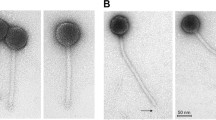
Abstract
Geobacter species may be important agents in the bioremediation of organic and metal contaminants in the subsurface, but as yet unknown factors limit the in situ growth of subsurface Geobacter well below rates predicted by analysis of gene expression or in silico metabolic modeling. Analysis of the genomes of five different Geobacter species recovered from contaminated subsurface sites indicated that each of the isolates had been infected with phage. Geobacter-associated phage sequences were also detected by metagenomic and proteomic analysis of samples from a uranium-contaminated aquifer undergoing in situ bioremediation, and phage particles were detected by microscopic analysis in groundwater collected from sediment enrichment cultures. Transcript abundance for genes from the Geobacter-associated phage structural proteins, tail tube Gp19 and baseplate J, increased in the groundwater in response to the growth of Geobacter species when acetate was added, and then declined as the number of Geobacter decreased. Western blot analysis of a Geobacter-associated tail tube protein Gp19 in the groundwater demonstrated that its abundance tracked with the abundance of Geobacter species. These results suggest that the enhanced growth of Geobacter species in the subsurface associated with in situ uranium bioremediation increased the abundance and activity of Geobacter-associated phage and show that future studies should focus on how these phages might be influencing the ecology of this site.
Similar content being viewed by others
Introduction
Viruses are known to have an important influence on the growth of bacterial populations in marine and freshwater environments (Bergh et al., 1989; Fuhrman and Noble, 1995; Noble and Fuhrman, 1999; Wilhelm and Suttle, 1999; Fischer and Velimirov, 2002; Bettarel et al., 2003; Suttle, 2007; Mouser et al., 2009; Weitz and Wilhelm, 2012; Weitz et al., 2013). However, their role in the microbial ecology of groundwater is less well understood. Viral lysis of groundwater bacteria may be particularly significant when designing bioremediation strategies to promote the growth of specific bacteria that can ameliorate contamination.
Geobacter species are often specifically enriched in subsurface environments contaminated with petroleum hydrocarbons or other organic contaminants because of their ability to couple the anaerobic oxidation of organic contaminants to the reduction of Fe(III) oxides that are generally abundant in the subsurface (Lovley et al., 2011). Furthermore, Geobacter growth in the subsurface is artificially promoted when simple organic substrates, such as acetate, are added to groundwater to stimulate the bioremediation of uranium and related contaminants (Anderson et al., 2003; Lovley et al., 2011; Handley et al., 2013).
Analysis of Geobacter growth during bioremediation of uranium-contaminated groundwater revealed that growth rates estimated from gene transcript abundance were much higher than the actual increase in cell numbers in the groundwater (Holmes et al., 2013a). Furthermore, rates of subsurface Geobacter growth were significantly lower than the rates predicted by genome-scale metabolic models (Scheibe et al., 2009; Fang et al., 2011; Lovley et al., 2011; Zhuang et al., 2011). Protozoa grazing on Geobacter species was one possible explanation for this discrepancy (Holmes et al., 2013b), as studies have shown that the specific growth rate of bacteria is higher in the presence of protozoa (Bloem et al., 1988; Verhagen et al., 1995; Strauss and Dodds, 1997; Biagini et al., 1998). In addition to protozoan grazing, it is also likely that phage activity has a substantial impact on bacterial growth in uranium-contaminated aquifers. Previous studies have shown that viral lysis can increase bacterial respiration and production rates in the subsurface by 27% (Middleboe and Lyck, 2002), and that viral lysis and protozoan grazing activities can have an additive effect on in situ bacterial growth (Berdjeb et al., 2011).
In this study, genomic, transcriptomic and proteomic tools were used to determine whether phages were active in an Fe(III)-reducing subsurface environment where Geobacter are dominant members of the bacterial community. Using these tools, we were able to identify a number of Geobacter-associated phage genes and show that many of them were being actively expressed during the Fe(III)-reducing phase of a uranium bioremediation field experiment.
Materials and methods
Site and description of field site
In 2011, a small-scale in situ bioremediation experiment was conducted on the grounds of a former uranium ore processing facility in Rifle, CO, USA, during the months of August–October as previously described (Giloteaux et al., 2013). This plot was biostimulated with acetate additions during the months of August–October. This research was part of the Uranium Mill Tailings Remedial Action (UMTRA) program of the US Department of Energy, and the plot used in this experiment was adjacent to a previously studied larger experimental plot at the site (Anderson et al., 2003; Vrionis et al., 2005). The monitoring array consisted of an injection gallery with six injection wells, nine down-gradient wells and one background monitoring well located upstream from the injection gallery (see Supplementary Material, Supplementary Figure S1). Groundwater for the experiments was collected from well CD-02.
During the field experiment, a concentrated acetate/bromide solution (50:20 mM) mixed with native groundwater was injected into the subsurface to provide ∼5 mM acetate to the groundwater over the course of 68 days as previously described (Anderson et al., 2003; Williams et al., 2011). Bromide was utilized as a nonreactive tracer.
Rifle sediment incubations
Background subsurface sediments were collected near the acetate-injection test plot with a back-hoe, placed in sterilized sealed mason jars and stored at 16 °C until use. Unfiltered background groundwater for sediment incubations was pumped to the surface into 5-gallon carboys with a peristaltic pump and stored at 4 °C.
For sediment incubations, 40 g of the background sediments described above, 6 ml groundwater and acetate (2 mM) were added to 60 ml serum bottles in an anaerobic chamber under an N2 atmosphere (Mouser et al., 2009). The bottles were sealed with a butyl rubber stopper and incubated at 20 °C. Three acetate-amended and three control (no acetate additions) incubations were monitored over the course of 20 days.
Analytical techniques
Samples for geochemical analyses were collected after purging 12 l of groundwater from the wells with a peristaltic pump. Ferrous iron was measured spectrophotometrically immediately after sampling using the phenanthroline method (AccuVac ampules; Hach Company, Loveland, CO, USA) for ferrous iron. After filtration through a 0.2 μm pore size polytetrafluoroethylene (PTFE (Teflon)) filter (Alltech Associates, Inc., Deerfield, IL, USA), acetate concentrations were measured with a Dionex ICS-1000 ion chromatograph equipped with a IonPac AS22 column, an ASRS 300 suppressor and 4.5 mM carbonate/1.4 mM bicarbonate eluent (Dionex Corporation, Sunnyvale, CA, USA).
