Abstract
Bleaching of Oculina patagonica has been extensively studied in the Eastern Mediterranean Sea, although no studies have been carried out in the Western basin. In 1996 Vibrio mediterranei was reported as the causative agent of bleaching in O. patagonica but it has not been related to bleached or healthy corals since 2003, suggesting that it was no longer involved in bleaching of O. patagonica. In an attempt to clarify the relationship between Vibrio spp., seawater temperature and coral diseases, as well as to investigate the putative differences between Eastern and Western Mediterranean basins, we have analysed the seasonal patterns of the culturable Vibrio spp. assemblages associated with healthy and diseased O. patagonica colonies. Two sampling points located in the Spanish Mediterranean coast were chosen for this study: Alicante Harbour and the Marine Reserve of Tabarca. A complex and dynamic assemblage of Vibrio spp. was present in O. patagonica along the whole year and under different environmental conditions and coral health status. While some Vibrio spp. were detected all year around in corals, the known pathogens V. mediteranei and V. coralliilyticus were only present in diseased specimens. The pathogenic potential of these bacteria was studied by experimental infection under laboratory conditions. Both vibrios caused diseased signs from 24 °C, being higher and faster at 28 °C. Unexpectedly, the co-inoculation of these two Vibrio species seemed to have a synergistic pathogenic effect over O. patagonica, as disease signs were readily observed at temperatures at which bleaching is not normally observed.
Similar content being viewed by others
Introduction
Over the past two decades, the increase in prevalence and severity of coral disease outbreaks has seriously impacted reef-building corals throughout the oceans worldwide (Goreau et al., 1998; Harvell et al., 1999). The most severe disease affecting coral reefs is bleaching, which is the loss of colour from affected corals caused by the disruption of symbiosis between coral hosts and photosynthetic microalgal endosymbionts, known as zooxanthellae (Brown, 1997). Since the early 1980s, mass coral bleaching events have increased in extent and frequency and have been directly correlated with increasing sea temperatures (Hoegh-Guldberg, 1999). This rise of global average temperatures has been about 0.74 °C in the later 100 years and approximately 0.13 °C/decade over the past 50 years (Salomon et al., 2007). Environmental changes associated with climate warming have been linked to an expansion of coral pathogen ranges and to changes in host susceptibility as a result of increasing environmental stress (Harvell et al., 2007). Several putative agents have been identified as the cause of mortality outbreaks worldwide, including eight coral pathogens that have been implicated in the onset of coral disease lesions. Half of these pathogens belong to the Vibrionaceae family (Rosenberg et al., 2007).
In the Northwestern Mediterranean Sea, two large mass mortality events of sessile epibenthic invertebrates were recorded in 1999 and 2003 (Cerrano et al., 2000; Garrabou et al., 2009) and two less marked events in 2006 and 2008 (Bensoussan et al., 2010; Vezzulli et al., 2010; Huete-Stauffer et al., 2011). This increasing of mass mortality of marine life may be linked with the warming trend in the Mediterranean Sea (Garrabou et al., 2009; Vargas-Yañez et al., 2010), where temperatures have risen from 1982 to 2005 at an average rate of 0.04±0.01 °C/year (Diaz-Almela et al., 2007). Together with the temperature, microbial pathogens have been associated with coral mass mortality episodes. For example, Vibrio coralliilyticus was linked to the disease of the purple gorgonian Paramuricea clavata (Ben-Haim et al., 2003; Bally and Garrabou, 2007; Vezzulli et al., 2010), being upregulated at higher temperatures (Kimes et al., 2012) and other Vibrio species with diseases of Oculina patagonica (see below). These two Vibrio spp. have been retrieved from Pocillopora damicornis, a major reef-building coral found throughout the Great Barrier Reef, suggesting that they may be opportunist pathogens (Bourne and Munn, 2005). The scleractinian coral O. patagonica is a putative alien species in the Mediterranean Sea where it was observed for the first time in 1966, on the Ligurian coast of Italy (Zibrowius, 1974). In the Spanish coast, it was first recorded in the Alicante Harbour in 1973 (Zibrowius and Ramos, 1983; Zibrowius 1992), and nowadays it is widely spread from Algeciras to Catalonia and the Balearic Islands (Zibrowius and Ramos, 1983; Ramos, 1985; Ballesteros, 1998; Izquierdo et al., 2007; Coma et al., 2011; Serrano et al., 2012, 2013; Rubio-Portillo et al., 2014). Bleaching of O. patagonica has been studied extensively, although there is a considerable controversy on the nature of its principal cause, due to the different existing hypothesis about whether microorganisms have a role or not in the bleaching process. O. patagonica bleaching was first observed along the Israeli coastline in the summer of 1993 and Vibrio shilonii (originally spelled shiloni), a later synonym of Vibrio mediteranei (Thompson et al., 2001), was identified as the causative agent of the disease (Kushmaro et al., 1996, 1997). In this case, increased seawater temperature was related to virulence, as this was the environmental factor that triggered the disease (Kushmaro et al., 1998). The adhesion of the pathogen to the coral mucus layer (Banin et al., 2001a), the production of toxin that inhibits algal photosynthesis (Banin et al., 2001b) and the expression of superoxide dismutase that protects Vibrio from oxidative stress into coral (Banin et al., 2003) are all temperature-dependent reactions. Accordingly, the infection occurred during summer (water temperatures from 25–30 °C), but not in winter (16–20 °C). Sussman et al. (2003) demonstrated that the marine fireworm Hermodice carunculata was a winter reservoir and spring–summer vector for V. mediteranei.
Although O. patagonica bleaching has continuously occurred in the Mediterranean sea during the summer months, V. mediterranei has not been related to bleached or healthy colonies of this coral since 2003 (Reshef et al., 2006) nor has caused experimental infection in aquaria. In fact, Ainsworth et al. (2008) stated that V. mediteranei was not involved or was not the primary cause of the annual bleaching of O. patagonica in the Eastern Mediterranean. These authors stated that bacteria do not have a primary but rather a secondary role during coral bleaching because of an increase in susceptibility to microbial attack experienced by corals during environmental stress. However, a recent study by (Mills et al., 2013), carried out in Israel, suggest that even though O. patagonica might have developed resistance to infection by V. mediteranei, bacteria are responsible for bleaching, being V. coralliilyticus one possible causative agent.
