Abstract
Kangaroos ferment forage material in an enlarged forestomach analogous to the rumen, but in contrast to ruminants, they produce little or no methane. The objective of this study was to identify the dominant organisms and pathways involved in hydrogenotrophy in the kangaroo forestomach, with the broader aim of understanding how these processes are able to predominate over methanogenesis. Stable isotope analysis of fermentation end products and RNA stable isotope probing (RNA-SIP) were used to investigate the organisms and biochemical pathways involved in the metabolism of hydrogen and carbon dioxide in the kangaroo forestomach. Our results clearly demonstrate that the activity of bacterial reductive acetogens is a key factor in the reduced methane output of kangaroos. In in vitro fermentations, the microbial community of the kangaroo foregut produced very little methane, but produced a significantly greater proportion of acetate derived from carbon dioxide than the microbial community of the bovine rumen. A bacterial operational taxonomic unit closely related to the known reductive acetogen Blautia coccoides was found to be associated with carbon dioxide and hydrogen metabolism in the kangaroo foregut. Other bacterial taxa including members of the genera Prevotella, Oscillibacter and Streptococcus that have not previously been reported as containing hydrogenotrophic organisms were also significantly associated with metabolism of hydrogen and carbon dioxide in the kangaroo forestomach.
Similar content being viewed by others
Introduction
The production of methane by domestic ruminants is well recognised as an important component of anthropogenic greenhouse gas emissions, contributing ∼17–25% of the global methane budget (Lassey, 2007; Conrad, 2009). Methane is generated by ruminants as a by-product of the enteric fermentation of forage material in the anaerobic conditions of the foregut. The hydrogen-dependent reduction of CO2 to methane by hydrogenotrophic methanogenic archaea is essential for normal function of the rumen microbial ecosystem, as accumulation of excess hydrogen in the system inhibits fermentation by preventing regeneration of NAD+. This leads in turn to reduced microbial production and forage digestion (Wolin et al., 1997).
Although hydrogenotrophy is predominately carried out by methanogens in the rumen, alternative pathways apparently exist in other comparable gut ecosystems. In Australia, kangaroos and wallabies graze on the same native pastures as cattle, and digest forage via microbial fermentation in an enlarged foregut analogous to the rumen, yet methane production by kangaroos is minimal or non-existent (Kempton et al., 1976; Englehardt et al., 1978; Dellow et al., 1988). Ouwerkerk et al. (2009) were unable to detect the presence of methanogenic archaea in the foreguts of kangaroos they surveyed, while Klieve et al. (2012) have recently demonstrated that the presence of methanogens in the kangaroo foregut is variable, with archaea apparently absent from many individuals.
The reasons why methanogenesis predominates in ruminants while alternative pathways for hydrogenotrophy apparently predominate in kangaroos are not well understood. It has been suggested (Attwood and McSweeney, 2008) that the hydrogen and CO2 produced by fermentation in the kangaroo foregut is utilised predominately by acetogenic bacteria, which reduce carbon dioxide to acetate via the acetyl-CoA pathway (Drake et al., 2008). Ouwerkerk et al. (2009) isolated several novel bacterial strains belonging to the family Lachnospiraceae from the foregut of kangaroos, which utilise hydrogen and produce acetate in in vitro cultures. Quantitative PCR assays confirmed that these bacteria are present in high numbers in the kangaroo foregut. The presence of a distinct acetogen population has also been observed in the forestomach of another macropod species (Macropus eugenii), which differs from the native acetogen population of the bovine rumen (Gagen et al., 2010).
If acetogenesis does predominate in the kangaroo foregut, the acetate produced could be absorbed by the host animal, thus supplying a significant proportion of the host’s energy needs (Henning and Hird, 1970; Sheppach, 1994). Reductive acetogenesis is therefore a pathway with the potential to reduce or completely eliminate methane production by the gut ecosystem, while simultaneously offering a significant energetic advantage to the host animal. If the nature of the interactions that allow acetogenesis to predominate in some enteric fermentation systems could be understood, it may be possible to promote reductive acetogenesis in the rumen ecosystem, thereby reducing livestock methane emissions while simultaneously improving the efficiency of feed utilisation.
The objective of this study was to investigate CO2 and hydrogen metabolism in the kangaroo foregut so that the mechanisms that allow kangaroos to ferment plant material without producing large quantities of methane could be better understood. This was done by using stable isotope tracing to track the biological fate of CO2 in the kangaroo foregut and the bovine rumen and by using RNA stable isotope probing (RNA-SIP) techniques to identify bacteria in the kangaroo foregut that are capable of hydrogen-dependent CO2 metabolism.
Materials and methods
Collection of gut samples
See Supplementary Materials 1 and 2 for full details of sample collection procedures.
Samples of the liquid portion of the forestomach contents of three wild male Eastern Grey kangaroos (M. giganteus) were collected in the field near Milmerran, Queensland. Samples of rumen fluid from four Bos taurus cattle that had been fed a diet consisting primarily of whole barley grain for 110 days were collected immediately after slaughter. Samples of rumen fluid of grass-fed cattle were collected from three rumen-fistulated B. taurus × B. indicus cattle housed at the Centre for Advanced Animal Studies at Gatton, Queensland. In all cases, liquid digesta was mixed with sterile anaerobic glycerol to give a final glycerol concentration of 30–40% and frozen on dry ice. During collection of all samples, a portion of liquid digesta was also collected into separate bottles that did not contain glycerol. These bottles were transported on ice and frozen at the earliest opportunity. This digesta was used for preparation of media for the incubation experiment.