Fe(III) reduction in the sediment incubations was monitored by measuring the formation of Fe(II) over time with a ferrozine assay in a split-beam dual-detector spectrophotometer (Spectronic Genosys2; Thermo Electron Corp., Mountain View, CA, USA) at an absorbance of 562 nm after 1 h of extraction with 0.5 N HCl (Lovley and Phillips, 1987, 1988). The remaining Fe(III) in the sediments that was not HCl extractable was then converted to Fe(II) by the addition of 0.25 M hydroxylamine (Lovley et al., 1987). After addition of hydroxylamine, samples were incubated for an additional 1 h, and then measured with a ferrozine assay. The percentage of Fe(II) in the sediments was then determined by dividing the HCl-extractable Fe(II) by the hydroxylamine-extractable Fe(II).
Phage particle and bacterial cell counts
Groundwater was collected at various time points from laboratory sediment incubations with a sterile syringe gassed out with N2. Bacterial cells and viral particles were quantified by counting labeled cells/particles with fluorescence microscopy on a Nikon (Chiyoda, Tokyo, Japan) Eclipse E600 microscope. Groundwater was diluted 10-fold in TE buffer (10 mM Tris-HCl and 1 mM EDTA, pH 7.5) and 900 μl of the diluted sample was added to 0.1 ml glutaraldehyde solution (final concentration, 2.5%; Sigma Chemical Company, St Louis, MO, USA). Cells/particles in the groundwater were fixed in the glutaraldehyde solution after a 5-min incubation at room temperature. Once cells were fixed, iron particles in the groundwater were dissolved by addition of 5.9 ml of filter-sterilized oxalate solution (197 mM ammonium oxalate and 119 mM oxalic acid) and 1.7 mM FeCl2. All samples were then filtered through a 0.45 μm polyvinylidene fluoridefilter (Millipore, Billerica, MA, USA) two times, and those analyzed for viral counts were also filtered through a 0.2 μm syringe filter (Corning Inc, Corning, NY, USA) to remove bacterial cells larger than 0.2 μm, leaving primarily viral particles for further analysis. Cells and particles were stained and enumerated immediately following this fixation step.
Bacterial cell suspensions were stained with an acridine orange solution (final concentration, 0.01%) and incubated at room temperature for 2 min as previously described (Coates et al., 1998). The sample was then vacuum filtered through a black Isopore membrane filter (pore diameter, 0.2 μm; Millipore) and examined under ultraviolet light. Viral suspensions were collected by vacuum filtration on an Anodisc filter (pore diameter, 0.02 μm; Whatman, Piscataway, NJ, USA) and stained by incubation with 100 μl 2 × SYBR Gold solution (Invitrogen, Carlsbad, CA, USA) for 15 min in the dark as previously described (Chen et al., 2001).
Extraction of nucleic acids from samples
DNA and RNA were extracted from groundwater collected from the U(VI)-contaminated aquifer during the bioremediation field experiments. In order to obtain sufficient biomass from the groundwater, it was necessary to concentrate 50 l of groundwater by impact filtration on 293 mm diameter Supor PES membrane disc filters (Pall Corporation, Cortland, NY, USA), and this took ∼3 min. All filters were placed into whirl-pack bags, flash frozen in a dry ice/ethanol bath and shipped back to the laboratory where they were stored at −80 °C. RNA was extracted from filters as previously described (Holmes et al., 2005) and DNA was extracted with the FastDNA SPIN Kit for Soil (MP Biomedicals, Santa Ana, CA, USA).
Analysis of nucleic acids by spectrophotometry (NanoDrop, Wilmington, DE, USA; Thermo Scientific, Waltham, MA, USA), microfluidic analysis (Experion, Bio-Rad, Hercules, CA, USA) and gel electrophoresis showed that high-quality DNA and RNA were extracted from the groundwater samples. In order to ensure that RNA samples were not contaminated with DNA, PCR amplification with primers targeting the 16S rRNA gene was conducted on RNA samples that had not undergone reverse transcription.
A DuraScript enhanced avian RT single-strand synthesis kit and random hexamers (Sigma-Aldrich, St Louis, MO, USA) were used to generate complementary DNA (cDNA) as previously described (Holmes et al., 2004).
Determination of bacterial community composition
In order to confirm that Geobacter species were enriched by acetate amendments to the groundwater in a manner similar to previous years (Holmes et al., 2002; Anderson et al., 2003; Holmes et al., 2005), 16S rRNA gene sequences present in the groundwater were analyzed with both clone library and terminal fragment length polymorphism analyses (Liu et al., 1997). Primers used for PCR amplification of bacterial 16S rRNA genes were the Bacteria-specific primers 8F and 1100R (Lane, 1991). Primer 8F was fluorescently labeled with FAM (6-carboxyfluoresceine) and the PCR products were purified with the Gel Extraction Kit (Qiagen, Venlo, Limburg, Germany). After gel purification, samples were cloned into the TOPO TA cloning vector, version M (Invitrogen), and 100 plasmid inserts from each clone library were sequenced with the M13F primer at the University of Massachusetts Sequencing Facility, and analyzed by terminal fragment length polymorphism analysis.
For terminal fragment length polymorphism studies, gel-purified 5′FAM-labeled 16S rRNA gene products were digested with MspI (New England Biolabs, Ipswich, MA, USA) at 37 °C for 3 h. All unincorporated dyes/nucleotides and salts were removed from the digested amplicons by centrifugation at 750 g in SigmaSpin Sequencing Reaction Clean-Up, Post-Reaction Clean-Up Columns (Sigma-Aldrich) according to the manufacturer’s instructions. Fluorescently labeled terminal restriction fragments were separated by capillary electrophoresis in a 3730xl DNA Analyzer (Applied Biosystems, Carlsbad, CA, USA) and analyzed with GeneMapper version 4.1 software (Applied Biosystems).
Design of Geobacter and phage-related primers
Geobacter-specific primers targeting citrate synthase (gltA; CS375F/CS598R) and the gene for DNA repair protein recombinase A (recA; recA-48F/recA-583R) were used to amplify Geobacter gene fragments from DNA extracted from groundwater collected from well CD-02 at the peak of Fe(III) reduction during the 2011 field experiment (Giloteaux et al., 2013) (Supplementary Table S1). Clone libraries were then assembled with these Geobacter-associated amplicons (Supplementary Table S1).
