Abstract
Bacteriophages encode auxiliary metabolic genes that support more efficient phage replication. For example, cyanophages carry several genes to maintain host photosynthesis throughout infection, shuttling the energy and reducing power generated away from carbon fixation and into anabolic pathways. Photodamage to the D1/D2 proteins at the core of photosystem II necessitates their continual replacement. Synthesis of functional proteins in bacteria requires co-translational removal of the N-terminal formyl group by a peptide deformylase (PDF). Analysis of marine metagenomes to identify phage-encoded homologs of known metabolic genes found that marine phages carry PDF genes, suggesting that their expression during infection might benefit phage replication. We identified a PDF homolog in the genome of Synechococcus cyanophage S-SSM7. Sequence analysis confirmed that it possesses the three absolutely conserved motifs that form the active site in PDF metalloproteases. Phylogenetic analysis placed it within the Type 1B subclass, most closely related to the Arabidopsis chloroplast PDF, but lacking the C-terminal α-helix characteristic of that group. PDF proteins from this phage and from Synechococcus elongatus were expressed and characterized. The phage PDF is the more active enzyme and deformylates the N-terminal tetrapeptides from D1 proteins more efficiently than those from ribosomal proteins. Solution of the X-ray/crystal structures of those two PDFs to 1.95 Å resolution revealed active sites identical to that of the Type 1B Arabidopsis chloroplast PDF. Taken together, these findings show that many cyanophages encode a PDF with a D1 substrate preference that adds to the repertoire of genes used by phages to maintain photosynthetic activities.
Similar content being viewed by others
Introduction
Bacteriophages (phages) frequently carry metabolic genes acquired from host genomes. Not being essential for replication per se, these are termed auxiliary metabolic genes (AMGs) (Breitbart et al., 2007). The list of AMGs identified in phage genomes and thought to benefit the phage during lytic replication now includes those functioning in photosynthesis (Mann et al., 2003; Sharon et al. 2009; Sharon et al., 2011), the pentose phosphate pathway (Thompson et al., 2011), phosphate acquisition (Goldsmith et al., 2011; Zeng and Chisholm, 2012), nucleotide metabolism (Rohwer et al., 2000; Mann et al., 2005; Sullivan et al., 2005; Dinsdale et al., 2008) and cytoskeletal construction (Kraemer et al., 2012), among others.
Marine cyanophages carry a rich array of AMGs, including genes encoding components of photosystem II (PSII). PSII is subject to photodamage, particularly the D1 and D2 proteins that compose the functional heterodimer at its core and that are responsible for the binding of pigments and cofactors necessary for primary photochemistry (Millard et al., 2004). To maintain sufficient active D1/D2, the proteins are protected by high-light-inducible proteins that assist by dissipating excess light energy, and are also regularly recycled and replaced (Millard et al., 2004).
During a cyanophage infection, continuing photosynthesis is required to provide both energy (ATP) and reducing power (NADPH) for phage replication (Mann et al., 2003; Lindell et al., 2005). Numerous cyanophages encode D1 proteins plus at least one other component of PSII (Lindell et al., 2005). During infection of Prochlorococcus MED4 by the T7-like cyanophage P-SSP7, phage D1 protein is expressed and partially compensates for the decline in host D1 synthesis (Lindell et al., 2005). Other factors likely contribute as well to the maintenance of photosynthetic activity during infection, for example, phage-encoded high-light inducible proteins. On the other hand, following infection of Synechococcus sp. WH7803 by S-PM2 phage, host D1 (psbA) mRNA initially increases 18-fold and remains slightly elevated even late in infection, while a high level of phage D1 transcripts was sustained throughout (Clokie et al., 2006). Clearly D1 synthesis, either from phage or host genes, is a priority during cyanophage infection.
During protein translation in bacteria, the first amino-acid residue inserted is N-formylmethionine. The formyl moiety is then co-translationally removed from at least 98% of bacterial proteins by a peptide deformylase (PDF) (personal communication; Sharon et al., 2011; reviewed in Meinnel and Giglione, 2008). Frequently the N-terminal methionine is also removed by methionine aminopeptidase. Both modifications take place immediately as the nascent polypeptide chain emerges from the portal of the ribosome exit tunnel, before protein folding blocks enzyme access to the N-terminus. In E. coli, deletion of the PDF gene is lethal (Mazel et al., 1994).