In order to shed some light about the relationship between Vibrio spp., seawater temperature and coral diseases, we have carried out an analysis of seasonal patterns of abundance and diversity in Vibrio spp. communities associated with healthy and diseased O. patagonica colonies. Two sampling points located in the Western Mediterranean sea were chosen for this study: Alicante Harbour and the Marine Reserve of Tabarca. Our results provide support for the bacterial bleaching hypothesis, showing that two species of Vibrio, namely V. mediteranei and V. coralliilyticus, are related to natural diseases of O. patagonica. The pathogenic potential of these two infectious bacteria was further investigated under laboratory conditions. Tissue damage increased with temperature and, remarkably, the co-inoculation of these two Vibrio species seemed to have a synergistic pathogenic effect over O. patagonica, as disease signs were readily observed at 20 °C.
Material and methods
Sample collection and environmental variable measurements
Samples were taken from two different locations in Alicante coast (Spain), separated by a distance of 22 kilometres: Alicante Harbour (38°20′11.1′′N, 00°29′11.8′′9 W) and the Marine Reserve of Tabarca in Alicante Bay (38°09′59″N, 0°28′56″W). Nine fragments of O. patagonica and three samples of surrounding water were randomly collected from each location every 3 months from September 2010 to December 2011. The colonies, located at 3 m depth, were removed by SCUBA diving using a hammer and chisel, placed in plastic bags underwater and transported to the laboratory in a cooler within the next 2 h.
Ambient seawater temperatures were recorded in situ with Madgetech Temperature Loggers (0.1 °C), with data points taken automatically every 1 h. Chlorophyll a (Chl a) concentration in water samples was determined by filtering 5 l of seawater onto a Whatman GF/F glass fibre filters (25 mm); pigment extraction was performed with 10 ml of 90% acetone, at 4 °C during 24 h in the dark, followed by centrifugation at 13 000 × g for 10 min (Biofuge-pico, Heraeus instruments) and absorbance reading at 750, 664, 647 and 630 nm according to Jeffrey and Humphrey (1975).
Circular sediment traps (86 mm in diameter and 96 mm high) were used to estimate sedimentation rates during 24 h. Collected sediments were separated by grain size using 2.0 and 0.0063 mm and GF/F glass fibre filters. The three sediment fractions obtained (gravel, sand and mud) were dried for 48 h at 110 °C to obtain the total suspended solids (g/l) and then were calcined at 550 °C for 1 h to get the organic matter (ESS method, 1993).
Bleaching estimations
The spatial extent of bleaching (white colouration) was visually estimated as a percentage of colony surface area. Colonies were grouped into five categories according to bleaching extent: 1 normally pigmented, 2 lightly bleached (<25% white), 3 moderately bleached (25–50% white), 4 severely bleached (>50% white) and 5 tissue necrosis (Bourne et al., 2008).
For Chl a measurements, 10 ml of the crushed tissue homogenate (obtained as described below) was centrifuged at 5000 × g for 10 min at 4 °C (Labofuge 400R, Heraeus instruments) and the supernatant was discarded leaving the algal pellet. Pigments were extracted from the algal pellet as described above.
Mucus and tissue extraction
Two coral fractions were used in this study: mucus layer and coral tissue. In order to separate these fractions, the colonies were gently washed three times with 50 ml of sterile filtered seawater (SFSW) to remove nonassociated bacteria, broken into ∼2 × 2 cm2 pieces, placed in 50-ml centrifuge tubes and centrifuged for 3 min at 2900 g (Labofuge 400R, Heraeus instruments) to obtain the secreted mucus from the supernatant. After centrifugation, the coral pieces were crushed in SFSW using a mortar and pestle, the CaCO3 skeleton was allowed to settle for 15 min and the supernatant (that is, crushed tissue) was removed and kept for further analyses.
Isolation of microorganisms
For plate counts of Vibrio spp., 10-fold serial dilutions of seawater, coral mucus and crushed tissue were prepared in SFSW and plated on thiosulphate citrate bile sucrose (TCBS) agar (Pronadisa, Spain) and marine agar (MA) (Pronadisa). All plates were incubated at 30 °C for 48 h. Different colony morphotypes were identified on the basis of colour, size and morphology, and were re-streaked onto fresh TCBS, incubated for further 18 h at 30 °C, and the process was repeated three times until pure cultures were obtained. Colonies isolated in MA were tested for Gram-negative staining and fermentative glucose metabolism by O/F test (Pronadisa, Spain), in order to analyse only the isolates, which could belong to Vibrio genus.
DNA extraction and PCR
DNA was extracted for each bacterial isolate by boiling. In brief, a colony was resuspended in 400 μl sterile Milli-Q water, heated to 99 °C in a dry block (Thermomoxer compact, Eppendorf) for 10 min and centrifuged at 13 000 × g for 10 min (Biofuge-pico, Heraeus instruments). DNA in the supernatant was used as a template for PCR amplification with primers 8F and 1492R, specific for bacterial 16S rRNA genes (Lane, 1991). The reaction mixtures (50 μl) contained 1.5 mM MgCl2, 50 mM KCl, 10 mM Tris-HCl (pH 9.0), each deoxyribonucleotide triphosphate (Invitrogen) at a concentration of 200 μM, 1 U of TaqI DNA polymerase (Invitrogen), each oligonucleotide primer at a concentration of 2 mM and 20 ng of template DNA. Amplification conditions included an initial denaturation step at 95 °C for 3 min followed by 30 cycles of 94 °C for 15 s, 55 °C for 30 s, 72 °C for 2 min, and a final extension step at 72 °C for 7 min. Negative controls to which no template DNA was added were included. PCR products were checked on 1% agarose gels (LE; SeaKem) in 1 × Tris-acetic acid-EDTA (TAE) buffer and visualised under UV light after ethidium bromide staining.
Analysis of isolates by amplified rDNA restriction analysis
PCR-amplified 16S rRNA genes were compared by amplified rDNA restriction analysis (Vaneechoutte et al., 1992) with enzymes HinfI and MboI (New England Biolabs). Enzymatic digestion was carried out by incubating (37 °C, 16 h) 10 μl of the PCR products with 5 U of enzyme and the corresponding enzyme buffer. Digestion products were analysed by electrophoresis on 2% agarose (LE; SeaKem) gels in 0.5 × Tris-boric acid-EDTA buffer (TBE), stained and visualised as described above. At least two isolates were selected from each restriction profile for sequencing.