The genomes of five different subsurface Geobacter species, G. daltonii, G. uraniireducens, strain M18, strain M21 and G. bemidjiensis obtained from the NCBI (National Center for Biotechnology Information) Genbank website (http://www.ncbi.nlm.nih.gov) were searched for phage-related genes. Once these phage genes were identified, metagenomic libraries targeting all organisms/particles in the groundwater constructed from groundwater concentrated by tangential flow filtration (Wrighton et al., 2012) were scanned for homologous genes on ggKbase (http://genegrabber.berkeley.edu/). Degenerate primers targeting Geobacter-related gp19, integrase, baseplate J and head morphogenesis gene fragments were designed from alignments of phage genomic and metagenomic sequences and clone libraries were assembled with gene fragments amplified from DNA extracted from groundwater using these primer sets (Supplementary Table S1).
For construction of all degenerate clone libraries, amplicons were first purified with the Gel Extraction Kit (Qiagen), and cloned into the TOPO TA cloning vector, version M (Invitrogen). From each of these clone libraries, 100 plasmid inserts were sequenced with the M13F primer at the University of Massachusetts Sequencing Facility and these sequences were used to target sequences for design of Geobacter and phage-related quantitative PCR (qPCR) primers.
All qPCR primer sets used in this study are available in Supplementary Table S1; Geobacter gltA was amplified with CS375F/598R, Geobacter recA was amplified with recA-2F/3R, Geobacter-related gp19 was amplified with gp19_5f/107r and Geobacter-related baseplate J was amplified with BP984F/1113R).
Quantification of gene and transcript abundance by qPCR
Quantitative PCR amplification and detection were performed with the 7500 Real Time PCR System (Applied Biosystems) using genomic DNA and cDNA made by reverse transcription from mRNA extracted from groundwater collected during the bioremediation experiment. All qPCR assays had triplicate biological and technical replicates. Each reaction mixture consisted of a total volume of 25 μl and contained 1.5 μl of the appropriate primers (stock concentrations, 1.5 μM), 5 ng cDNA and 12.5 μl Power SYBR Green PCR Master Mix (Applied Biosystems). Standard curves covering 8 orders of magnitude were constructed with serial dilutions of known amounts of purified cDNA quantified with a NanoDrop ND-1000 spectrophotometer at an absorbance of 260 nm. Transcript abundances and qPCR efficiencies (90–99%) were calculated from appropriate standard curves and all qPCR experiments followed MIQE guidelines (Bustin et al., 2009). Optimal thermal cycling parameters consisted of an activation step at 50 °C for 2 min, an initial 10 min denaturation step at 95 °C followed by 40 cycles of 95 °C for 15 s and 58–60 °C for 1 min. After 40 cycles of PCR amplification, dissociation curves were made for all qPCR products by increasing the temperature from 58 °C to 95 °C at a ramp rate of 2%. The curves all yielded a single predominant peak, further supporting the specificity of the PCR primer pairs.
Phylogenetic analysis
Many of the phage genes were identified in the various Geobacter genomes through comparison with previous annotations with the blastp and PSI-BLAST algorithms (Altschul and Lipman, 1990; Altschul et al., 1997) with an E-value cutoff of 1 × 10−5. The programs PHAST (Zhou et al., 2011) and/or Prophinder were also used to find phage-related sequences. Others were determined by the presence of known phage domains (http://pfam.sanger.ac.uk/, http://blast.jcvi.org/web-hmm/) or the word ‘phage’ in protein coding features.
The nucleotide sequences of gp19 tail tube, integrase, head morphogenesis and baseplate J genes amplified with PCR from the uranium-contaminated aquifer have been deposited in the GenBank database under accession numbers KJ572864–KJ572873.
Protein extraction and quantification
Total protein was extracted from membrane disc filters by first crushing filters into a fine powder. This powder (∼0.5 g per tube) was then added to 10 different 2 ml screw cap tubes containing lysing matrix B (MP Biomedicals) and 1 ml lysis buffer (50 mM Tris-HCl, pH 7.5, 1% SDS) with protease inhibitor (Complete Mini EDTA-free Protease Inhibitor Cocktail Tablet; Roche Diagnostics, Indianapolis, IN, USA). Cells were lysed in the FastPrep-24 Tissue and Cell Homogenizer (MP Biomedicals) for 45 s at 6 m s−1. Cellular debris was then removed by centrifugation at 16 100 g for 30 min at 4 °C and the supernatant from all 10 lysis tubes was combined and transferred to an Amicon Ultra centrifugal filter unit with a molecular weight cutoff of 3000 g mol−1 (Millipore). Proteins were concentrated at room temperature by centrifugation at 2000 g for 1 h.
Proteins were quantified with the bicinchoninic acid assay (Bio-Rad) using bovine serum albumin as a standard. Equal amounts of protein (25 μg) from all 9 groundwater samples were boiled for 5 min with loading buffer and separated by electrophoresis in glycine-buffered 12.5% polyacrylamide gels. Total proteins were stained with Coomassie Blue solution (0.2% Coomassie blue, 7.5% acetic acid and 50% ethanol) and destained in a solution consisting of 30% ethanol and 10% acetic acid.
Overexpression and purification of Gp19 protein
The G. bemidjiensis gene that codes for a putative phage Gp19 tail tube protein (Gbem_2546) and is 100% identical to a sequence detected in metagenomic and proteomic libraries assembled from this site was expressed in Rosetta (DE3) pLysS competent cells (Novagen, Madison, WI, USA). Primers spanning the open reading frame of Gbem_2546 were designed with the NdeI restriction sequence added to the 5′ end of the forward primer and the XhoI restriction sequence added to the 5′ end of the reverse primer. PCR products were ligated into the PCR2.1 TOPO vector and transformed into chemically competent Top10 Escherichia coli cells (Invitrogen). Plasmids were purified from 10 different clones and sequenced with M13F and M13R to ensure that no errors were present in the gp19 open reading frame that had been ligated into the TOPO vector. The purified PCR2.1 TOPOgp19 vector and the pet29a vector were digested with NdeI and XhoI at 37 °C for 3 h. Restriction products were then concentrated by isopropanol precipitation as previously described (Aklujkar et al., 2013).