Inhibition of PDFs by actinonin, a naturally occurring antimicrobial, inhibits bacterial growth (Chen et al., 2000), and actinonin treatment in vivo also leads to decline of photosynthetic function of the plastids in plants and green algae. In the unicellular alga Chlamydomonas, actinonin destabilizes D2, shunting it to a degradative pathway, thus interfering with the assembly of PSII (Giglione et al., 2003). In vascular plants, where the translation rate of D1 is 50–100 times greater than those of other PSII proteins (Kyle et al., 1984), the effect of actinonin on D1 synthesis and assembly is most pronounced (Hou et al., 2004). Therefore, the chloroplast PDF is essential for maintenance of PSII. If this N-terminal methionine excision system is overwhelmed, as might be expected by the high level of protein synthesis during phage replication, improperly processed and non-functional proteins would result. Cyanobacteria, important aquatic primary producers, share high functional similarity to chloroplasts (Giovannoni et al., 1988), but it is not known if their PDF shares functional or structural similarity to the PDF of chloroplasts.
PDFs are a subclass of the metalloprotease superfamily of enzymes known as the ‘clan MA and MB’ metalloproteases. Proteins from this family share a common structure containing a three-stranded β strand facing a catalytic metal and a HEXXH motif-containing α helix (Giglione et al., 2004). Although PDFs display variability in amino-acid sequence and overall length, they share three absolutely conserved and unique motifs that together form the entire active site: the G-motif (GΦGΦAAXQ where Φ is any hydrophobic amino acid and X is any amino acid), H-motif (QHEXDHLXG) and C-motif (EGCXS) (Giglione et al., 2004). PDFs are classified into three types based on sequence homologies and structural distinctions. Type 1 PDFs (PDF1) are found in bacteria and bacterial-derived organelles of eukaryotes (plastids, mitochondria and apicoplasts), while Type 2 PDFs are restricted to the Gram-positive Bacteria. These PDFs all possess the three conserved motifs and all act on the same substrates (N-formyl methionine polypeptides), although some variation in substrate specificity has been found in that the high-turnover D1 protein from PSII is a preferred substrate for the PDF in plant plastids (Dirk et al., 2008). These active PDFs differ in the secondary structure of their C-terminal domain; it is an α helix in Type 1 proteins and a supplementary β strand in Type 2 proteins. Within the PDF1, the C-terminal α-helix displays variable length and is not required for catalytic activity (Meinnel et al., 1996).
The less-studied Type 3 PDFs, found in Archaea and in the mitochondria of trypanosomatids, differ in their ‘conserved’ motifs. Archaea do not use N-formylmethionine for translation initiation; the formylated substrate for the archaeal PDFs is currently unknown (Bouzaidi-Tiali et al., 2007).
Evidence that phages encode AMGs has often come from the identification of metabolic homologs in sequenced phage genomes. Recently a novel method was used to search marine metagenomes for virally-encoded microbial metabolic genes (Sharon et al., 2011). By recognizing phage scaffolds in these marine metagenomes and then identifying metabolic genes within them, this approach is not limited to sequenced genomes. The most abundant metabolic genes found by this survey were PDFs, thereby predicting that PDFs are a significant part of the metabolic repertoire carried by marine phages.
Bringing all these threads together, we hypothesized that encoding and expressing a PDF could be one of the strategies used by cyanophages to manipulate host metabolism during infection. Here, we report identification of a PDF gene in the genome of a marine cyanophage (S-SSM7) and solution of its crystal structure. The PDF encoded by cyanophage S-SSM7 belongs to the Type 1B PDF subgroup, which is widely distributed among marine phage. We also determined the enzymatic activity and structure of the previously annotated PDF protein from Synechococcus elongatus PCC 6301. Comparison of these enzymes demonstrates that the S-SSM7 enzyme is highly active with a substrate preference for the high-turnover PSII protein D1, consistent with our hypothesis that phage encode PDFs to manipulate host metabolism and help sustain photosynthesis during infection.
Materials and methods
Target selection
The gene for the Synechococcus phage PDF, YP_004324347, was identified in the genome of Synechococcus phage S-SSM7 during a bioinformatic survey looking for unknown phage proteins that are abundant in environmental metagenomes but under-represented in the PhAnToMe database phage genome database (http://www.phantome.org/). Protein sequences identified in phage genomes using Glimmer V3.02 were compared against all publicly available metagenomes (from http://edwards.sdsu.edu/cgi-bin/mymgdb/show.cgi) using tBLASTn (E value cutoff 10−5). ORFS were annotated using the PhAnToMe database with BLASTp (E value cutoff 10−5). The encoded protein is referred to throughout the text as the phage PDF.