16S rRNA gene sequencing and analysis
PCR products were purified using the GeneJET PCR purification kit (Fermentas, EU) and sequenced using an ABI 3730xl sequencer (Applied Biosystems). All sequences were preliminary classified using BLAST (Basic Local Alignment Search Tool) and the reference NCBI database (http://www.ncbi.nlm.nih.gov). Sequence anomaly tests, clustering and rarefactions were carried out with MOTHUR (Schloss et al., 2009). More sequence analyses were conducted using the ARB software package (Ludwig et al., 2004) with the reference databases SSU Ref 111 (SILVA project, Quast et al., 2013; http://www.arb-silva.de) and LTP s111 (LTP project, Munoz et al., 2011; http://www.arb-silva.de/projects/living-tree). All sequences were automatically aligned using the SINA software (Pruesse et al., 2012), followed by a manual inspection of misplaced bases using the ARB sequence editor and taking into account the secondary structure of the rRNA. Three phylogenetic reconstructions were performed using Neighbour Joining, Maximum Likelihood (RaxML, Stamatakis, 2006; model: GTRGAMMA) and Maximum Parsimony methods. In all cases, a 40% maximum frequency filter was applied to remove noise from the alignment and to further guarantee positional orthology. The partial sequences were added a posteriori using the ARB parsimony tool. The selected tree represents a consensus topology between the different reconstructions; multifurcations have been manually introduced where the phylogeny could not be unambiguously resolved according to the current data.
Laboratory infection experiments
Fragments (about 5 cm of diameter) of healthy O. patagonica were collected as described above from the Marine Reserve of Tabarca in June 2013. Each fragment was transferred to the laboratory and acclimated for 3 days in aquaria (20 l) before being placed in separate aquaria (500 ml) for inoculation experiments. Afterward, fragments were slowly acclimated to the experimental temperature by increasing the temperature by 0.5 °C per day, until the final temperature was reached. Fragments showing disease signs were immediately removed from the experiment. Before inoculation, the corals were maintained at the experimental temperature for 3 days in SFSW. Water was replaced every 3 days during the infection experiment.
Corals were infected with two representative Vibrio strains isolated from diseased O. patagonica colonies collected in June 2011: isolate Vic-Oc-068 that showed a 99.5% 16S rRNA gene sequence identity to V. coralliilyticus and isolate Vic-Oc-097 that showed a 99.5% sequence identity to V. mediteranei. A third Vibrio strain, Vic-Oc-027, isolated from healthy corals and showing a 100% 16S rRNA gene sequence identity to V. gigantis was used as a control. These bacteria were grown at 30 °C in Luria Bertani broth with 3% NaCl for 4 h under agitation, collected by centrifugation (4000 × g, 10 min) and washed twice with SFSW. Infections were carried out at three different temperatures: 20 °C, 24 °C and 28 °C. For each temperature, four replicate aquaria (500 ml) were inoculated with 0.5 ml of each Vibrio strain and a mixture of the three strains at a final concentration of 105 cfu ml−1 in the innoculum. In addition, four replicate uninfected corals were inoculated with 0.5 ml of SFSW and kept in the same conditions as the infected specimens. Thus, a total of 20 aquaria (including the 4 controls) were set. All infection experiments lasted for 10 days.
During the course of all experiments, the percentage of damaged tissue was estimated visually and recorded every day for each colony in the infected and control aquaria. At the end of the experiments, Vibrio spp. abundance and Chl a concentration were measured as described for the environmental samples.
Data analysis
Environmental data (temperature, seawater Chl a, sedimentation rate, organic matter and mud) were taken as the independent variables, whereas biological parameters (Vibrio spp. diversity, Vibrio spp. plate counts, bleaching and Chl a from tissue) were taken as dependent variables. First, a principal component analysis (PCA) (using correlation) was carried out to determine which environmental variables were important in driving differences between samples taken from every sampling location. Then, ordination methods were used to analyse the variation of the phylotypes according to the environmental data using canonical correspondence analysis (CCA) to study the relationship between environmental variables and biological parameters. The resulting ordination biplots approximated the weighted average of each phylotypes with respect to each of the environmental variables, which were represented as arrows. The length of these arrows indicated the relative weight of that environmental factor, while the angle between arrows indicated the degree of correlation between two environmental factors. A Monte Carlo test with 999 permutations was carried out to ensure the significance of the canonical axes. These analyses were performed using the software package CANOCO 4.5 (ter Braak and Smilauer, 2002).
Differences among Vibrio assemblages were assessed with UniFrac analysis (Lozupone and Knight, 2005), which is a β-diversity measure (differentiation at diversity level among habitats) that uses phylogenetic information to compare environmental samples, which were plotted in two dimensions with the UniFrac-based principal coordinate analysis (PCoA) To further identify environmental factors explaining differences among Vibrio assemblages, each sample was classified into four different categories based on sampling location (Harbour and Tabarca) and coral bleaching status (healthy and unhealthy). A relatively small UniFrac distance implies that two communities are similar, consisting of lineages sharing a common evolutionary history.
For each temperature (20, 24 and 28 °C) in the infection experiment, differences in Vibrio abundance and Chl a concentration were tested using two-factor (temperature and infection treatment) ANOVA (analysis of the variance). Before calculating ANOVA, heterogeneity of variance was tested using Cochran’s C-test, and vibrio abundance data were transformed (Ln(x)) as the variances were significantly different at P=0.05. When ANOVA indicated a significant difference for a certain factor, the source of differences was identified using Student–Newman–Keul (SNK) tests (Underwood, 1997).
Results
Environmental parameters, coral health status and seasonal variations of Vibrio spp.
The distribution of the samples according to their physicochemical characteristics was studied by PCA of the environmental data. Two components were required to explain 89.1% of the total variance among the samples. The first component, C1, had a very strong contribution of sedimentation rate and mud fraction and, to a lesser extent, of organic matter and Chl a concentration, while C2 was mainly related with temperature (Table 1). According to these two components, both Harbour and Tabarca samples were organised according to C1, which could be an indicator of trophic status, whereas C2 (that is, the temperature) determined the segregation of summer, spring-autumn and winter samples (Figure 1).
The state of O. patagonica colonies (Figure 2) in the sampling sites was monitored every 3 months from September 2010 to December 2011. The highest bleaching indexes were recorded in September 2011 in Tabarca when the sea temperature reached 28.2 °C. Bleached colonies showed an extensive tissue loss, leading to necrosis and mortality of affected areas. In the Harbour samples, the bleaching indexes were higher in December 2010, after sea temperature had reached 28.8 °C during the summer; tissue loss was not detected in these bleached colonies (Figure 3a). In December 2010, the Harbour sampling site was shaded by a large ship, and this could have an effect on coral disease as we checked experimentally that the lack of light could induce bleaching (Rubio-Portillo et al., submitted).