After precipitation, pellets from both the pet29a vector and the gp19 insert were resuspended in 10 μl nuclease free water (Ambion, Austin, TX, USA) and ligated together with T4 ligase (New England Biolabs) for 16 h at 16 °C. The ligation mixture was transformed into E. coli Top10 cells and transformants were selected on LB medium supplemented with kanamycin (50 μg ml−1). Purified plasmids were screened for insert by digestion with NdeI and XhoI, and plasmids containing the gp19 gene were transformed into E. coli Rosetta (DE3) pLysS cells. Protein induction with isopropyl β-D-1-thiogalactopyranoside was optimal after 3 h of incubation (Supplementary Figure S2A).
The Gp19 protein assembles into protomers. Therefore, to get a single monomer of Gp19, it was necessary to purify the recombinant Gp19 protein under denaturing conditions (8 M urea) with the Ni-NTA Spin kit (Qiagen) (Supplementary Figure S2B). Isolation of a Gp19 monomer was confirmed by a western blot with an antibody raised against the recombinant protein’s histidine tag (Supplementary Figure S2C).
Urea was removed from the purified protein by dialysis with a 3.5 kDa membrane in phosphate-buffered saline solution (pH 7.6) at 4 °C. After dialysis, the protein was concentrated in a Centricon filter unit with a molecular weight cutoff of 3.5 kDa (Millipore). Purified and concentrated protein was used to raise anti-Gp19 antibodies in rabbits (Thermo Scientific Pierce Protein Research, Thermo Scientific Pierce Protein Biology Products, Rockford, IL, USA).
Western blot analysis
Protein samples extracted as described above were loaded onto an SDS–polyacrylamide gel electrophoresis gel and transferred to a polyvinylidene fluoride membrane (Millipore). The membrane was incubated in a rotary shaker with MaNa buffer (0.10 M maelic acid, 0.15 M NaCl; pH 7.5) with 10% Blocking Solution (Roche) at room temperature for 45 min. The antibodies (GltA or Gp19) were diluted (1:1000 and 1:5000) in MaNa+blocking solution and added to the membrane. After 2 h of incubation with the antibody solution, the membrane was washed once (15 min) with MaNa buffer, and a secondary donkey anti-rabbit antibody conjugated with alkaline phosphatase diluted 1:5000 in MaNa buffer was added to the membrane and incubated for 1 h. Following incubation with the secondary antibody, the membrane was washed two times with MaNa buffer and once with MaNa buffer +0.3% Tween. The blot was then equilibrated for 5 min in detection buffer (0.1 M Tris-HCl, 0.1 M NaCl, pH 9.5) and bands were visualized by the addition of nitro-blue tetrazolium chloride/5-bromo-4-chloro-3′-indolyphosphate p-toluidine salt according to the manufacturer’s instructions (Roche Applied Science, Indianapolis, IN, USA).
Results and discussion
Phage genes in subsurface Geobacter isolates and subsurface metagenomes
In order to determine the potential role of phage in Geobacter-dominated subsurface environments, the genomes of Geobacter species isolated from contaminated subsurface environments (G. uraniireducens, G. daltonii, G. bemidjiensis, strain M21 and strain M18) were analyzed for evidence of viral infection. Each contained numerous phage-related genes (Supplementary Table S2). G. uraniireducens, strain M21 and strain M18, which were isolated from the uranium bioremediation site in Rifle (Shelobolina et al., 2008), had the greatest number of phage genes. There were 151, 126 and 181 phage genes detected in G. uraniireducens, strain M21 and strain M18, respectively. G. bemidjiensis, which was isolated from a hydrocarbon-contaminated aquifer (Nevin et al., 2005), had 70 phage-related genes, and G. daltonii, isolated from a uranium and nitrate-contaminated aquifer (Prakash et al., 2010), had 56. Many of these genes coded for structural proteins associated with tailed bacteriophage such as capsid, tail tube, tail fiber, tail sheath and baseplate proteins (Figure 1).
It appears that many of the phages associated with these subsurface Geobacter are temperate. It has been previously established that when ⩾3 phage genes are found within a 10 open reading frame window, they are likely to have been acquired from phage insertion into the bacterial chromosome rather than from horizontal gene transfer (Paul, 2008). Many of the Geobacter subsurface phage genes were found in genomic clusters with ⩾3 genes (Figure 2), making it likely that they were acquired from prophage. Further evidence that some of the subsurface Geobacter phage are lysogenic comes from the fact that multiple phage attachment sites (att) were detected in the Geobacter chromosomes, and the programs PHAST (Zhou et al., 2011 PHAST) and/or Prophinder (http://aclame.ulb.ac.be/Tools/Prophinder/) identified prophage in all five of the genomes.
Distribution of phage gene clusters within five subsurface Geobacter genomes. Supplementary Figure S3 provides a more detailed representation of the phage-related gene clusters found in the different Geobacter genomes.
The largest prophage gene cluster was found in strain M18 (Figure 3). It spans a 155-kb region, contains all of the genes necessary for survival and assembly of a Mu-like prophage (Paul, 2008) and was flanked by the phage attachment sites attL and attR (Figure 3). The strain M18 genome also had five other phage gene clusters ranging in size from 13.6 to 39.1 kb. The strain M21 genome had 8 phage gene clusters ranging in size from 4.97 to 46.82 kb (Figure 3). The G. uraniireducens genome had 10 phage gene clusters ranging in size from 6.35 to 45.49 kb. The G. bemidjiensis genome had six phage gene clusters ranging in size from 6.15 to 17.9 kb. The G. daltonii genome only had 4 gene clusters ranging in size from 1.64 to 18.68 kb (Supplementary Figure S3).
All 5 genomes contain a number of genes coding for putative integrase proteins, and as many as 37 integrase genes are present in the genome of G. uraniireducens (Figure 1 and Supplementary Table S2). All five of the subsurface Geobacter genomes also contain a number of prophage repressor proteins that allow the phage to reside inactive in the chromosome of its host bacterium (Los and Wegrzyn, 2012), and genes coding for the NusB antitermination factor, which allows phage to switch from the lysogenic to the lytic cycle (Supplementary Table S2). In addition, many of the genomes contain genes coding for prophage addiction proteins (Phd and Doc), which make the host cell dependent on the phage for survival, and genes for antirestriction proteins, which allow the prophage to evade destruction by the host cell (Supplementary Table S2).