The gene for the cyanobacterial PDF, YP_170923, in the genome sequence of S. elongatus PCC 6301, was identified based on gene annotations in the NCBI Reference Sequence database (http://www.ncbi.nlm.nih.gov/RefSeq/). (The original host strain for S-SSM7, Synechococcus WH8109, has not been sequenced.) The encoded protein is referred to throughout the text as the bacterial PDF.
Gene design
Starting from the amino-acid sequences for these two PDF proteins, gene sequences were designed for expression in E. coli using Gene Composer (Emerald BioSystems, Bainbridge Island, WA, USA) (Lorimer et al., 2009; Lorimer et al., 2011). Back-translation of the amino-acid sequences employed a Universal Codon Usage Table designed to accommodate expression in E. coli with a minimum usage threshold of 2%. Restriction enzyme recognition sequences for BamHI and HindIII were excluded from the sequence to facilitate cloning. The engineered gene sequences were synthesized by DNA2.0 (Menlo Park, CA). The genes were sub-cloned into an expression vector containing an isopropyl--D-thiogalactopyranoside-inducible (IPTG-inducible) T7 promoter using the PIPE cloning method (Klock et al., 2008; Raymond et al., 2009; Raymond et al., 2011). The vector provides an N-terminal hexahistidine-Smt tag; the Smt tag is specifically and efficiently removed by UlpI protease (Mossessova and Lima, 2000).
Gene expression and protein purification
E. coli BL21(DE3) cells expressing the engineered PDF gene from Synechococcus phage S-SSM7 or S. elongatus were cultured at 37 °C to an A600 of ∼0.6 in TB medium (Teknova, Hollister, CA, USA). Cultures were induced with 1 mM IPTG and incubated overnight at 25 °C. The bacterial cells were harvested by centrifugation at 4 °C and the resulting cell paste was stored at −80 °C. For protein purification, the cells were thawed in 25 mM Tris-HCl (pH 8.0), 200 mM NaCl, 50 mM arginine, 10 mM imidiazole, 0.02% CHAPS detergent, 0.5% glycerol, 1 mM Tris(2-carboxyethyl)phosphine (TCEP), 100 mg lysozyme, 250 U μl−1 Benzonase (Novagen, Madison, WI, USA) and one complete EDTA-free Protease Inhibitor Cocktail tablet (Roche, Indianapolis, IN, USA), and then lysed by sonication using a Misonix S-4000 sonicator (Qsonica, Newtown, CT, USA) at 70% power (67–69 W), 2 s on/1 s off, 3 min total. Immediately after sonication, the crude lysate was clarified by centrifugation at 18 000 g for 35 min at 4 °C. The tagged proteins were initially purified using the protein maker (Smith et al., 2011). Briefly, lysates were applied to a 5-ml HisTrap FF nickel-chelate column (GE Healthcare, Waukesha, WI, USA) in 25 mM Tris-HCl (pH 8.0), 200 mM NaCl, 50 mM arginine, 10 mM imidazole, 0.25% glycerol and 1 mM TCEP. The column was washed with three column volumes of the same buffer, then eluted in three steps: a 5-ml elution with 30 mM, a 5-ml elution with 206 mM and two 5 ml elutions at 500 mM imidazole. Elution fractions containing partially purified PDF proteins were identified based on molecular weight by SDS polyacrylamide gel electrophoresis (SDS–PAGE). The fraction containing the target protein was treated with 50 ml of 1 mg ml−1 of 6 × His-tagged ubiquitin-like protease 1 overnight to cleave the Smt–PDF fusion and remove the His tag. The resulting protein was dialyzed against 1 l of 25 mM Tris-HCl (pH 8.0), 200 mM NaCl, 50 mM arginine, 10 mM imidazole, 0.25% glycerol and 1 mM TCEP, and then run over a second nickel-chelate column as described above. The flow-through, wash, and elution fractions were analyzed by SDS–PAGE and the fraction containing PDF protein was concentrated using an Amicon 10 kDa MWCO concentrator (Millipore, Billerica, MA, USA) to ∼5 ml and further purified by size exclusion chromatography on a Sephacryl S-100 10/300 GL column (GE Healthcare) in 25 mM Tris-HCl (pH 8.0), 200 mM NaCl, 1.0% glycerol and 1 mM TCEP. Peak fractions (3.0 ml) were concentrated to ∼10 mg ml−1 and flash frozen in liquid nitrogen in 100 μl aliquots. Selenomethionine-labeled proteins were produced in E. coli BL21(DE3) cells grown in M9 minimal medium supplemented with 5 mg ml−1 thiamine, 60 mg l−1 selenomethionine and other metabolites to inhibit methionine biosynthesis (Doublié, 1997), followed by purification as described above.
