ mechanisms may ensure a rapid response when algal polymers become available.
On overall, this proteogenomic study of the marine Bacteroidetes ‘G. forsetii’ allowed a detailed functional characterization of a series of enzymes, transporters and sensing proteins that concertedly participate in the consumption of typical algal polysaccharides. It can serve as a useful starting point in assessing substrate utilization capacities and metabolic versatilities of other Bacteroidetes representatives, such as members of the genera Polaribacter and Formosa, which have been identified as key organisms in the bacterioplankton during an algae bloom (Teeling et al., 2012), or related bacteria in the human intestine (Martens et al., 2011). Such functional analyses of model bacteria have the potential to establish protein biomarkers with well-characterized specificities that will be valuable for the interpretation of future metaproteomic in situ studies and thereby will ultimately deepen our understanding of the concerted polysaccharide breakdown by complex natural microbial communities.
References
Antelmann H, Tjalsma H, Voigt B, Ohlmeier S, Bron S, van Dijl JM et al (2001). A proteomic view on genome-based signal peptide predictions. Genome Res 11: 1484–1502.
Azam F, Malfatti F . (2007). Microbial structuring of marine ecosystems. Nat Rev Microbiol 5: 782–791.
Bairoch A . (2000). The ENZYME database in 2000. Nucleic Acids Res 28: 304–305.
Bauer M, Kube M, Teeling H, Richter M, Lombardot T, Allers E et al (2006). Whole genome analysis of the marine Bacteroidetes ‘Gramella forsetii‘ reveals adaptations to degradation of polymeric organic matter. Environ Microbiol 8: 2201–2213.
Beattie A, Hirst EL, Percival E . (1961). Studies on the metabolism of the Chrysophyceae. Biochem J 79: 531–537.
Candiano G, Bruschi M, Musante L, Santucci L, Ghiggeri GM, Carnemolla B et al (2004). Blue silver: a very sensitive colloidal Coomassie G-250 staining for proteome analysis. Electrophoresis 25: 1327–1333.
Cantarel BL, Coutinho PM, Rancurel C, Bernard T, Lombard V, Henrissat B . (2009). The carbohydrate-active enzymes database (CAZy): an expert resource for glycogenomics. Nucleic Acids Res 37: D233–D238.
Cho KH, Salyers AA . (2001). Biochemical analyis of interactions between outer membrane proteins that contribute to starch utilization by Bacteroides thetaiotaomicron. J Bacteriol 183: 7224–7230.
Davis TA, Volesky B, Mucci A . (2003). A review of the biochemistry of heavy metal biosorption by brown algae. Water Res 37: 4311–4330.
Drummond AJ, Ashton B, Buxton S, Cheung M, Cooper A, Duran C et al (2010), Geneious v5.3 Available from http://www.geneious.com/.
Eilers H, Pernthaler J, Peplies J, Glöckner FO, Gerdts G, Amann R . (2001). Isolation of novel pelagic bacteria from the German bight and their seasonal contributions to surface picoplankton. Appl Environ Microbiol 67: 5134–5142.
Eymann C, Dreisbach A, Albrecht A, Bernhardt J, Becher D, Gentner S et al (2004). A comprehensive proteome map of growing Bacillus subtilis cells. Proteomics 4: 2849–2876.
Field CB, Behrenfeld MJ, Randerson JT, Falkowski P . (1998). Primary production of the biosphere: integrating terrestrial and oceanic components. Science 281: 237–240.
Fernández-Gómez B, Richter M, Schüler M, Pinhassi J, Acinas SG, González JM et al (2013). Ecology of marine Bacteroidetes: a comparative genomics approach. ISME J 7: 1026–1037.
Gomez-Pereira PR, Fuchs BM, Alonso C, Oliver MJ, van Beusekom JEE, Amann R . (2010). Distinct flavobacterial communities in contrasting water masses of the North Atlantic Ocean. ISME J 4: 472–487.
Gomez-Pereira PR, Schüler M, Fuchs BM, Bennke C, Teeling H, Waldmann J et al (2012). Genomic content of uncultured Bacteroidetes from contrasting oceanic provinces in the North Atlantic Ocean. Environ Microbiol 14: 52–66.
González JM, Fernández-Gómez B, Fernández-Guerra A, Gómez-Consarnau L, Sánchez O, Coll-Lladó M et al (2008). Genome analysis of the proteorhodopsin-containing marine bacterium Polaribacter sp. MED152 (Flavobacteria). Proc Natl Acad Sci USA 105: 8724–8729.