Media
Anaerobic media containing 33% (v/v) of the appropriate digesta type (kangaroo, grain-fed rumen or grass-fed rumen) were prepared in the manner described previously (Klieve et al., 1989). The media was supplemented with buffered mineral salts, bicarbonate and trace elements at concentrations designed to approximately simulate the natural conditions of the bovine rumen (Erdman, 1988) and kangaroo forestomach (Beal, 1984; Dellow et al., 1988) (see Supplementary Material 3 for full details of media composition). All media were prepared under an anaerobic gas mix consisting of 95% CO2 and 5% H2. Aliquots (100 ml) of the media were dispensed into 500-ml bottles, with the headspace filled with 95% CO2/5% H2, which were sealed and autoclaved.
Minimal carbon anaerobic salts medium (MCAS, Supplementary Material 3) was designed in-house to simulate the major ion concentrations found in the kangaroo forestomach (Beal, 1984; Dellow et al., 1988). Bicarbonate medium for isotope labelling incubations was identical to MCAS, except that it contained 100 mM of stable carbon isotope labelled (13C) NaHCO3 with the NaCl concentration adjusted to give the same sodium concentration as MCAS (Supplementary Material 3).
Isotope tracing to track the biological fate of CO2 in simulated bovine and kangaroo foregut fermentations
The gut content samples collected as described above were thawed, then 50-ml aliquots were centrifuged to pellet the microbial biomass and solid digesta particles. The supernatant containing glycerol was discarded, then the pellets were washed twice by centrifuging and resuspending in MCAS medium. The pellet was resuspended in 50 ml MCAS medium and added to a 500-ml Wheaton bottle containing 100 ml of the appropriate digesta-based medium (kangaroo, grain-fed rumen or grass-fed rumen). All washing and inoculation steps were performed in an atmosphere of 95% CO2/5% H2 in an anaerobic chamber (Coy, Grass Lake, MI, USA). After inoculation, each 500-ml bottle was supplemented with 1.0 ml of 538 mM solution of 13C-labelled NaHCO3 (99 atom % 13C, Sigma-Aldrich, St Louis, MO, USA), which had been flushed with nitrogen to remove dissolved oxygen, to produce a final concentration of 105 mM bicarbonate that consisted of 5% 13C-labelled bicarbonate (equivalent to δ 13CVPDB 3,683‰). Control incubations were identical but were supplemented with 1.0 ml of 538 mM unlabelled (12C) NaHCO3 solution. After addition of NaHCO3, the bottles were sealed and incubated at 39 °C.
Five-millilitre samples of the fluid portion and gas headspace were taken immediately after inoculation (time 0 samples), and at 3 h, 6 h, 24 h, 48 h and 7 days post inoculation. Samples were taken by piercing the butyl rubber stopper of the culture bottle with a 21-gauge needle with a syringe attached. Gas samples were transferred to evacuated 4.5-ml glass vials (Labco, Lampeter, UK). Fluid samples to be used for analysis of volatile fatty acids (VFAs) were transferred to sterile polypropylene vials to which 20 μl of saturated HgCl2 solution was added and mixed to stop microbial activity. These samples were stored frozen at −20 °C prior to further processing. On the day of extraction, the frozen samples were thawed, and the internal standard 3-methylvaleric acid (3 mV) was added to give a final concentration of 4.0 mM. VFAs were extracted and purified from the thawed fluid samples by vacuum distillation (Vreman et al., 1978).
Gas chromatography-isotope ratio mass spectrometry
Gas chromatography combustion isotope ratio mass spectrometry (GC-c-IRMS) was used to determine the concentrations and carbon stable isotope ratios (δ13CVPDB) of actetate, propionate, butyrate and valerate in the fluid portion of the gut fermentations. For the gas headspace samples, both the concentration and carbon stable isotope ratio of methane was determined. For carbon dioxide, only the carbon stable isotope ratio was determined as the very high initial concentration of 95% carbon dioxide prevented accurate measurement of changes in concentration of carbon dioxide in the headspace. Details of the GC-c-IRMS procedures used are provided in Supplementary Material 4.
Variability between sample categories in both concentration and carbon isotope ratios of methane and VFAs were tested for significance using repeated measures analysis of variance (RM-ANOVA). All data analysis was conducted in GenStat version 14.2.0.6286 (VSN International, Hempstead, UK).
Incubation of gut samples for RNA stable isotope labelling
An RNA-SIP experiment was conducted separately from the isotope-tracing experiment. This experiment was performed under enrichment conditions designed to encourage growth of hydrogenotrophic organisms. A sample from a single female Eastern Grey kangaroo (Macropus giganteus) was incubated at 39 °C under an equimolar mixture of 206 kpa H2/CO2 for 3 days to enrich the sample for micro-organisms that were capable of utilising H2/CO2 for growth. One hundred millilitres of fluid was then centrifuged to pellet the biomass within the sample. This pellet was washed twice in MCAS. The final washed pellet was resuspended in 30 ml MCAS.
Aliquots (1 ml) of the washed and resuspended pre-incubated foregut content were used to inoculate 20 ml Hungate tubes containing 10 ml 13C-bicarbonate (99 atom % 13C) medium under an atmosphere of nitrogen gas. The tubes were then pressurised with 13CO2 gas (99 atom % 13C, Sigma-Aldrich) to 103 kPa above atmospheric pressure. H2 gas was then added to a final pressure of 206 kPa to give a total of 0.2 mmol each of 13CO2 and H2 in the headspace. Unlabelled control incubations were identical except that the medium contained unlabelled (12C) bicarbonate, and the tubes were pressurised with 12CO2 and H2. Additional controls were also set up which contained 13C-labelled bicarbonate medium and 13CO2, but no hydrogen. These controls were used to identify organisms that were able to assimilate the 13C-label without hydrogen present.