The temporal variation of culturable Vibrio counts in seawater, mucus and coral tissue during the study period is shown in Figure 3. Vibrio spp. ranged from 1 × 10° to 1.8 × 103 CFU l−1 in seawater and from 9.8 × 101 to 2 × 105 and from 3 × 101 to 7.6 × 105 CFU g−1 in the mucus and coral tissue, respectively. In seawater (Figure 3b), culturable Vibrio spp. were only detected from June to December, while they could be detected in all the coral samples taken along the year (Figure 3c), although the counts were lower from January to March. In both localities, culturable counts in coral samples collected during the bleaching event were consistently higher in diseased (1 × 105±2 × 104 CFU g−1) than in healthy corals (1 × 103±1 × 102 CFU g−1) (t-test, P<0.05). Culturable Vibrio accounted for less than 0.02‰ of the total DAPI counts in mucus and coral tissue (unpublished results).
Diversity and dynamics of Vibrio community
The screening of the 296 Vibrio isolates obtained by using amplified rDNA restriction analysis yielded a total of 53 different patterns. At least one isolate per restriction pattern was selected for complete sequencing, producing in total 133 sequences of adequate quality (1458 bp and 0% ambiguities as median values) (See Supplementary Figure S1). The operational taxonomic unit saturation observed in the rarefaction curves (Supplementary Figure S2) indicated that the sequencing effort undertaken was sufficient to detect nearly all the distinct species (98.7% similarity in the whole 16S rRNA gene, Stackebrandt and Ebers, 2006) and genera (94.5% similarity, Yarza et al., 2008) present in the samples. The analysis of the available 19 complete genomes of Vibrio species (Yarza, unpublished) indicated that the number of ribosomal operons varies from 6 to 12, with an average of 8. The maximum divergence among copies of 16S rRNA genes present in the different operons ranges from 98.3% in Vibrio splendidus LGP32 to 99.9% in Vibrio spp. EJY3, with an average of 99.3%. In particular, only 2 of the 19 species have similarities above 98.7% among the different 16S rRNA gene copies. Thus, our cutoff for species delineation seems appropriate for Vibrio strains. This is in agreement with previous studies (Yarza et al., 2010) that showed that interoperonic 16S rRNA differences are very seldom higher than 2%.
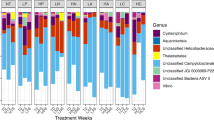
For classifying purposes, we considered a phylotype as a monophylethic group of sequences with similarities of 98.7% or above. Among the 133 sequenced 16S rRNA genes, 22 distinct phylotypes were observed (Table 1), which were named according to the known taxa found within the clades (Supplementary Figure S1). Vibrio communis—Vibrio owensii (22.22% of samples) and Vibrio harveyi—Vibrio rotiferianus (11.11%) were the phylotypes most frequently retrieved in the seawater, appearing both in the Harbour and Tabarca. Some phylotypes like Providencia vermicola, Photobacterium lutimaris and Shewanella fidelis were only found in the Harbour water. Most coral samples harboured strains closely related to V. splendidus—V. gigantis—V. atlanticus—V. pomeroyi (50%), V. harveyi-like—V. rotiferianus (24.07% of the samples) and Vibrio comitans–Vibrio rarus–Vibrio breoganii (17.59%), which were detected all year around in corals. In contrast, some phylotypes like the known pathogens V. mediteranei and V. coralliilyticus were only present in warm months (Supplementary Figure S1 and Supplementary Table S1).
CCA was used to investigate the relationship between environmental factors and the presence of different phylotypes in the coral samples. Environmental variables appear in the CCA biplot as arrows; their length and orientation indicate the relative influence of each axis, and the angle between arrows indicates the degree of correlation between variables. All arrows represent a gradient, with the mean value located at the origin and the arrow pointing to the direction of increase. For clarity, results from mucus and coral tissue are shown together as they displayed very similar trends (data not shown). As shown in Figure 4, there was a succession of phylotypes along the organic matter gradient that was accompanied by a decrease in diversity. More Vibrio phylotypes were retrieved from Tabarca (16 phylotypes, 4 unique of this site) than from Harbour samples (14 phylotypes). Three phylotypes, V. splendidus—V. gigantis—V. atlanticus—V. pomeroyi (number 1 in the figure, we will refer to this group as V. splendidus super clade), V. harveyi-like (10) and V. comitans (14) appeared at almost every sampling time and location, and thus could be part of the ‘constitutive’ coral microbiota. Phylotypes appearing together in the CCA space (such as 15 and 17 in Tabarca, and 20 and 21 in the Harbour) displayed the same dependence of physicochemical parameters (Figure 4a) and could thus have the same growth requirements. Finally, the potential pathogens V. mediteranei (2) and V. coralliilyticus (4) were only present from June to December and always in diseased corals. As shown in Figure 4a, V. mediterranei was more frequently retrieved from Harbour corals and V. coralliilyticus, from Tabarca’s.
Biplot of the CCA axes 1 and 2 for Vibrio phylotypes and environmental parameters. Every phylotype is represented by a circle that is divided into different sections according to their proportions in the sampling locations (a), coral status (b) and sampling times (c). Phylotypes are labelled as in Table 3.
The Vibrio community was different in healthy and diseased corals (Supplementary Figure S1) as was also indicated by UniFrac-based principal coordinate analysis (Figure 5). Vibrio communities in healthy corals were similar between the two locations, both dominated by V. splendidus super clade (70.37% of the healthy corals in Harbour and 72.22% in Tabarca). Bleaching events seemed to disrupt this equilibrium leading to a decrease in V. splendidus super clade (below 30% in Harbour and 20% in Tabarca diseased corals) and an increase of potential pathogens. Thus, V. mediterranei could be isolated from 59.25% of diseased corals in the Harbour, while V. coralliilyticus was detected in 81.25% of the unhealthy corals in Tabarca. The different distribution of these pathogens, together with their different locations in the CCA space (Figure 4), suggests that they may have different growth requirements in natural conditions.