Metagenomic and proteomic analysis of Geobacter-associated phage in the groundwater during U(VI) bioremediation
Analysis of metagenomic data assembled from groundwater collected during a field experiment conducted in 2008 in which acetate was added to the subsurface to stimulate metal reduction and facilitate U(VI) bioremediation (Giloteaux et al., 2013) also revealed the presence of a number of Geobacter-associated phage genes (Figure 4 and Supplementary Table S2). In all, 189 different Geobacter phage genes were detected, and many of these genes were similar to phage genes from subsurface isolates and genes found in clone libraries assembled from groundwater collected from this site. For example, two gp19 genes detected in the Rifle metagenome (ACDPHA_C00010G00002 and ACD55_50296.5307.13G0001) were identical to Gp19 tail tube proteins (Gbem_2546 and Gbem_2547) found in G. bemidjiensis, and the metagenomic sequence ACDUNK_1878.11196.24G0012 was 100% identical to an integrase protein from G. bemidjiensis (Gbem_1749).
Proteomic data assembled from groundwater collected during U(VI) bioremediation field experiments conducted in 2007, 2008 and 2010 (Wilkins et al., 2009; Wrighton et al., 2012; Giloteaux et al., 2013) were also analyzed to determine whether Geobacter-associated phage proteins were being actively translated during the bioremediation process. A total of 87 different Geobacter-associated phage proteins were detected, many of which were involved in phage assembly such as tail, head or baseplate subunits (Figure 5 and Supplementary Table S3). Another protein that was detected in the groundwater proteome was a Geobacter-associated prophage antirepressor protein that is involved in prophage induction. Although Geobacter-associated phage-like proteins were being expressed in the subsurface, it is not yet clear whether these proteins are being assembled into intact, infectious phages. It is possible that some of these proteins are just being expressed by in situ Geobacter and serve a different function within the bacterial cell. For example, pyocins expressed by Pseudomonas species and several proteins associated with type IV secretion systems in Gram-negative bacteria are homologous to bacteriophage tail proteins (Michel-Briand and Baysse, 2002; Leiman et al., 2009). However, expression of such a wide diversity of phage structural proteins, many of which do not have homologs in bacteria, suggests that Geobacter-associated phages were active in the subsurface during uranium bioremediation.
Quantitative and transcriptomic analysis of in situ phage activity
The expression of some of the predominant Geobacter-associated phage genes that were detected in Rifle metagenomes, proteomes and/or pure culture genomes was monitored over the course of 53 days during a bioremediation experiment conducted in 2011.
Geochemical analyses of groundwater collected from well CD-02 revealed an increase in Fe(II) concentrations at the beginning of the field experiment (days 0–16), followed by an increase in sulfide concentrations (days 18–44) (Figures 6a and b). Previous studies have shown that these reduced products are formed biologically by dissimilatory Fe(III) and sulfate-reducing bacteria primarily from the genera Geobacter, Desulfobacter and Desulfosporosinus (Anderson et al., 2003; Holmes et al., 2013b).
(a) Fe(II) and acetate concentrations detected in groundwater collected from well CD-02 over the course of 53 days; (b) Fe(II) and hydrogen sulfide concentrations detected in groundwater; (c) the proportion of Geobacter 16S rRNA gene sequences detected in bacterial libraries and the abundance of Geobacter gltA and recA gene copies in the groundwater at various time points.
In this experiment, when Fe(III) reduction was the primary electron accepting process, Geobacter accounted for up to 65% of the 16S rRNA gene sequences (Figure 6b) and the dominant terminal fragment length polymorphism fragment size (395 bp) corresponded with a clone sequence that was 97% identical to Geobacter bemidjiensis (Supplementary Figure S4A). Quantitative PCR and quantitative reverse transcriptase-PCR also revealed a significant increase in the number of Geobacter gltA and recA gene copies (Figure 6c) and gltA mRNA transcripts (Figure 7) during the Fe(III)- reducing phase of the experiment. This indicated that, similar to previous studies (Holmes et al., 2005; Wilkins et al., 2011; Yun et al., 2011), Geobacter were not only more abundant but were also more metabolically active during this phase of the experiment.
In situ analysis of groundwater collected over the course of the uranium bioremediation field experiment. (a) The number of Geobacter gltA gene copies relative to the number of phage gp19 and baseplate J gene copies detected in the groundwater; (b) the number of Geobacter gltA mRNA transcripts relative to the number of phage gp19 and baseplate J mRNA transcripts detected in the groundwater.
Associated with this increase in Geobacter abundance and activity was a dramatic increase in the number of gene copies and transcripts for two important phage assembly proteins, baseplate J and tail tube gp19 proteins (Figure 7a). Phage genes and transcripts were highest between days 3 and 11 when Geobacter activity and abundance was also greatest. However, whereas gp19 and baseplate J gene copies and transcripts started to decline after day 11, Geobacter gltA gene copies remained relatively high until day 30 when sulfate reduction became the primary electron accepting process.
Similar results were obtained from Rifle sediment incubations (Figure 8). Fluorescently labeled phage particles and Geobacter-associated gp19 gene copies were greatest on day 9 (6.9 × 107 phage particles per ml and 4.7 × 107 gp19 gene copies per μg total DNA) when overall bacterial cell numbers and Geobacter gltA gene copies were also at their peak (4.6 × 106 bacterial cells per ml and 2.24 × 106 gltA gene copies per μg total DNA; Figures 8a and b). After day 9, the number of Geobacter-associated gp19 gene copies dropped significantly, whereas Geobacter gltA gene copies remained relatively high until day 16 when Fe(II) concentrations also started to decline (Figure 8c) and sulfate reduction started (data not shown). The overall abundance of phage particles remained high until day 13 and total bacterial abundance did not start to decrease until day 16 when electron donors became limiting.
Analysis of phage and bacterial growth in groundwater collected from laboratory sediment incubations. (a) The number of bacterial cells and phage particles monitored over time by fluorescence microscopy; (b) the number of Geobacter gltA and Geobacter-associated phage gp19 gene copies per μg total DNA monitored over time by qPCR; (c) changes in Fe(II) concentrations over time.