Grossart HP, Levold F, Allgaier M, Simon M, Brinkhoff T . (2005). Marine diatom species harbour distinct bacterial communities. Environ Microbiol 7: 860–873.
Haug A, Larsen B, Smidsrod O . (1966). A study of the constitution of alginic acid by partial acid hydrolysis. Acta Chem Scand 20: 183–190.
Hehemann JH, Correc G, Barbeyron T, Helbert W, Czjzek M, Michel G . (2010). Transfer of carbohydrate-active enzymes from marine bacteria to Japanese gut microbiota. Nature 464: 908–914.
Hehemann JH, Kelly AG, Pudlo NA, Martens EC, Boraston AB . (2012). Bacteria of the human gut microbiome catabolize red seaweed glycans with carbohydrate-active enzyme updates from extrinsic microbes. Proc Natl Acad Sci USA 109: 19786–19791.
Janse I, van Rijssel M, van Hall PJ, Gerwig GJ, Gottschal JC, Prins RA . (1996). The storage glucan of Phaeocystis globosa (Prymnesiophyceae) cells. J Phycol 32: 382–387.
Jing H, Takagi J, Liu JH, Lindgren S, Zhang RG, Joachimiak A et al (2002). Archael surface layer proteins contain beta propeller, PKD, and beta helix domains and are related to metazoan cell surface proteins. Structure 10: 1453–1464.
Kirchman DL . (2002). The ecology of Cytophaga-Flavobacteria in aquatic environments. FEMS Microbiol Ecol 39: 91–100.
Koebnik R . (2005). TonB-dependent trans-envelope signalling: the exception or the rule? Trends Microbiol 13: 343–347.
Kraan S . (2012). Algal polysaccharides, novel applications and outlook. In Chang CF (ed) Carbohydrates–Comprehensive Studies on Glycobiology and Glycotechnology. InTech.
Krewulak KD, Vogel HJ . (2011). TonB or not TonB: is that the question? Biochem Cell Biol 89: 87–97.
MacCoss MJ, Wu CC, Liu H, Sadygov R, Yates JR III . (2003). A correlation algorithm for the automated quantitative analysis of shotgun proteomics data. Anal Chem 75: 6912–6921.
Magrane M, UniProt consortium (2011), UniProt Knowledgebase: a hub of integrated protein data. Database: The Journal of Biological Databases and Curation: bar009, doi:10.1093/database/bar009.
Martens EC, Koropatkin NM, Smith TJ, Gordon JI . (2009). Complex glycan metabolism by the human gut microbiota: the Bacteroidetes Sus-like paradigm. J Biol Chem 284: 24673–24677.
Martens EC, Lowe EC, Chiang H, Pudlo NA, Wu M, McNulty NP et al (2011). Recognition and degradation of plant cell wall polysaccharides by two human gut symbionts. PLoS Biol 9: e1001221.
Morris RM, Nunn BL, Frazar C, Goodlett DR, Ting YS, Rocap G . (2010). Comparative metaproteomics reveals ocean-scale shift in microbial nutrient utilization and energy transduction. ISME J 4: 673–685.
Otto A, Bernhardt J, Meyer H, Schaffer M, Herbst FA, Siebourg J et al (2010). Systems-wide temporal proteomic profiling in glucose-starved Bacillus subtilis. Nat Commun 1: 137.
Park SK, Venable JD, Xu T, Yates JR III . (2008). A quantitative analysis software tool for mass spectrometry-based proteomics. Nat Methods 5: 319–322.
Pinhassi J, Sala MM, Havskum H, Peters F, Guadayol O, Malits A et al (2004). Changes in bacterioplankton composition under different phytoplankton regimens. Appl Environ Microbiol 70: 6753–6766.
Preiss J, Ashwell G . (1962). Alginic acid metabolism in bacteria. II. The enzymatic reduction of 4-deoxy-L-erythro-5-hexoseulose uronic acid to 2-keto-3-deoxy-D-gluconic acid. J Biol Chem 237: 317–321.
Richter M, Lombardot T, Kostadinov I, Kottmann R, Duhaime MB, Pelpies J et al (2008). JCoast-A biologist-centric software tool for data mining and comparison of prokaryotic (meta-)genomes. BMC Bioinformatics 9: 177.
Romine MF . (2011). Genome-wide protein localization prediction strategies for gram negative bacteria. BMC Genomics 12 ((Suppl 1)): S1.