Analysis of two potential Vibrio pathogens by experimental infections
Experimental infections were carried out at three different temperatures (20, 24 and 28 °C) simulating the seasonal fluctuations observed in the study sites, at the end of spring and during summer. Together with their respective uninfected controls, infections with four different inocula were set for every temperature: V. mediterranei, V. coralliilyticus, V. gigantis and a mixture of the three of them. The final concentration of vibrios in water was similar to that found in the environmental samples when seawater temperatures were between 15 and 20 °C (around 102UFC ml−1). Tissue damage on O. patagonica increased with temperature in all infection treatments (Figure 7), including controls, while concentrations of photosynthetic pigments decreased significantly (ANOVA, P<0.01; Table 2 and Figure 6a). In addition, significant differences were detected in the number of culturable vibrios from corals, with samples maintained at 28 °C showing higher abundances (ANOVA, P<0.01; Table 2 and Figure 6b).
At 20 °C, corals coinoculated with the mixture of Vibrio spp. developed signs of disease after 7 days and reached 78.3±10.4% of tissue damage after 10 days based on visual assessment (Figure 6), together with reduced Chl a concentrations compared with the controls treatments (ANOVA, P<0.01; Table 2 and Figure 6a). Corals inoculated with either pathogen did not show significant differences in tissue damage at the end of the experiment compared with the controls (Table 2 and Figure 6b).
Corals maintained at 24 °C underwent a more pronounced development of disease signs, mainly in aquaria inoculated with the mixed culture in which all samples showed 100% of tissue damaged. Corals infected only with V. mediteranei or V. coralliilyticus reached 80±7.6% and 36.6±5.8% of tissue damaged, respectively. Accordingly, significant differences in Chl a concentration were also detected between the two distinct infections (ANOVA, P<0.01; Table 2 and Figure 6a). At this temperature, V. mediteranei seemed thus to be more virulent than V. coralliilyticus (Figure 7).
Finally, at 28 °C all the corals were damaged indicating that this temperature could be lethal to O. patagonica if maintained for a long time. Damage extent reached 70±10% and 76.6±5.8% in uninfected corals and in those inoculated with V. gigantis, respectively. In contrast, while no significant differences were detected between single infections with either pathogen at this temperature; their mixture was more deleterious for the corals (Figure 7).
Discussion
Our results indicate that a complex and dynamic assemblage of Vibrio spp. is present in O. patagonica along the whole year and under different environmental conditions and coral health status. Thus, Vibrio spp. could be considered as a part of the autochthonous coral microbiota albeit, in our case, the culturable Vibrio spp. constitutes a very small fraction of the total microbiota. The association of Vibrio with healthy O. patagonica had been demonstrated before in specimens taken from the Israeli coast (Koren and Rosenberg, 2006). These authors showed that by 16S rRNA gene clone library analysis that V. splendidus was the most abundant bacteria in O. patagonica mucus both in summer and in winter. Later, Sharon and Rosenberg (2010) showed that O. patagonica taken from the Eastern Mediterranean maintained Vibrio spp. in the viable but nonculturable state. These authors speculate that this had a probiotic effect conferring protection against further infection. In other Mediterranean corals, such as the gorgonian Paramuricea clavata, Vibrio spp. constitute less than 1% of the healthy coral bacterial community as determined by partial 16S rRNA gene partial pyrosequencing (Vezzulli et al., 2013).
Changes in environmental parameters were not only accompanied by changes in health coral status but also in the numbers and composition of their associated Vibrio spp. assemblages. Certainly, the occurrence of vibrios during the studied time frame was highly correlated with seawater temperatures, with a sharp increase in Vibrio abundances during summer months when the temperatures reached around 28 °C. Vibrio concentrations were also linked to organic matter concentration and Chl a (which is an index of phytoplankton biomass), which could be due to the fact that phytoplankton blooms may support the proliferation of Vibrio spp. (Asplund et al., 2011) and thus would be an indirect effect of temperature.
Together with the changes in Vibrio spp. numbers along the year, there was a succession of phylotypes with time, as well as differences in the vibrios recovered from the two sampling sites (see Figure 4). Most importantly, Unifrac analysis indicated that diseased and healthy corals harboured distinct Vibrio spp. assemblages. The majority of isolates from healthy samples was closely related to V. splendidus super clade, while vibrios related to V. mediterranei and to V. coralliilyticus were recovered only from diseased samples (see Table 3) and were not detected in healthy colonies (although some diseased corals did not harbour either of these vibrios). There was a clear spatial distribution of these phylotypes, with V. mediterraei mainly found in the Harbour and V. coralliilyticus in Tabarca. Accordingly, and considering the different disease signs experienced by corals in both locations, these two Vibrio spp. seem to be related with O. patagonica diseases.
The presence of isolates associated with V. mediterranei in diseased corals contrasts with the results of Ainsworth et al. (2008) and Mills et al. (2013) that reported the absence of this bacterium from bleached O. patagonica in the Eastern Mediterranean. This is especially noteworthy if we consider that our sampling overlapped in time with that of Mills et al. (2013). Furthermore, while we detected V. coraliilyticus-like organisms only in diseased corals, it has been detected in healthy corals from the Eastern Mediterranean (Pollock et al., 2010). As differences between Eastern and Western Mediterranean basins are well known for planktonic microbes (Siokou-Frangou et al., 2010), it would not be surprising that corals from both sides of this sea could be harbouring different microbiotas. Caution should then be exerted when drawing general conclusions based on locally restricted studies.
The data on seasonal Vibrio spp. dynamics discussed above show the temporal coincidence between the presence in the corals of certain species of Vibrio and the development of disease symptoms that were clearly correlated with an increase in water temperature. The effect of temperature on coral disease and Vibrio numbers increase in marine environments has been widely reported before (Kushmaro et al., 1998; Ben-Haim et al., 2003; Bourne et al., 2008; Vezzulli et al., 2010), and there is still much controversy on the role of Vibrio spp. in causing disease (see above). We tried to get a new insight into this issue by analysing the response of O. patagonica to experimental infection under different conditions. This experimental approach has well-known limitations (Ainsworth and Hoegh-Guldberg, 2009). Furthermore, the use of SFSW in the experimental aquaria hampers the possibility of adaptive bleaching (the switching of associated zooxanthelae, Buddemeier and Fautin, 1993), and thus limits the response of corals to stressful conditions. However, experimental infection can still be useful for comparing coral responses to infection under different conditions.