These results could indicate that some of the phages are becoming lysogenic or the fact that the population of Geobacter species changed over the course of the Fe(III)-reducing phase of the experiment (Supplementary Figure S4B). The qPCR experiments targeted sequences most similar to gp19 and baseplate J sequences detected in G. bemidjiensis and strain M18. In the early stages of Fe(III) reduction, both of these species were predominant; however, after day 11, these species were no longer detected and sequences most similar to G. psychrophilus predominated. It is possible that the phages were not infecting the other Geobacter species that bloomed later in the experiment; however, further investigation into this possibility is required.
Proteomic analysis of in situ phage activity
In order to further evaluate the potential presence of Geobacter phage in the groundwater during bioremediation, the abundance of a phage tail tube Gp19 protein was monitored over time with an antibody to a protein encoded by a metagenomic sequence (ACDPHA_C00010G00002) that was identical to Gbem_2546 from G. bemidjiensis. The abundance of tail tube Gp19 tracked with the abundance of Geobacter species as detected with a Geobacter-specific citrate synthase antibody (Figure 9). Results from the western blot were similar to those observed with quantitative reverse transcriptase-PCR; Geobacter GltA and phage Gp19 proteins were most abundant between days 3 and 11 and Geobacter GltA proteins continued to be expressed at low levels until day 62.
Implications
These studies demonstrate that subsurface Geobacter species at the Rifle site are susceptible to bacteriophage infection and that many Geobacter-associated phage proteins are being expressed during uranium bioremediation. The genomes of the 3 species that were isolated from this site (strain M18, strain M21 and G. uraniireducens) contain as many as 181 different phage-related genes, and the chromosome of strain 18 may be harboring a Mu-like temperate phage. Proteomic and transcriptomic studies of groundwater collected at the site also demonstrated that Geobacter-associated phage genes required for host cell lysis and phage assembly are being actively transcribed and translated in the subsurface during uranium bioremediation.
The presence of a number of phage-related integrase and prophage addiction genes in the Geobacter genomes may indicate that they have a history of being infected by temperate phage or other mobile genetic elements. Lysogeny is common in environments with low nutrient concentrations and is thought to be an adaptation of viruses that allows them to survive adverse conditions such as periods when resources are scarce and host abundance is low (Ripp and Miller, 1997; Williamson et al., 2002; Thomas et al., 2011; Los and Wegrzyn, 2012).
Before the addition of acetate to stimulate the growth of Geobacter in the subsurface, Geobacter species grow slowly and are not abundant at the Rifle site (Holmes et al., 2004; Mouser et al., 2009; N’Guessan et al., 2010). Therefore, a lysogenic phase for phage would be favored. However, when growth is stimulated with acetate, it is possible that cell lysis by prophage may be induced. Further studies will need to be done to verify whether cell lysis is being induced by Geobacter prophage in the subsurface.
The impact of Geobacter-associated phage on Geobacter growth during bioremediation could be significant, but is difficult to quantify. In aquatic environments, viruses are responsible for the mortality of up to 50% of the bacterial population (Fuhrman and Noble, 1995; Guixa-Boixareu et al., 1996; Weinbauer and Hoefle, 1998a,1998b; Noble and Fuhrman, 2000). Lysis of Geobacter species can be expected to release organic carbon compounds that are more complex than acetate, and this could explain the increased abundance of microorganisms that appear to have a fermentative metabolism during uranium bioremediation (Wrighton et al., 2012). Thus, more studies need to be done to determine the impact of cell lysis by Geobacter-associated phage on microbial community dynamics, and future attempts to predictively model the dynamics of microbial growth and activity during in situ uranium bioremediation (Scheibe et al., 2009; Mahadevan et al., 2011; Zhuang et al., 2011) should include a viral lysis component.
References
Aklujkar M, Coppi MV, Leang C, Kim BC, Chavan MA, Perpetua LA et al. (2013). Proteins involved in electron transfer to Fe(III) and Mn(IV) oxides by Geobacter sulfurreducens and Geobacter uraniireducens. Microbiology 159: 515–535.
Altschul SF, Lipman DJ . (1990). Protein database searches for multiple alignments. Proc Natl Acad Sci USA 87: 5509–5513.
Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W et al. (1997). Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25: 3389–3402.
Anderson RT, Vrionis HA, Ortiz-Bernad I, Resch CT, Long PE, Dayvault R et al. (2003). Stimulating the in situ activity of Geobacter species to remove uranium from the groundwater of a uranium-contaminated aquifer. Appl Environ Microbiol 69: 5884–5891.
Berdjeb L, Pollet T, Domaizon I, Jacquet S . (2011). Effect of grazers and viruses on bacterial community structure and production in two contrasting trophic lakes. BMC Microbiol 11: 88.
Bergh O, Borsheim KY, Bratbak G, Heldal M . (1989). High abundance of viruses found in aquatic environments. Nature 340: 467–468.
Bettarel Y, Sime-Ngando T, Amblard C, Carrias JF, Portelli C . (2003). Virioplankton and microbial communities in aquatic systems: a seasonal study in two lakes of differing trophy. Freshwater Biol 48: 810–822.
Biagini GA, Finlay BJ, Lloyd D . (1998). Protozoan stimulation of anaerobic microbial activity: enhancement of the rate of terminal decomposition of organic matter. FEMS Microbiol Ecol 27: 1–8.
Bloem J, Starink M, Bargilissen MJB, Cappenberg TE . (1988). Protozoan grazing, bacterial activity and mineralization in 2-stage continuous cultures. Appl Environ Microbiol 54: 3113–3121.
Bustin SA, Benes V, Garson JA, Hellemans J, Huggett J, Kubista M et al. (2009). The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin Chem 55: 611–622.
Chen F, Lu JR, Binder BJ, Liu YC, Hodson RE . (2001). Application of digital image analysis and flow cytometry to enumerate marine viruses stained with SYBR gold. Appl Environ Microbiol 67: 539–545.
Coates JD, Councell T, Ellis DJ, Lovley DR . (1998). Carbohydrate oxidation coupled to Fe(III) reduction, a novel form of anaerobic metabolism. Anaerobe 4: 277–282.