The tubes were incubated at 39 °C, with replicate samples removed at 3 h, 16 h and 7 days post inoculation. Cells were recovered from the medium by centrifugation, then immediately flash-frozen in liquid nitrogen.
RNA extraction
Frozen cell pellets were resuspended and thawed in 10 mM Tris, 1 mM EDTA buffer (pH 8.0) containing 15 mg ml−1 lysozyme and incubated for 10 min at room temperature. Total nucleic acid was then extracted and precipitated according to Yu and Forster’s (2005) protocol. The total nucleic acid pellets were resuspended in nuclease-free water and processed with the RNeasy kit (Qiagen, Hilden, Germany) following the manufacturer’s instructions to remove contaminating DNA and residual protein. RNA was eluted in nuclease-free water and quantified using the RiboGreen RNA quantification kit (Life Technologies, Carlsbad, CA, USA).
Isopycnic centrifugation and fractionation
13C-labelled RNA was resolved from unlabelled RNA by isopycnic centrifugation in caesium trifluoroacetate (CsTFA) using a modified version of the protocol of Whiteley et al. (2007) (see Supplementary Material 5) for detailed protocol). A total of 600 ng of RNA was loaded per gradient, then each gradient was fractionated after ultracentrifugation. Each ultracentrifuge run included a control gradient containing an equimolar mixture of 13C and 12C RNA from Eschericia coli (Supplementary Material 6). The RNA was recovered from each gradient fraction by precipitation in isopropanol, using glycogen as a carrier and resuspended in 30-μl nuclease-free water.
Quantification of CsTFA gradient fractions and reverse transcription
The quantity of RNA in each gradient fraction was measured using Ribogreen according to the manufacturer’s recommendations. For each gradient, the three fractions closest to the ‘heavy’ and ‘light’ RNA peaks were identified by comparison to the E. coli control gradient from the same ultracentrifuge run and selected for further processing. For each fraction, 200 pg of RNA was reverse-transcribed using random hexamers and Thermoscript reverse transcriptase (Life Technologies) according to the manufacturer’s instructions. Preliminary comparisons of the bacterial communities represented in the ‘heavy’ and ‘light’ fractions of CsTFA gradients were performed using denaturing gradient gel electrophoresis (Supplementary Material 7). The results of these denaturing gradient gel electrophoresis comparisons were used to determine the incubation time that resulted in differential incorporation of isotope label by the primary utilisers of CO2/HCO3−, rather than ‘downstream’ incorporation of labelled metabolites by all the bacteria present in the sample (Supplementary Material 7).
Amplicon pyrosequencing
The cDNA produced by reverse transcription of the three ‘heavy’ and three ‘light’ CsTFA fractions obtained from samples incubated for 16 h was pooled to produce one ‘heavy’ and one ‘light’ cDNA solution for each gradient. This cDNA was then used as template for the production of 16S rRNA gene amplicons for pyrosequencing (see Supplementary Material 8 for detailed PCR protocol). Sequence data were processed, clustered into operational taxonomic units at the 97% identity level and classified as described previously (Eigeland et al., 2012; Gulino et al., 2013). All data processing was performed in QIIME 1.5.0 (Caporaso et al., 2010).
Identification of 13C-labelled operational taxonomic units
Operational taxonomic units (OTUs) representing organisms that incorporated the 13C-label from bicarbonate/CO2 in the presence of hydrogen were identified using a χ2 analysis (Sokal and Rohlf, 1969) to identify the OTUs whose distribution between ‘heavy’ and ‘light’ gradient fractions was significantly affected by the presence of 13C-label in the media used for incubation. OTUs that were significantly (P<0.001) associated with CO2/H2 utilisation were identified by comparison to the Greengenes (DeSantis et al., 2006) and Genbank (Benson et al., 2011) databases. Full details of the statistical procedure used are given in Supplementary Material 9.
Mining kangaroo foregut and bovine rumen data sets
To obtain estimates of the prevalence of bacteria associated with H2 and CO2 metabolism in the kangaroo forestomach and bovine rumen the 16S rRNA sequences of OTUs that were identified as representatives of H2- and CO2-utilising organisms in the in vitro SIP experiment were used to query a database of bacterial 16S rRNA gene sequences that was generated in a separate study ((Gulino et al., 2013) MG-RAST project IDs 3340 and 3374). This database includes bacterial 16S rRNA gene sequences obtained from the rumen of grass-fed cattle, grain-fed cattle, grey kangaroos (M. giganteus), red kangaroos (M. rufus) and wallaroos (M. robustus).
Results
Isotope tracing to compare the fates of carbon dioxide/bicarbonate in the bovine rumen and kangaroo forestomach
The in vitro fermentations performed with kangaroo forestomach and bovine rumen contents showed very clear differences between kangaroos and cattle in the pathways of CO2 metabolism. In bovine rumen fermentations, both grain-fed and grass-fed, methane was detectable in the headspace 3 hours after inoculation, and continued to accumulate for the following 7 days of the experiment (Figures 1a and 2a). In contrast, kangaroo fermentations produced a measurable quantity of methane only after 7 days incubation, and this quantity was significantly (P<0.01) lower than in rumen fermentations (Figures 1a and 2a).
Mean (±s.e.) concentration (a) and δ13CVDPB (b) of methane and δ13CVDPB of CO2 (c) in headspace gas of in vitro gut content fermentations spiked with H13CO3−. Fermentations performed using kangaroo forestomach content produced significantly less methane than fermentations performed with bovine rumen fluid, and the small quantity of methane that was produced remained relatively unlabelled, indicating that it was largely not derived from CO2. ▴ Bovine rumen fluid (grain-fed) (n=4). • Bovine rumen fluid (grass-fed) (n=4). ▪ Kangaroo forestomach content (n=3). Early time points for δ13C of methane are missing because carbon isotope ratios could not be determined until sufficient methane had accumulated to allow measurement.