Disease signs developed faster and to a higher extent in corals inoculated with ‘pathogenic’ vibrios than in untreated controls, with higher/faster disease signs in the corals inoculated with the mixed inoculum. These results are in agreement with those of Cervino et al. (2004). These authors observed that when four Vibrio spp. isolated from diseased specimens of the Caribbean coral Montastrea spp. were inoculated together onto healthy specimens, yellow band disease (YBD) symptoms developed faster than when they were inoculated individually, leading to the suggestion that these four vibrios were acting as a consortium. Subsequently, these authors confirmed that a consortium of different Vibrio species caused YBD in both Caribbean and Pacific Sea. The infection could be initiated at 25 °C and increased with rising seawater temperature (29–30 °C) (Cervino et al. 2008). Another coral disease, black band, is caused by a consortium of microorganism that includes cyanobacteria, sulphur reducers and oxidisers, as well as Vibrio spp. (Frias-Lopez and Klaus, 2004; Arotsker et al., 2009). However, Vezzulli et al. (2010) observed that, when they inoculated three different Vibrio strains together on Paramuricea clavata, disease signs developed more slowly than when strains were inoculated separately, and therefore a clear role cannot be assigned to bacteria in the onset of coral diseases.
As expected, an increase in temperature produced an increase in disease symptoms. Indeed, at 28 °C, coral underwent bleaching even in the absence of inoculum. Although this could mean that temperature by itself is enough to induce disease, it does not rule out the possibility of damage being caused by infection as corals can harbour Vibrio spp. in viable but nonculturable state (Sharon and Rosenberg, 2010; Vezzulli et al., 2013). However, for these authors, viable but nonculturable vibrios would be acting as a protection against pathogens, which is not the case shown here. Again, this would not solve the question of whether Vibrio spp. are primary or opportunistic pathogens (that is not a clear cut classification). Furthermore, although our results show a clear implication of Vibrio spp. in coral infection in aquaria, they do not rule out the possibility that bleaching symptoms developed at the different assayed temperatures correspond to different aetiologies. Indeed, it has been proposed (Lesser et al., 2007) that coral diseases should rather be called syndromes.
Overall the most striking result from the experimental infections described above is the development of disease signs at 20 °C when mixed cultures of V. mediterranei, V. coralliilyticus and V. gigantis were inoculated into healthy corals. The fact that only the mixture of vibrios was inducing disease signs at low temperature indicated that these bacteria could be harmful to coral under conditions in which the individual (putative) pathogens would not have any deleterious effect. In our environmental survey, V. coralliilyticus and V. mediterranei were seldom together in the same sampling site and seem to have different growth requirements. However, the experimental infection results show the pathogenic power of the mixture, which raises concerns about the possible deleterious effects of the dispersal of pathogens among different locations. This risk, if proven real such as for human pathogens transported by ballast water (Ruiz et al., 2000), could have consequences for coral health worldwide.
References
Ainsworth T, Hoegh-Guldberg O . (2009). Bacterial communities closely associated with coral tissues vary under experimental and natural reef conditions and thermal stress. Aquat Biol 4: 289–296.
Ainsworth TD, Fine M, Roff G, Hoegh-Guldberg O . (2008). Bacteria are not the primary cause of bleaching in the Mediterranean coral Oculina patagonica. ISME J 2: 67–73.
Arotsker L, Siboni N, Ben-Dov E, Kramarsky-Winter E, Loya Y, Kushmaro A . (2009). Vibrio sp. as a potentially important member of the Black Band Disease (BBD) consortium in Favia sp. corals. FEMS Microbiol Ecol 70: 515–524.
Asplund ME, Rehnstam-Holm AS, Atnur V, Raghunath P, Saravanan V, Härnström Collin B et al (2011). Water column dynamics of Vibrio in relation to phytoplankton community composition and environmental conditions in a tropical coastal area. Environ Microbiol 13: 2738–2751.
Ballesteros E . (1998). Addicions a la fauna d'invertebrats bentonics marins de l'Arxipelag de Cabrera (Illes Balears, Mediterrania Occidental). Boll Soc Hist Nat Illes Balears 41: 41–48.
Bally M, Garrabou J . (2007). Thermodependent bacterial pathogens and mass mortalities in temperate benthic communities: a new case of emerging disease linked to climate change. Glob Change Biol 13: 2078–2088.
Banin E, Israely T, Fine M, Loya Y, Rosenberg E . (2001a). Role of endosymbiotic zooxanthellae and coral mucus in the adhesion of the coral-bleaching pathogen Vibrio shiloi to its host. FEMS Microbiol Lett 199: 33–37.
Banin E, Sanjay KH, Naider F, Rosenberg E . (2001b). Proline-rich peptide from the coral pathogen Vibrio shiloi that inhibits photosynthesis of zooxanthellae. Appl Environ Microbiol 67: 1536–1541.
Banin E, Vassilakos D, Orr E, Martinez RJ, Rosenberg E . (2003). Superoxide dismutase is a virulence factor produced by the coral bleaching pathogen Vibrio shiloi. Curr Microbiol 46: 418–422.
Ben-Haim Y, Thompson FL, Thompson CC, Cnockaert MC, Hoste B, Swings J et al (2003). Vibrio coralliilyticus sp. nov., a temperature-dependent pathogen of the coral Pocillopora damicornis. Int J Syst Evol Microbiol 53: 309–315.
Bensoussan N, Romano JC, Harmelin JG, Garrabou J . (2010). High-resolution characterization of northwest Mediterranean coastal waters thermal regimes: to better understand responses of benthic communities to climate change. Estuar Coast Shelf S 87: 431–441.
Bourne D, Iida Y, Uthicke S, Smith-Keune C . (2008). Changes in coral-associated microbial communities during a bleaching event. ISME J 2: 350–363.
Bourne DG, Munn CB . (2005). Diversity of bacteria associated with the coral Pocillopora damicornis from the Great Barrier Reef. Environ Microbiol 7: 1162–1174.
Brown BE . (1997). Coral bleaching: causes and consequences. Coral Reefs 16: 129–138.
Buddemeier RW, Fautin DG . (1993). Coral bleaching as an adaptive mechanism. Bioscience 43: 320–326.
Cerrano C, Bianchi CN, Cattaneo-vietti R, Bava S, Morri C, Sara G et al (2000). A catastrophic mass mortality episode of gorgonians and other organisms in the Ligurian Sea (North western Mediterranean), summer 1999. Ecol Lett 3: 284–293.
Cervino JM, Hayes RL, Polson SW, Polson SC, Goreau TJ, Martinez RJ et al (2004). Relationship of Vibrio species infection and elevated temperatures to yellow blotch/band disease in Caribbean corals. Appl Environ Microbiol 70: 6855–6864.