Fang YL, Scheibe TD, Mahadevan R, Garg S, Long PE, Lovley DR . (2011). Direct coupling of a genome-scale microbial in silico model and a groundwater reactive transport model. J Contam Hydrol 122: 96–103.
Fischer UR, Velimirov B . (2002). High control of bacterial production by viruses in a eutrophic oxbow lake. Aq Microb Ecol 27: 1–12.
Fuhrman JA, Noble RT . (1995). Viruses and protists cause similar bacterial mortality in coastal seawater. Limnol Oceanogr 40: 1236–1242.
Giloteaux L, Holmes DE, Williams KH, Wrighton KC, Wilkins MJ, Montgomery AP et al. (2013). Characterization and transcription of arsenic respiration and resistance genes during in situ uranium bioremediation. ISME J 7: 370–383.
Guixa-Boixareu N, Calderon-Paz J, Heldal M, Bratbak G, Pedros-Alio C . (1996). Viral lysis and bacterivory as prokaryotic loss factors along a salinity gradient. Aq Microb Ecol 11: 215–227.
Handley KM, VerBerkmoes NC, Steefel CI, Williams KH, Sharon I, Miller CS et al. (2013). Biostimulation induces syntrophic interactions that impact C, S and N cycling in a sediment microbial community. ISME J 7: 800–816.
Holmes DE, Finneran KT, O’Neil RA, Lovley DR . (2002). Enrichment of members of the family Geobacteraceae associated with stimulation of dissimilatory metal reduction in uranium-contaminated aquifer sediments. Appl Environ Microbiol 68: 2300–2306.
Holmes DE, Giloteaux L, Barlett M, Chavan MA, Smith JA, Williams KH et al. (2013a). Molecular analysis of the in situ growth rates of subsurface Geobacter species. Appl Environ Microbiol 79: 1646–1653.
Holmes DE, Giloteaux L, Williams KH, Wrighton KC, Wilkins MJ, Thompson CA et al. (2013b). Enrichment of specific protozoan populations during in situ bioremediation of uranium-contaminated groundwater. ISME J 7: 1286–1298.
Holmes DE, Nevin KP, Lovley DR . (2004). In situ expression of nifD in Geobacteraceae in subsurface sediments. Appl Environ Microbiol 70: 7251–7259.
Holmes DE, Nevin KP, O’Neil RA, Ward JE, Adams LA, Woodard TL et al. (2005). Potential for quantifying expression of the Geobacteraceae citrate synthase gene to assess the activity of Geobacteraceae in the subsurface and on current-harvesting electrodes. Appl Environ Microbiol 71: 6870–6877.
Lane DJ . (1991). rDNA sequencing. In: Stachenbrady E (ed) Nucleic Acid Techniques in Bacterial Systematics. Wiley: Chichester, pp 115–175.
Leiman PG, Basler M, Ramagopal UA, Bonanno JB, Sauder JM, Pukatzki S et al. (2009). Type VI secretion apparatus and phage tail-associated protein complexes share a common evolutionary origin. Proc Natl Acad Sci USA 106: 4154–4159.
Liu W-T, Marsh TL, Cheng H, Forney LJ . (1997). Characterization of microbial diversity by determining terminal restriction fragment length polymorphisms of genes encoding 16S rRNA. Appl Environ Microbiol 63: 4516–4522.
Los M, Wegrzyn G . (2012). Pseudolysogeny. In: Lobocka M, Szybalski WT (eds) Advances in Virus Research, Volume 82: Bacteriophages, Part A pp 339–349.
Lovley DR, Phillips EJ . (1988). Novel mode of microbial energy metabolism: organic carbon oxidation coupled to dissimilatory reduction of iron or manganese. Appl Environ Microbiol 54: 1472–1480.
Lovley DR, Phillips EJP . (1987). Rapid asay for microbially reducible ferric iron in aquatic sediments. Appl Environ Microbiol 53: 1536–1540.
Lovley DR, Stolz JF, Nord GL, Phillips EJP . (1987). Anaerobic production of magnetite by a dissimilatory iron-reducing microorganism. Nature 330: 252–254.
Lovley DR, Ueki T, Zhang T, Malvankar NS, Shrestha PM, Flanagan KA et al. (2011). Geobacter: the microbe electric’s physiology, ecology, and practical applications. In: Poole RK (ed) Advances in Microbial Physiology vol. 59 Elsevier Ltd, Academic Press: Amsterdam, The Netherlands, pp 1–100.
Mahadevan R, Palsson BO, Lovley DR . (2011). In situ to in silico and back: elucidating the physiology and ecology of Geobacter spp. using genome-scale modelling. Nat Rev Microbiol 9: 39–50.
Michel-Briand Y, Baysse C . (2002). The pyocins of Pseudomonas aeruginosa. Biochimie 84: 499–510.
Middleboe M, Lyck PG . (2002). Regeneration of dissolved organic matter by viral lysis in marine microbial communities. Aq Microb Ecol 27: 187–194.
Mouser PJ, Holmes DE, Perpetua LA, DiDonato R, Postier B, Liu A et al. (2009). Quantifying expression of Geobacter spp. oxidative stress genes in pure culture and during in situ uranium bioremediation. ISME J 3: 454–465.
N’Guessan AL, Elifantz H, Nevin KP, Mouser PJ, Methe B, Lwoodard T et al. (2010). Molecular analysis of phosphate limitation in Geobacteraceae during the bioremediation of a uranium-contaminated aquifer. ISME J 4: 253–266.
Nevin KP, Holmes DE, Woodard TL, Hinlein ES, Ostendorf DW, Lovley DR . (2005). Geobacter bemidjiensis sp nov and Geobacter psychrophilus sp nov., two novel Fe(III)-reducing subsurface isolates. Int J Syst Evol Micr 55: 1667–1674.
Noble RT, Fuhrman JA . (1999). Breakdown and microbial uptake of marine viruses and other lysis products. Aq Microb Ecol 20: 1–11.
Noble RT, Fuhrman JA . (2000). Rapid virus production and removal as measured with fluorescently labeled viruses as tracers. Appl Environ Microbiol 66: 3790–3797.
Paul JH . (2008). Prophages in marine bacteria: dangerous molecular time bombs or the key to survival in the seas? ISMEJ 2: 579–589.