Mean (±s.e.) concentration (a) and δ13CVDPB (b) of methane and δ13CVDPB of CO2 (c) in headspace gas of in vitro gut content fermentations spiked with unlabelled (12C) HCO3−. Both grain-fed and grass-fed bovine rumen fermentations produced methane that was strongly depleted in 13C relative to CO2, consistent with H2/CO2-dependent methanogenesis. Fermentations performed with kangaroo forestomach content produced significantly less methane than bovine rumen fluid fermentations, and the small quantity of methane that was produced was not depleted in 13C relative to CO2, indicating that it was produced by a pathway other than H2/CO2-dependent methanogenesis. ▴ Bovine rumen fluid (grain-fed) (n=4). • Bovine rumen fluid (grass-fed) (n=3). ▪ Kangaroo forestomach content (n=3). Early time points for δ13C of methane are missing because carbon isotope ratios could not be determined until sufficient methane had accumulated to allow measurement.
Analysis of the carbon isotope ratios of CO2 and methane in the headspace gases of the in vitro fermentations spiked with 13C-labelled bicarbonate showed that the bicarbonate in solution rapidly established equilibrium with CO2 in the headspace. The δ13C value of CO2 rose rapidly during the first 6 h of the experiment, then remained relatively stable (Figure 1c). The methane produced by the rumen bacterial community in the presence of labelled (13C) bicarbonate was strongly labelled for both grass-fed and grain-fed fermentations, indicating that it was derived from CO2/HCO3− (Figure 1b). In kangaroo fermentations however, the small quantity of methane that was produced remained relatively unlabelled, indicating that it was derived from substrates other than CO2 (Figure 1b).
Clear differences between the kangaroo forestomach and bovine rumen were also observed in fermentations using unlabelled (12C) bicarbonate. The methane produced by bovine rumen fermentations consistently had a δ13C value 40–50‰ lower than the δ13C of the CO2 in the headspace (Figure 2). The small quantity of methane produced in kangaroo fermentations however had a δ13C value only 2–8‰ lower than the CO2 in the headspace (Figure 2).
Analysis of the stable carbon isotope ratios of the VFAs produced in the in vitro fermentations clearly showed that conversion of CO2 to acetate is much more active in the kangaroo foregut than in the bovine rumen. Kangaroo foregut fermentations spiked with 13C-labelled bicarbonate produced strongly labelled acetate, whereas bovine rumen content fermentations produced only slightly labelled acetate (Figure 3b). Repeated measures ANOVA showed that this observed difference in the extent of acetate labelling between kangaroos and cattle was significant (P<0.05). Production of 13C-labelled propionate occurred in all three systems (Figure 3d), and the extent of labelling did not differ significantly between sample types.
Mean (±s.e.) changes relative to time 0 in concentration and δ13CVDPB of acetate (a, b) and propionate (c, d) in in vitro gut content fermentations spiked with H13CO3−. Fermentations performed with kangaroo forestomach content resulted in significantly more incorporation of 13C label than fermentations performed with bovine rumen fluid, indicating that reductive acetogenesis contributes to a larger proportion of the acetate pool in kangaroos than it does in cattle. Additional data for butyrate and valerate are presented in Supplementary Figure S4. ▴ Bovine rumen fluid (grain-fed) (n=4). • Bovine rumen fluid (grass-fed) (n=3). ▪ Kangaroo forestomach content (n=3).
RNA-SIP to identify bacteria associated with metabolism of carbon dioxide/bicarbonate and hydrogen.
Figure 4 illustrates the distribution of OTUs significantly associated with hydrogen/CO2 utilisation between the ‘heavy’ and ‘light’ fractions of CsTFA gradients derived from samples incubated in the presence of either 13CO2/H13CO3− or 12CO2/H12CO3−. Although many OTUs were detected in both the ‘heavy’ and ‘light’ gradient fractions, χ2 analysis allowed the identification of those OTUs for which the distribution of RNA between the ‘heavy’ and ‘light’ fractions changed significantly when 13CO2/H13CO3− was present.
Number of 16S rRNA amplicon reads per OTU obtained from ‘heavy’ and ‘light’ fractions of CsTFA gradients. Only OTUs for which a statistically significant labelling effect was observed are shown. The CsTFA gradients were loaded with RNA extracted from kangaroo forestomach contents that had been incubated for 16 h in the presence of hydrogen and either labelled (13C, upper figure) or unlabelled (12C, lower figure) carbon dioxide and bicarbonate. Number of reads for each OTU are expressed as a percentage of the total number of reads obtained for each gradient fraction to normalise for variation in total read number between samples.
Bacterial OTUs that were identified by RNA-SIP and χ2 analysis as CO2/H2 metabolisers under the conditions used in the in vitro isotope labelling incubations are presented in Table 1. Sequences representing each OTU have been submitted to the Genbank database under accession numbers (KC632738–KC633091). Raw sequence reads have been submitted to the NCBI Sequence Read Archive under accession numbers (SRP033591, SRS512720, SRS512721, SRS512722, SRS512723, SRS513566 and SRS513567).