Cervino JM, Thompson FL, Gomez-Gil B, Lorence E a, Goreau TJ, Hayes RL et al (2008). The Vibrio core group induces yellow band disease in Caribbean and Indo-Pacific reef-building corals. J Appl Microbiol 105: 1658–1671.
Coma R, Serrano E, Linares C, Ribes M . (2011). Sea urchins predation facilitates coral invasion in a marine reserve. PloS One 6: 22017.
Diaz-Almela E, Marbà N, Duarte CM . (2007). Consequences of Mediterranean warming events in seagrass (Posidonia oceanica) flowering records. Glob Change Biol 13: 224–235.
ESS: Environmental Science Section (1993) ESS Method 340.2: Total Suspended Solids, Mass Balance, Volatile Suspended Solids vol. 3. Environmental Sciences Section of Wisconsin State Lab of Hygiene: ESS Methods: WI, USA, pp 185–192 Chapter 2.
Frias-Lopez J, Klaus J . (2004). Bacterial community associated with black band disease in corals. Appl Environ Microbiol 70: 5955–5962.
Garrabou J, Coma R, Bensoussan N, Bally M, Chevaldonné P, Cigliano M et al (2009). Mass mortality in Northwestern Mediterranean rocky benthic communities: effects of the 2003 heat wave. Glob Change Biol 15: 1090–1103.
Goreau TJ, Cervino J, Goreau M, Hayes R, Hayes M, Richardson L et al (1998). Rapid spread of Caribbean coral reef diseases. Rev Biol Trop 46: 157–171.
Harvell CD, Kim K, Burkholder JM, Colwell RR, Epstein PR, Grimes DJ et al (1999). Emerging marine diseases—climate links and anthropogenic factors. Science 285: 1505–1510.
Harvell D, Jordán-Dahlgren E, Merkel S, Rosenberg E, Raymundo L, Smith G et al (2007). Coral disease, environmental drivers, and the balance between coral and microbial associates. Oceanography 20: 172–195.
Hoegh-Guldberg O . (1999). Climate change, coral bleaching and the future of the world’s coral reefs. Mar Freshwater Res 50: 839–866.
Huete-Stauffer C, Vielmini I, Palma M, Navone A, Panzalis P et al (2011). Paramuricea clavata (Anthozoa, Octocorallia) loss in the marine protected area of Tavolara (Sardinia, Italy) due to a mass mortality event. Mar Ecol 32: 107–116.
Izquierdo A, Loya A, Díaz-Valdés M, Ramos-Esplá AA . (2007). Non-indigenous species at the Alicante harbour (SE-Spain): Oculina patagonica de Angelis, 1908, and Botrycapulus aculeatus (Gmelin, 1791). Rapp Comm Int Mer Medit 38: 506.
Jeffrey SW, Humphrey GF . (1975). New spectrophotometric equations for determining chlorophylls a, b, c1 and c2 in higher plants, algae and natural phytoplankton. Biochem Physiol Pflanz 167: 191–194.
Kimes NE, Grim CJ, Johnson WR, Hasan NA, Tall BD, Kothary MH et al (2012). Temperature regulation of virulence factors in the pathogen Vibrio coralliilyticus. ISME J 6: 835–846.
Koren O, Rosenberg E . (2006). Bacteria associated with mucus and tissues of the Coral Oculina patagonica in summer and winter. App Env Microbiol 72: 5254–5259.
Kushmaro A, Loya Y, Fine M, Rosenberg E . (1996). Bacterial infection and coral bleaching. Nature 380: 396.
Kushmaro A, Rosenberg E, Fine M, Ben-Haim Y, Loya Y . (1998). Effect of temperature on bleaching of the coral Oculina patagonica by Vibrio shiloi AK-1. Mar Ecol Prog Ser 171: 131–137.
Kushmaro A, Rosenberg E, Fine M, Loya Y . (1997). Bleaching of the coral Oculina patagonica by Vibrio AK-1. Mar Ecol Prog Ser 147: 159–165.
Lane DJ . (1991). 16S/23S rRNA sequencing. In: Stackebrandt E, Goodfellow M (eds) Nucleic Acid Techniques in Bacterial Systematics. Wiley: Chichester, UK, pp 115–175.
Lesser M, Bythell J, Gates R, Johnstone R, Hoeghguldberg O . (2007). Are infectious diseases really killing corals? Alternative interpretations of the experimental and ecological data. J Exp Mar Bio Ecol 46: 36–44.
Lozupone C, Knight R . (2005). UniFrac: a new phylogenetic method for comparing microbial communities. App Env Micro 71: 8228–8235.
Ludwig W, Strunk O, Westram R, Richter L, Meier H, Buchner A et al (2004). ARB: a software environment for sequence data. Nucleic Acids Res 32: 1363–1371.
Mills E, Shechtman K, Loya Y, Rosenberg E . (2013). Bacteria appear to play important roles in both causing and preventing the bleaching of the coral Oculina patagonica. Mar Ecol Prog Ser 489: 155–162.
Munoz R, Yarza P, Ludwig W, Euzéby J, Amann R, Schleifer KH et al (2011). Release LTPs104 of the All-Species Living Tree. Syst Appl Microbiol 34: 169–170.
Pollock FJ, Wilson B, Johnson WR, Morris PJ, Willis BL, Bourne DG . (2010). Phylogeny of the coral pathogen Vibrio coralliilyticus. Environ Microbiol Rep 2: 172–178.
Pruesse E, Peplies J, Glöckner FO . (2012). SINA: accurate high-throughput multiple sequence alignment of ribosomal RNA genes. Bioinformatics 28: 1823–1829.
Quast C, Pruesse E, Yilmaz P, Gerken J, Schweer T, Yarza P et al (2013). The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res 41: 590–596.
Ramos AA . (1985) La reserva marina de la Isla Plana o Nuevo Tabarca (Alicante). Universidad de Alicante: Alicante.
Reshef L, Koren O, Loya Y, Rosenberg IZ, Rosenberg E . (2006). The coral probiotic hypothesis. Env Micro 8: 2068–2073.
Rosenberg E, Koren O, Reshef L, Efrony R, Zilber-Rosenberg I . (2007). The role of microorganisms in coral health, disease and evolution. Nat. Rev. Microbiol 5: 355–362.
Rubio-Portillo E, Vazquez-Luis M, Izquierdo Muñoz A, Ramos-Esplá AA . (2014). Distribution patterns of alien coral Oculina patagonica De Angelis D’Ossat, 1908 in western Mediterranean Sea. J Sea Res 85: 372–378.