Enzyme assays
PDF enzyme activity was assayed using a coupled PDF/FDH (formate dehydrogenase) assay. The formate released by PDF from the fMAS substrate is oxidized by FDH with the concomitant reduction of NAD+ to NADH (Lazennec and Meinnel, 1997). NADH was measured by absorbance at 340 nm using a SpectraMax M2 96-well microplate reader (Molecular Devices, Sunnyvale, CA, USA). The substrate was 1 mM fMAS. Initial rates of reaction (vapp) were calculated from the initial linear portion of plots of NADH produced versus time. Km and Vmax were extrapolated from plots of vapp versus substrate concentration (GraphPad Prism, La Jolla, CA, USA). kcat was calculated by dividing Vmax by enzyme concentration. The inhibitor data were obtained using 0.2 nM enzyme.
N-terminal tetrapeptides
The N-terminal tetrapeptide sequences encoded by annotated ribosomal protein genes in S. elongatus PCC 6301 (Genbank: AP008231.1), Synechococcus sp. CC931 (Genbank: NC_008319.1), Synechococcus sp. CC9605 (Genbank: NC_007516.1) and Synechococcus sp. CC9902 (Genbank: NC_007513.1) were tabulated to yield a total 259 proteins (64, 65, 66 and 64 from each genome). The most frequently occurring tetrapeptides consisted of one with six occurrences (fMAKK) and 17 with four occurrences (including fMSRV and fMARI). The peptides fMAKK, fMSRV and fMARI were used as substrates in the PDF specificity assays, compared against the encoded N-terminal tetrapeptides of four representative D1 proteins: the D1 of Synechococcus phage S-SSM7 (Genbank: GU071098.1) and three bacterial D1 proteins from S. elongatus PCC 6301 (Table 1).
Phylogenetic analyses
For phylogenetic analysis, in addition to the phage and bacterial PDFs, 49 representative annotated PDF genes were selected from the sequenced genomes in The SEED database (http://theseed.uchicago.edu/FIG/) (Overbeek et al., 2005). A multiple sequence alignment was generated at the amino-acid level using ClustalW (Chenna et al., 2003) and the tree was drawn using FigTree v1.31 (http://tree.bio.ed.ac.uk/software/figtree/).
Global PDF distribution
The 70 phage-encoded PDFs reported by Sharon et al. (Sharon et al., 2011) were compared against all 12 672 581 sequencing reads in the Global Ocean Survey (GOS) metagenomes (Yooseph et al., 2007) using BLASTX (Altschul et al., 1990) (parameters: –e 1e-5 –F F –r 1 –q -1 –v 5 –b 5). GOS sequences with at least 30% amino-acid identity and 50% similarity to any of the 70 PDFs were tallied as phage-encoded PDF homologs. The normalized relative abundance was calculated for each sampled location as the number of phage-encoded PDF homologs divided by the total number of sequencing reads in that data set. The map was generated using ArcGIS version 10.1. The mapped chlorophyll concentrations are the September, 2012, values from the NASA Earth Observations site (http://neo.sci.gsfc.nasa.gov).
Crystallization
Crystallization trials of the phage PDF (native and Se-Met) were carried out using sitting drop vapor diffusion at 16 °C in a drop of a 1:1 mixture of protein (10.17 mg ml−1) and reservoir solution. Crystals grew from 0.1 M HEPES, pH 7.5; 70% (v/v) (+/−)-2-methyl-2,4-pentanediol. For co-crystallization with the inhibitor, actinonin, the native PDF protein was incubated at 4 °C overnight with a five-fold molar excess of actinonin (Santa Cruz Biotechnology, Santa Cruz, CA, USA) before crystallization. Crystals were frozen in liquid nitrogen in a solution of 30% (w/v) ethylene glycol for cryoprotection.
Crystallization of the native bacterial PDF was achieved as described above except that the crystals grew in 0.15 M KBr, 30% PEG-MME 2000.