Among the OTUs identified as CO2/H2 metabolisers in kangaroos one is of particular interest (OTU 236, Table 1) because of its very close 16S rRNA gene sequence similarity to the known reductive acetogen Blautia coccoides (Kamlage et al., 1997; Liu et al., 2008). Comparison to our database obtained by 16S rRNA gene amplicon pyrosequencing of the forestomach contents of kangaroos and the rumens of cattle showed that sequences sharing ⩾97% 16S rRNA gene similarity to this OTU were present in all macropods sampled (22 individual animals) and represented an average of 0.30% (±0.21%) of total 16S reads. In the rumen of cattle however, sequences sharing ⩾97% 16S rRNA gene similarity to this OTU were only present in 40% of animals sampled (9 of 22 individual animals), where they represented an average of 0.05% (±0.11%) of total 16S reads.
Discussion
While the isotope tracer experiments were conducted under conditions designed to approximately simulate in situ conditions in the bovine rumen and kangaroo forestomach, there were technical and logistical limitations that prevented all conditions from being mimicked exactly. The collection of kangaroo forestomach material from remote sites necessitated cold storage and preservation of samples in glycerol during transportation. This glycerol was removed by centrifugation and washing of sample biomass prior to inoculation of the incubations. Previous work in our laboratory has demonstrated that a diverse range of viable bacteria and archaea can be recovered from gut samples stored frozen in glycerol solution (Ouwerkerk et al., 2005); however, it is likely that the viability of some organisms, especially fungi and protozoans, with potentially important ecological roles would have been adversely affected by cold storage. The temperature of incubation may have also affected the results to some extent. The typical temperature of the kangaroo forestomach is slightly lower than that of the bovine rumen (36–37°C versus 39 °C). For the sake of consistency and to avoid introducing additional sources of variation between the kangaroo and bovine systems, all of the incubations for both the chemical isotope-tracing experiment and the RNA-SIP experiment were conducted at 39 °C. Despite these limitations, very clear and significant differences in formation of fermentation end products were observed between the kangaroo forestomach and bovine rumen systems.
Reductive acetogenesis is more active in the kangaroo forestomach than in the bovine rumen
The significantly reduced production of methane in kangaroo forestomach incubations supports previous studies that found that kangaroos produce little or no methane in comparison to ruminants (Kempton et al., 1976; Englehardt et al., 1978; Dellow et al., 1988). This reduced methane output and the fact that the small quantity of methane produced in the kangaroo forestomach fermentations remained relatively unlabelled in the presence of 13CO2/H13CO3− indicate that methanogenesis is not only less active in the foregut of kangaroos than in the rumen of cattle but also occurs by a fundamentally different pathway. It is clear from these observations that methane is not a major sink for CO2 in the foregut of eastern grey kangaroos.
The results of the isotope analysis of the VFAs produced in the three fermentation systems in the presence of 13CO2/H13CO3− clearly indicate that the conversion of CO2 to acetate via the reductive acetogenesis pathway is markedly more active in the kangaroo forestomach than in the bovine rumen. When considered in combination with the results of the gas headspace analysis, these results very strongly suggest that reductive acetogenesis is an important sink for carbon dioxide and hydrogen in the kangaroo foregut.
Analysis of the fractionation of carbon isotopes of methane produced in fermentations using unlabelled (12C) bicarbonate provided further evidence for a difference in the dominant methanogenic pathways occurring in the foregut of kangaroos. The strong isotope fractionation observed between headspace CO2 and methane in the bovine rumen fermentations (Figure 2) was consistent with isotopic fractionation that occurs during reduction of CO2 to methane (Krzycki et al., 1987; Botz et al., 1996). In contrast, relatively little difference was observed in the δ13C values of methane and CO2 in the kangaroo fermentations (Figure 2). When the carbon isotope ratios of acetate and methane in the kangaroo fermentations are compared however, the observed difference of −17 to −21‰ is more consistent with what would be expected for acetoclastic methanogenesis (Krzycki et al., 1987; Conrad et al., 2002; Penning et al., 2006).
The incorporation of 13C label into propionate in all three systems (Figure 3) was unexpected. The most likely explanation for this production of labelled propionate is that 13C-labelled CO2 was incorporated during the conversion of pyruvate to oxaloacetate, which is subsequently converted to propionate via malate, fumarate and succinate. The fact that the extent to which this process occurred did not differ significantly between the three systems suggests that it is a less important factor than reductive acetogenesis in explaining the reduced methane output of kangaroos relative to ruminants.
Identification of CO2/H2 utilising bacteria by RNA-SIP
RNA-SIP combined with 16S amplicon pyrosequencing and statistical comparison of the sequences obtained from ‘heavy’ and ‘light’ gradient fractions proved to be an effective approach for the identification of organisms involved in CO2/H2 metabolism in the kangaroo forestomach. This approach allowed OTUs corresponding to labelled RNA to be distinguished from the background ‘smear’ of RNA, which results in the generation of PCR product from all CsTFA gradient fractions. As has been the case with many other SIP studies (for example, Neufeld et al., 2007; DeRito and and Madsen, 2009; Bertram et al., 2013), in this study it was necessary to conduct the incubations under enrichment conditions that were not entirely representative of in situ conditions in order to achieve sufficient amounts of isotope labelling to allow separation of labelled and unlabelled biomolecules. In order to verify the presence and abundance of the organisms identified as CO2/H2 utilisers in the SIP experiment in live kangaroos, the 16S rRNA sequences of OTUs identified were used to query a database of bacterial 16S rRNA gene sequences that had been generated in a previous study (Gulino et al., 2013).