Ruiz GM, Rawlings TK, Dobbs FC, Drake LA, Mullady T, Huq A et al (2000). Global spread of microorganisms by ships. Nature 408: 49–50.
Salomon S, Qin D, Manning M, Maquis M, Averys K . (2007) Climate change 2007: synthesis report. IPCC: Geneva.
Schloss PD, Westcott SL, Ryabin T, Hall J R, Hartmann M, Hollister EB et al (2009). Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol 75: 7537–7541.
Serrano E, Coma R, Ribes M, Weitzmann B, García M, Ballesteros E . (2013). Rapid northward spread of a zooxanthellate coral enhanced by artificial structures and sea warming in the Western Mediterranean. PloS One 8: e52739.
Serrano E, Coma R, Ribes M . (2012). A phase shift from macroalgal to coral dominance in the Mediterranean. Coral Reefs 31: 1199.
Sharon G, Rosenberg E . (2010). Healthy corals maintain Vibrio in the VBNC state. Environ Microbiol Rep 2: 116–119.
Siokou-Frangou I, Christaki U, Mazzocchi MG, Montresor M, Ribera d’Alcalá M, Vaqué D et al (2010). Plankton in the open Mediterranean Sea: a review. Biogeosciences 7: 1543–1586.
Stackebrandt E, Ebers J . (2006). Taxonomic parameters revisited: tarnished gold standards. Microbiol Today 8: 6–9.
Stamatakis A . (2006). ‘RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22: 2688–2690.
Sussman M, Loya Y, Fine M, Rosenberg E . (2003). The marine fireworm Hermodice carunculata is a winter reservior and spring-summer vector for the coral- bleaching pathogen Vibrio shiloi. Environ Microbiol 5: 250–255.
ter Braak CJF, Smilauer P . (2002). CANOCO reference manual and CanoDraw for Windows user’s guide: software for canonical community ordination (version 4.5). Microcomputer Power: Ithaca, NY, USA, p 500.
Thompson FL, Hoste B, Thompson CC, Huys G, Swings J . (2001). The coral bleaching Vibrio shiloi Kushmaro et al., 2001 is a later synonym of Vibrio mediterranei Pujalte and Garay 1986. Syst Appl Microbiol 24: 516–519.
Underwood AJ . (1997) Experiments in Ecology: Their Logical Design and Interpretation Using Analysis of Variance. Cambridge University Press, p 504.
Vaneechoutte M, Rossau R, De Vos P, Gillis M, Janssens D, Paepe N et al (1992). Rapid identification of bacteria of the Comamonadaceae with amplified ribosomal DNA-restriction analysis (ARDRA). FEMS Microbiol Lett 93: 227–233.
Vargas-Yañez M, Moya F, García-Martínez MC, Tel E, Zunino P et al (2010). Climate change in the Western Mediterranean Sea 1900–2008. J Marine Syst 82: 171–176.
Vezzulli L, Colwell RR, Pruzzo C . (2013). Ocean warming and spread of pathogenic Vibrios in the aquatic environment. Microb Ecol 65: 817–825.
Vezzulli L, Previati M, Pruzzo C, Marchese A, Bourne DG, Cerrano C . (2010). Vibrio infections triggering mass mortality events in a warming Mediterranean Sea. Environ Microbial 12: 2007–2019.
Yarza P, Ludwig W, Euzéby J, Amann R, Schleifer K H, Glöckner FO et al (2010). Update of the All-Species Living Tree Project based on 16S and 23S rRNA sequence analyses. Syst Appl Microbiol 33: 291–299.
Yarza P, Richter M, Peplies J, Euzeby J, Amann R, Schleifer KH et al (2008). The All-Species Living Tree project: a 16S rRNA-based phylogenetic tree of all sequenced type strains. Syst Appl Microbiol 31: 241–250.
Zibrowius H . (1974). Oculina patagonica, Scléractiniaire hermatypique introduit en Méditerranée. Helgo Wissenschaftliche Meeresuntersuchungen 26: 153–173.
Zibrowius H . (1992). Ongoing modification of the Mediterranean marine fauna and flora by establishment of exotic species. Mesogée 51: 83–107.
Zibrowius H, Ramos A . (1983). Oculina patagonica, scléractiniaire exotique en Méditerranée—nouvelles observations dans le Sud-Est de l'Espagne. Rapp Comm int Expl. Sci Mer Medit 28: 297–301.
Acknowledgements
This work was supported by the projects CGL2009-12651-C02-01 and CGL2012-39627-C03-01 (to JA) of the Spanish Ministry of Economy and Competitiveness, which were also co-financed with FEDER support from the European Union (EU), and the EU project SYMBIOMICS (www.symbiomics.de) grant no.264774 (to AAR). We thank Felio Lozano and Andrés Izquierdo for their assistance during sample collection, and Maria Gomariz and Manuel Martínez-García for their help with data analysis. We acknowledge the friendly cooperation of Reserve Marine of Tabarca guards, Autoridad Portuaria of Alicante and the Secretaría General de Pesca Marítima.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
Esther Rubio-Portillo holds a pre-doctorate grant from the University of Alicante. The remaining authors declare no conflict of interest.
Additional information
Supplementary Information accompanies this paper on The ISME Journal website
Rights and permissions
About this article
Cite this article
Rubio-Portillo, E., Yarza, P., Peñalver, C. et al. New insights into Oculina patagonica coral diseases and their associated Vibrio spp. communities. ISME J 8, 1794–1807 (2014). https://doi.org/10.1038/ismej.2014.33
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ismej.2014.33
Keywords
This article is cited by
-
The coral pathogen Vibrio coralliilyticus kills non-pathogenic holobiont competitors by triggering prophage induction
Nature Ecology & Evolution (2022)
-
Improving environmental monitoring of Vibrionaceae in coastal ecosystems through 16S rRNA gene amplicon sequencing
Environmental Science and Pollution Research (2022)
-
Shifts in marine invertebrate bacterial assemblages associated with tissue necrosis during a heat wave
Coral Reefs (2021)
-
Stenotrophomonas maltophilia AHL-Degrading Strains Isolated from Marine Invertebrate Microbiota Attenuate the Virulence of Pectobacterium carotovorum and Vibrio coralliilyticus
Marine Biotechnology (2019)
-
Seasonal Stability in the Microbiomes of Temperate Gorgonians and the Red Coral Corallium rubrum Across the Mediterranean Sea
Microbial Ecology (2018)