Saeed AI, Sharov V, White J, Li J, Liang W, Bhagabati N et al (2003). TM4: a free, open-source system for microarray data management and analysis. Biotechniques 34: 374.
Schauer K, Rodionov DA, de Reuse HD . (2008). New substrates for TonB-dependent transport: do we only see the ‘tip of the iceberg’? Trends Biochem Sci 33: 330–338.
Shipmann JA, Berleman JE, Salyers AA . (2000). Characterization of four outer membrane proteins involved in binding starch to the cell surface of Bacteroides thetaiotaomicron. J Bacteriol 182: 5365–5372.
Schut F, de Vries EJ, Gottschal JC, Robertson BR, Harder W, Prins RA et al (1993). Isolation of typical marine bacteria by dilution culture: growth, maintenance, and characteristics of isolates under laboratory conditions. Appl Environ Microbiol 59: 2150–2160.
Sonnenburg ED, Zheng H, Joglekar P, Higginbottom SK, Firbank SJ, Bolam DN et al (2010). Specificity of polysaccharide use in intestinal bacteroides species determines diet-induced microbiota alterations. Cell 141: 1241–1252.
Tang K, Jiao N, Liu K, Zhang Y, Li S . (2012). Distribution and functions of TonB-dependent transporters in marine bacteria and environments: implications for dissolved organic matter utilization. PloS One 7: e41204.
Taniguchi N, Honke K, Fukuda M . (2002) Handbook of Glycosyltransferase and Related Genes. Springer: Tokyo.
Teeling H, Fuchs BM, Becher D, Klockow C, Gardebrecht A, Bennke CM et al (2012). Substrate-controlled succession of marine bacterioplankton populations induced by a phytoplankton bloom. Science 336: 606–611.
Thomas F, Hehemann JH, Rebuffet E, Czjzek M, Gurvan M . (2011). Environmental and gut Bacteroidetes: the food connection. Front Microbiol 2: 93.
Thomas F, Barbeyron T, Tonon T, Génicot S, Czjzek M, Michel G . (2012). Characterization of the first alginolytic operons in a marine bacterium: from their emergence in marine Flavobacteriia to their independent transfers to marine Proteobacteria and human gut Bacteroides. Environ Microbiol 14: 2379–2394.
Williams TJ, Long E, Evans F, DeMaere MZ, Lauro FM, Raftery MJ et al (2012). A metaproteomic assessment of winter and summer bacterioplankton from Antarctic Peninsula coastal surface waters. ISME J 6: 1883–1900.
Acknowledgements
We are grateful to S Grund and M Moche for mass spectrometry analyses. We express our thanks to M Richter for assistance with the JCoast program, to BM Fuchs for provision of the ‘G. forsetii’ strain, to F Simonato for experimental growth studies with ‘G. forsetii’ and appreciate the support in genomic analysis by JF Kabisch. The Federal Ministry of Education and Research (BMBF) funded this work within the ‘Microbial Interactions in Marine Systems’ project (MIMAS, project 03F0480A, http://mimas-project.de).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies this paper on The ISME Journal website
Supplementary information
Rights and permissions
This work is licensed under a Creative Commons Attribution-NonCommercial-NoDerivs 3.0 Unported License. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-nd/3.0/
About this article
Cite this article
Kabisch, A., Otto, A., König, S. et al. Functional characterization of polysaccharide utilization loci in the marine Bacteroidetes ‘Gramella forsetii’ KT0803. ISME J 8, 1492–1502 (2014). https://doi.org/10.1038/ismej.2014.4
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ismej.2014.4
Keywords
This article is cited by
-
Epiphytic common core bacteria in the microbiomes of co-located green (Ulva), brown (Saccharina) and red (Grateloupia, Gelidium) macroalgae
Microbiome (2023)
-
Three marine species of the genus Fulvivirga, rich sources of carbohydrate-active enzymes degrading alginate, chitin, laminarin, starch, and xylan
Scientific Reports (2023)
-
Niche partitioning of the ubiquitous and ecologically relevant NS5 marine group
The ISME Journal (2022)
-
Glycoside hydrolase from the GH76 family indicates that marine Salegentibacter sp. Hel_I_6 consumes alpha-mannan from fungi
The ISME Journal (2022)
-
Gramella crocea sp. nov., isolated from activated sludge of a seafood processing plant
Antonie van Leeuwenhoek (2022)