The identification of several putative CO2/H2-utilising bacteria, including the known reductive acetogen Blautia coccoides, and the confirmation that these bacteria occur in large numbers in at least three large kangaroo species (Figure 5) represent important steps towards a more complete understanding of the bacterial community involved in hydrogenotrophy in the kangaroo forestomach. The association of Blautia spp. with H2/CO2 utilisation in the kangaroo forestomach supports the hypothesis that reductive acetogens have an important role as hydrogenotrophs in macropods (Attwood and McSweeney, 2008; Ouwerkerk et al., 2009). Gagen et al. (2010) detected sequences of the Acetyl-CoA synthase gene (which is specific to the reductive acetogenesis pathway) in the forestomach of the Tammar wallaby (Macropus eugenii) that shared >99% similarity to the draft genome of Blautia hydrogenotrophica. It is interesting to note that the RNA-SIP experiment did not detect any other known reductive acetogens, despite the fact that Gagen et al. (2010) and Ouwerkerk et al. (2009) have demonstrated through both culture-based and culture-independent means that a considerable diversity of reductive acetogens exists in the macropod forestomach. It is possible that not all of these acetogens are metabolically active in situ, or that the incubation conditions used in the SIP experiment did not favour their activity. It is also possible that some of the other phylogenetic groups identified by RNA-SIP as H2/CO2 utilisers contain previously unrecognised acetogens or other hydrogenotrophs.
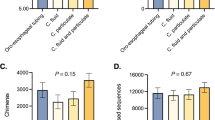
Heatmap representing the relative abundances in kangaroos and cattle of bacterial OTUs identified as H2/CO2 utilisers in the kangaroo foregut. For each sample, the colours represent the percentage of all bacterial 16S rRNA gene sequences obtained in 16S amplicon pyrosequencing surveys that share ⩾97% identity to representative sequences of H2/CO2 utilising OTUs identified in the RNA-SIP experiment.
Some OTUs identified by RNA-SIP were of particular interest because of their relatively high abundance in the forestomach of kangaroos. Foremost among these is OTU 24 (Table 1), which shared 98% 16S rRNA gene sequence similarity with a Prevotella sp. that was present in all individuals of all species of kangaroos previously surveyed, and comprised up to 9.8% of all 16S sequence reads in the forestomach of some animals (Figure 5). Comparison of the 16S rRNA gene sequences of this OTU to the Greengenes (DeSantis et al., 2006) and GenBank (Benson et al., 2011) databases indicated that its closest cultured relative was a Prevotella sp. cultured from the human oral cavity (Table 1), while its closest uncultured relative was a Prevotella sequence recovered from a metagenomic survey of the foregut of the Tammar wallaby (Pope et al., 2010). Unfortunately, it is difficult to speculate about the role of this OTU purely on the basis of its relatively distant phylogenetic relationships to other organisms. Members of the genus Prevotella have diverse functions in a variety of gut ecosystems, including the human colon and the bovine rumen, but have not been previously reported as being involved in the metabolism of H2 and CO2.
OTUs classified within the genera Streptococcus and Oscillibacter were also identified as H2/CO2 utilisers (Table 1), and 16S rRNA gene sequences of these OTUs were detected in high numbers in previously surveyed kangaroos (Figure 5). These genera have not been previously reported as containing hydrogenotrophic organisms, and again, it is difficult to speculate about the ecological roles of these OTUs in the kangaroo forestomach on the basis of their phylogenetic relationships. Work is currently focussed on isolating these bacteria in pure culture so that their potential roles as hydrogenotrophs can be investigated further.
Comparison of the 16S rRNA gene sequences of CO2/H2-utilising bacteria obtained from the RNA-SIP experiment to 16S rRNA gene sequences of the bacterial communities present in the bovine rumen led to an unexpected observation. Sequences closely related to many of the bacteria identified as being involved in CO2/H2 metabolism in the kangaroo forestomach were also present in the rumen of cattle that had been fed a grain-based diet, but were absent or only present in much lower numbers in the rumen of cattle fed a grass-based diet (Figure 5). The reason for this apparent similarity in the composition of the CO2/H2-utilising bacterial community in the kangaroo forestomach and the rumen of grain-fed cattle is not yet clear. It is known that cattle raised on a grain-based diet produce less methane than cattle on a grass-based diet (Harper et al., 1999; Doreau et al., 2011). It is possible that the reduced methane production observed in grain-fed cattle and kangaroos is related to the presence of hydrogenotrophic bacteria that provide an alternative hydrogen sink to methanogenesis.
While it is now clear that reductive acetogenesis is more active in the kangaroo forestomach than the bovine rumen, the channelling of CO2 and hydrogen into acetate production does not entirely explain the drastically reduced methane output of kangaroos. Our data also provides some evidence that acetoclastic methanogens are active in the kangaroo forestomach, so it is possible, in principle at least, that methane production could still occur via the acetoclastic pathway. While some acetate was apparently converted to methane in our in vitro fermentations, it not clear how much acetate would be diverted into this pathway in situ. Acetoclastic methanogenesis has a major role in carbon cycling in many anaerobic systems such as wetland and marine sediments (Conrad et al., 2002). In gut ecosystems however the continual export of acetate limits its recycling through the microbial community. While it is known that the epithelial cells of the mammalian gut are efficient scavengers of acetate and other VFAs (Pennington, 1952; Henning and Hird, 1970; Dijkstra, 1994), there are currently little experimental data available to indicate whether or not the host might be able to outcompete acetoclastic methanogens in rates of acetate uptake. Further studies using isotopically labelled acetate tracers could shed light on these questions.