Prakash O, Gihring TM, Dalton DD, Chin K-J, Green SJ, Akob DM et al. (2010). Geobacter daltonii sp. nov., an Fe(III)- and uranium(VI)-reducing bacterium isolated from a shallow subsurface exposed to mixed heavy metal and hydrocarbon contamination. Int J Syst Evol Micr 60: 546–553.
Ripp S, Miller RV . (1997). The role of pseudolysogeny in bacteriophage-host interactions in a natural freshwater environment. Microbiology 143: 2065–2070.
Scheibe TD, Mahadevan R, Fang YL, Garg S, Long PE, Lovley DR . (2009). Coupling a genome-scale metabolic model with a reactive transport model to describe in situ uranium bioremediation. Microb Biotech 2: 274–286.
Shelobolina ES, Vrionis HA, Findlay RH, Lovley DR . (2008). Geobacter uraniireducens sp nov., isolated from subsurface sediment undergoing uranium bioremediation. Int J Syst Evol Micr 58: 1075–1078.
Strauss EA, Dodds WK . (1997). Influence of protozoa and nutrient availability on nitrification rates in subsurface sediments. Microbial Ecol 34: 155–165.
Suttle CA . (2007). Marine viruses—major players in the global ecosystem. Nat Rev Microbiol 5: 801–812.
Thomas R, Berdjeb L, Sime-Ngando T, Jacquet S . (2011). Viral abundance, production, decay rates and life strategies (lysogeny versus lysis) in Lake Bourget (France). Environ Microbiol 13: 616–630.
Verhagen FJM, Laanbroek HJ, Woldendorp JW . (1995). Competition for ammonium between plant roots and nitrifying and heterotrophic bacteria and the effects of protozoan grazing. Plant Soil 170: 241–250.
Vrionis HA, Anderson RT, Ortiz-Bernad I, O’Neill KR, Resch CT, Peacock AD et al. (2005). Microbiological and geochemical heterogeneity in an in situ uranium bioremediation field site. Appl Environ Microbiol 71: 6308–6318.
Weinbauer MG, Hoefle MG . (1998a). Size-specific mortality of lake bacterioplankton by natural virus communities. Aq Microb Ecol 15: 103–113.
Weinbauer MG, Hofle MG . (1998b). Significance of viral lysis and flagellate grazing as factors controlling bacterioplankton production in a eutrophic Lake. Appl Environ Microbiol 64: 431–438.
Weitz JS, Poisot T, Meyer JR, Flores CO, Valverde S, Sullivan MB et al. (2013). Phage-bacteria infection networks. Trends Microbiol 21: 82–91.
Weitz JS, Wilhelm SW . (2012). Ocean viruses and their effects on microbial communities and biogeochemical cycles. F1000 Biol Rep 4: 17.
Wilhelm SW, Suttle CA . (1999). Viruses and nutrient cycles in the sea. Bioscience 49: 781–788.
Wilkins MJ, Callister SJ, Miletto M, Williams KH, Nicora CD, Lovley DR et al. (2011). Development of a biomarker for Geobacter activity and strain composition; Proteogenomic analysis of the citrate synthase protein during bioremediation of U(VI). Microb Biotech 4: 55–63.
Wilkins MJ, VerBerkmoes NC, Williams KH, Callister SJ, Mouser PJ, Elifantz H et al. (2009). Proteogenomic monitoring of Geobacter physiology during stimulated uranium bioremediation. Appl Environ Microbiol 75: 6591–6599.
Williams KH, Long PE, Davis JA, Wilkins MJ, N’Guessan AL, Steefel CI et al. (2011). Acetate availability and its influence on sustainable bioremediation of uranium-contaminated groundwater. Geomicrobiol J 28: 519–539.
Williamson SJ, Houchin LA, McDaniel L, Paul JH . (2002). Seasonal variation in lysogeny as depicted by prophage induction in Tampa Bay, Florida. Appl Environ Microbiol 68: 4307–4314.
Wrighton KC, Thomas BC, Sharon I, Miller CS, Castelle CJ, VerBerkmoes NC et al. (2012). Fermentation, hydrogen, and sulfur metabolism in multiple uncultivated bacterial phyla. Science 337: 1661–1665.
Yun J, Ueki T, Miletto M, Lovley DR . (2011). Monitoring the metabolic status of Geobacter species in contaminated groundwater by quantifying key metabolic proteins with Geobacter-specific antibodies. Appl Environ Microbiol 77: 4597–4602.
Zhou Y, Liang Y, Lynch KH, Dennis JJ, Wishart DS . (2011). PHAST: a fast phage search tool. Nucleic Acids Res 39: W347–W352.
Zhuang K, Izallalen M, Mouser P, Richter H, Risso C, Mahadevan R et al. (2011). Genome-scale dynamic modeling of the competition between Rhodoferax and Geobacter in anoxic subsurface environments. ISME J 5: 305–316.
Acknowledgements
Research at the University of Massachusetts was funded by the Office of Science (BER), US Department of Energy, Award No. DE-SC0006790. Additional support for field research was equally supported through the Integrated Field Research Challenge Site (IFRC) at Rifle, CO, USA, and the Lawrence Berkeley National Laboratory’s Sustainable Systems Scientific Focus Area. Research at the Lawrence Berkeley National Laboratory was supported by the US Department of Energy (DOE), Office of Science (BER) under contract DE-AC02-05CH11231, the Subsurface Biogeochemistry Program (SBR) Grant Number DE-SC0004733, and by the Office of Basic Energy Sciences of the US Department of Energy under Contract No. DE-AC02-05CH11231. We thank Jillian Banfield for allowing us to analyze metagenomic and proteomic data generated by her laboratory at the University of California, Berkeley.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies this paper on The ISME Journal website
Rights and permissions
About this article
Cite this article
Holmes, D., Giloteaux, L., Chaurasia, A. et al. Evidence of Geobacter-associated phage in a uranium-contaminated aquifer. ISME J 9, 333–346 (2015). https://doi.org/10.1038/ismej.2014.128
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ismej.2014.128
This article is cited by
-
Mercury alters the rhizobacterial community in Brazilian wetlands and it can be bioremediated by the plant-bacteria association
Environmental Science and Pollution Research (2020)
-
Meta-proteomic analysis of protein expression distinctive to electricity-generating biofilm communities in air-cathode microbial fuel cells
Biotechnology for Biofuels (2018)