Strains closely related to at least some of the bacteria involved in reductive acetogenesis in the kangaroo forestomach (for example, Blautia coccoides) were found to be already present in the rumens of some but not all cattle (Figure 5). The ecological factors that allow these organisms to proliferate in kangaroos and remain competitive against hydrogenotrophic methanogens while carrying out the thermodynamically less favourable process of reductive acetogenesis are not well understood. It appears that the growth of methanogens is suppressed in the kangaroo foregut ecosystem, as evidenced by the significantly lower quantities of methane produced in the kangaroo forestomach fermentations relative to the bovine rumen fermentations, and the drastically different structure of archaeal communities observed in the kangaroo forestomach by Klieve et al. (2012). The mechanisms of this suppression are currently unknown, and future work should focus on investigating the possibility of both host-mediated and intermicrobial mechanisms of methanogen suppression in kangaroos. If the factors underlying the suppression of methanogens and the proliferation of reductive acetogens in the kangaroo forestomach can be elucidated, it may be possible to design strategies to mitigate ruminant methane production by manipulating these factors in the rumen.
Conclusions
The data presented here provide strong evidence that reductive acetogenesis is an important sink for carbon dioxide and hydrogen in the kangaroo foregut. The dominance of reductive acetogenesis as a pathway for hydrogen removal in the kangaroo foregut explains the relatively low production of methane by kangaroos, and suggests that kangaroos are able to utilise feed energy more efficiently than ruminants. Blautia spp. appear to have a key role as reductive acetogens in the kangaroo forestomach, whereas the role of other bacteria including Prevotella, Oscillibacter and Streptococcus spp. as hydrogenotrophs is less clear.
References
Attwood G, McSweeney C . (2008). Methanogen genomics to discover targets for methane mitigation technologies and options for alternative H2 utilisation in the rumen. Austral J Exp Agri 48: 28–37.
Beal AM . (1984). Electrolyte composition of parotid saliva from sodium-replete red kangaroos (Macropus rufus). J Exp Biol 111: 225–237.
Benson DA, Karsch-Mizrachi I, Lipman DJ, Ostell J, Sayers EW . (2011). GenBank. Nucleic Acids Res 39: D32–D37.
Bertram S, Blumenberg M, Michaelis W, Siegert M, Krüger M, Seifert R . (2013). Methanogenic capabilities of ANME-archaea deduced from 13C-labelling approaches. Environ Microbiol 15: 2384–2393.
Botz R, Pokojski H-D, Schmitt M, Thomm M . (1996). Carbon isotope fractionation during bacterial methanogenesis by CO2 reduction. Org Geochem 25: 255–262.
Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK et al (2010). QIIME allows analysis of high-throughput community sequencing data. Nat Methods 7: 335–336.
Conrad R . (2009). The global methane cycle: recent advances in understanding the microbial processes involved. Environ Microbiol Rep 1: 285–292.
Conrad R, Klose M, Claus P . (2002). Pathway of CH4 formation in anoxic rice field soil and rice roots determined by 13C-stable isotope fractionation. Chemosphere 47: 797–806.
Dellow DW, Hume ID, Clarke RTJ, Bauchop T . (1988). Microbial activity in the forestomach of free-living macropodid marsupials; comparisons with laboratory studies. Austral J Zool 36: 383–395.
DeRito CM, Madsen EL . (2009). Stable isotope probing reveals Trichosporon yeast to be active in situ in soil phenol metabolism. ISME J 3: 477–485.
DeSantis TZ, Hugenholtz P, Larsen N, Rojas M, Brodie EL, Keller K et al (2006). Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl Environ Microbiol 72: 5069–5072.
Dijkstra J . (1994). Production and absorption of volatile fatty acids in the rumen. Livestock Production Sci 39: 61–69.
Doreau M, van der Werf HMG, Micol D, Dubroeucq H, Agabriel J, Rochette Y et al (2011). Enteric methane production and greenhouse gases balance of diets differing in concentrate in the fattening phase of a beef production system. J Animal Sci 89: 2518–2528.
Drake HL, Göβner AS, Daniel SL . (2008). Old acetogens, new light. Ann NY Acad Sci 1125: 100–128.
Eigeland KA, Lanyon JM, Trott DJ, Ouwerkerk D, Blanshard W, Milinovich GJ et al (2012). Bacterial community structure in the hindgut of wild and captive dugongs (Dugong dugon). Aquatic Mammals 38: 402–411.
Englehardt W, Wolter S, Lawrenz H, Hemsley JA . (1978). Production of methane in two non-ruminant herbivores. Compar Biochem Physiol 60: 309–311.
Erdman RA . (1988). Dietary buffering requirements of the lactating dairy cow: a review. J Dairy Sci 71: 3246–3266.
Gagen EJ, Denman SE, Padmanabha J, Zadbuke S, Al Jassim R, Morrison M et al (2010). Functional gene analysis suggests different acetogen populations in the bovine rumen and tammar wallaby forestomach. Appl Environ Microbiol 76: 7785–7795.
Gulino L, Ouwerkerk D, Kang A, Maguire AJ, Kienzle M, Klieve AV . (2013). Shedding the light on the macropod foregut using 454-amplicon pyrosequencing. PLos One 8: e1463.
Harper LA, Denmead OT, Freney JR, Byers FM . (1999). Direct measurements of methane emissions from grazing and feedlot cattle. J Animal Sci 77: 1392–1401.
Henning SJ, Hird FJR . (1970). Concentrations and metabolism of volatile fatty acids in the fermentative organs of two species of kangaroo and the guinea-pig. Br J Nutr 24: 145–155.
Kamlage B, Gruhl B, Blaut M . (1997). Isolation and characterization of two new homoacetogenic hydrogen-utilizing bacteria from the human intestinal tract that are closely related to Clostridium coccoides. Appl Environ Microbiol 63: 1732–1738.
Kempton TJ, Murray RM, Leng RA . (1976). Methane production and digestibility measurements in grey kangaroo and sheep. Austral J Biol Sci 29: 209–214.
Klieve AV, Hudman JF, Bauchop T . (1989). Inducible bacteriophages from ruminal bacteria. Appl Environ Microbiol 55: 1630–1634.
Klieve AV, Ouwerkerk D, Maguire AJ . (2012). Archaea in the foregut of macropod marsupials: PCR and amplicon sequence-based observations. J Appl Microbiol 113: 1065–1075.
Krzycki JA, Kenealy WR, DeNiro MJ, Zeikus JG . (1987). Stable carbon isotope fractionation by Methanosarcina barkeri during methanogenesis from acetate, methanol or carbon dioxide-hydrogen. Appl Environ Microbiol 53: 2597–2599.
Lassey KR . (2007). Livestock methane emission: from the individual grazing animal through national inventories to the global methane cycle. Agri Forest Meteorol 142: 120–132.
Liu C, Finegold SM, Song Y, Lawson PA . (2008). Reclassification of Clostridium coccoides, Ruminococcus hansenii, Ruminococcus hydrogenotrophicus, Ruminococcus luti, Ruminococcus productus and Ruminococcus schinkii as Blautia coccoides gen. nov., comb. nov., Blautia hansenii comb. nov., Blautia hydrogenotrophica comb. nov., Blautia luti comb. nov., Blautia producta comb. nov., Blautia schinkii comb. nov. and description of Blautia wexlerae sp. nov., isolated from human faeces. Int J Syst Evol Microbiol 58: 1896–1902.
Neufeld JD, Schäfer H, Cox MJ, Boden R, McDonald IR, Murrell JC . (2007). Stable-isotope probing implicates Methylophaga spp and novel Gammaproteobacteria in marine methanol and methylamine metabolism. ISME J 1: 480–491.
Ouwerkerk D, Klieve AV, Forster RJ, Templeton JM, Maguire AJ . (2005). Characterization of culturable anaerobic bacteria from the forestomach of an eastern grey kangaroo, Macropus giganteus. Lett Appl Microbiol 41: 327–333.
Ouwerkerk D, Maguire AJ, McMillen L, Klieve AV . (2009). Hydrogen utilising bacteria from the forestomach of eastern grey (Macropus giganteus) and red (Macropus rufus) kangaroos. Animal Production Sci 49: 1043–1051.
Penning H, Claus P, Casper P, Conrad R . (2006). Carbon isotope fractionation during acetoclastic methanogenesis by Methanosaeta concilii in culture and a lake sediment. Appl Environ Microbiol 72: 5648–5652.
Pennington R . (1952). The metabolism of short-chain fatty acids in the sheep. 1. Fatty acid utilization and ketone body production by rumen epithelium and other tissues. Biochem J 51: 251.
Pope PB, Denman SE, Jones M, Tringe SG, Barry K, Malfatti SA et al (2010). Adaptation to herbivory by the Tammar wallaby includes bacterial and glycoside hydrolase profiles different from other herbivores. Proc Natl Acad Sci USA 107: 14793–14798.
Sheppach W . (1994). Effects of short chain fatty acids on gut morphology and function. Gut 35: S35–S38.
Sokal RR, Rohlf FJ . (1969) Biometry. WH Freeman and Company: San Francisco, CA, USA.
Vreman HJ, Dowling JA, Raubach RA, Weiner MW . (1978). Determination of acetate in biological material by vacuum microdistillation and gas chromatography. Anal Chem 50: 1138–1141.
Whiteley AS, Thomson B, Lueders T, Manefield M . (2007). RNA stable-isotope probing. Nat Protoc 2: 838–844.
Wolin MJ, Miller TL, Stewart CS . (1997). Microbe-microbe interactions. In: Hobson PN, Stewart CS, (eds) The Rumen Microbial System. Chapman and Hall: London, UK, pp 467–488.
Yu Z, Forster RJ . (2005). Nucelic acid extraction, oligonucleotide probes and PCR methods. In: Makkar HPS, McSweeny CS, (eds) Methods in Gut Microbial Ecology for Ruminants. Springer: Dordrecht, Netherlands, pp 81–104.
Acknowledgements
We wish to acknowledge the assistance of Anita Maguire in sample collection and processing. We also thank Professor Sue Golding and Kim Baublys of the Stable Isotope Geochemistry laboratory at the University of Queensland for their assistance with stable isotope analysis. This work was funded by the Queensland Government.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
The state of Queensland (through the Department of Agriculture, Fisheries and Forestry).
Supplementary Information accompanies this paper on The ISME Journal website
Rights and permissions
About this article
Cite this article
Godwin, S., Kang, A., Gulino, LM. et al. Investigation of the microbial metabolism of carbon dioxide and hydrogen in the kangaroo foregut by stable isotope probing. ISME J 8, 1855–1865 (2014). https://doi.org/10.1038/ismej.2014.25
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ismej.2014.25
Keywords
This article is cited by
-
Formate-induced CO tolerance and methanogenesis inhibition in fermentation of syngas and plant biomass for carboxylate production
Biotechnology for Biofuels and Bioproducts (2023)
-
Isolation and characterization of reductive acetogens from rumen fluid samples of Murrah buffaloes
3 Biotech (2023)
-
Dietary selection of metabolically distinct microorganisms drives hydrogen metabolism in ruminants
The ISME Journal (2022)
-
Mathematical modelling supports the existence of a threshold hydrogen concentration and media-dependent yields in the growth of a reductive acetogen
Bioprocess and Biosystems Engineering (2020)
-
Do different livestock dwellings on single grassland share similar faecal microbial communities?
Applied Microbiology and Biotechnology (2019)