Crystallography: phage protein
For the apo Se-Met phage protein data set (PDB 3UWA), X-ray diffraction data were collected at the Advanced Photon Source, beamline 19ID (Argonne, IL, USA) at a wavelength of 0.9791750 Å. The structure was solved by Se-SAD (Supplementary Tables S2 and S3). Crystals display a symmetry of P212121 with cell dimensions a=47.5 Å, b=58.3 Å, c=62.4 Å. For the actinonin-bound native phage protein data set (PDB 3UWB), the data were obtained using a Rigaku SuperBright FR-E+ rotating-anode X-ray generator with Osmic VariMax HF optics and a Saturn 944+ CCD detector. Data were collected to 1.7 Å resolution and the structure was solved by molecular replacement using the phage structure, PDB 3UWA, as the search model (Supplementary Tables S2 and S3). In both cases, a single crystal was used for each complete data set.
Crystallography: bacterial protein
A data set for the apo native bacterial crystal (PDB 4DR8) was collected at the Stanford Synchrotron Radiation Lightsource, beamline 7–1. Data were collected to 1.55 Å resolution and the structure was solved by molecular replacement using PDB 1LRY as the search model (Supplementary Tables S4 and S5). PDB 1LRY, the PDF from Pseudomonas aeruginosa, was the PDF with a solved structure that had the highest similarity to the Synechococcus protein. Crystals display P1 symmetry with cell dimensions of a=43.40 Å, b=65.36 Å, c=69.03 Å. To generate the actinonin-bound bacterial protein, the apo crystals were soaked overnight with actinonin (1 mM in crystallization mother liquor). The actinonin-bound data set was collected in-house as described for the phage data sets. Data were collected to 1.9 Å resolution and the structure (PDB 4DR9) was solved by molecular replacement using the native structure (PDB 4DR8) as the search model (Supplementary Tables S4 and S5).
Results
Global distribution of phage PDF homologs
The initial report of abundant phage-encoded PDFs in marine environments (Sharon et al., 2011) did not address their biological function or the question of their global distribution. Therefore, we first searched all available GOS metagenomes for homologs of those phage-encoded PDFs using BLASTX. Mapping of the relative abundance of these homologs at all the GOS-sampling locations makes their widespread distribution apparent (Figure 1, Supplementary Table S1), thus demonstrating their importance in the environment. Additional studies will be required to correlate their pattern of distribution with ecological factors such as host distribution and nutrient levels; however, we have extensively characterized the structure and function of a phage-encoded PDF.
Global distribution of phage-encoded PDFs. GOS-sampling locations (blue dots) and relative abundances of phage-encoded PDFs (white circles) in the GOS metagenomes are plotted on a global map of marine chlorophyll concentrations. See Supplementary Table S1 for the data plotted.
Synechococcus phage PDF activity
Polypeptide deformylation activity of the PDF protein from Synechococcus phage S-SSM7 (referred to throughout as the ‘phage PDF’) was compared with the activity of the PDF from S. elongatus PCC 6301 (referred to throughout as the ‘bacterial PDF’) using a real-time PDF/FDH enzyme-coupled assay (see Methods). The assay was optimized to measure initial rates accurately and produced z’ values >0.8 (data not shown). This assay was first used to evaluate the sensitivity of the phage PDF to inhibition by actinonin, a naturally occurring antibiotic demonstrated to be a potent inhibitor of bacterial PDFs (Chen et al., 2000). Results in Figure 2a show that actinonin is a potent inhibitor of phage and bacterial PDFs with an apparent IC50 value of 10 nM against both enzymes, suggesting that both enzymes have similar catalytic active sites.
(a) Inhibition of the Synechococcus phage S-SSM7 PDF (blue) and the PDF from Synechococcus elongatus PCC 6301 (red) by actinonin. Enzyme: 0.2 nM PDF. Substrate: 1.5 mM fMLIS, the N-terminal tetrapeptide of the D1 protein encoded by the phage. (b) Kinetic assays comparing the activity of the Synechococcus phage S-SSM7 PDF (blue) with that from S. elongatus PCC 6301 (red) on three different N-terminal tetrapeptide substrates derived from phage or bacterial D1 proteins (see Table 1). Red square=phage D1, fMLIS; solid red circle=bacterial D1, fMTSI; open red circle=bacterial D1, fMTTA.
The deformylation activity of the phage PDF was assayed using formylated N-terminal tetrapeptides derived from D1 proteins from cyanophage S-SSM7 and S. elongatus PCC 6301 as well as the most frequently occurring N-terminal tetrapeptides of ribosomal proteins from four Synechococcus genomes (Table 1 and Methods). The phage PDF has significantly higher specific activity than the bacterial enzyme on the substrates used above (average kcat values of 615 versus 192 s−1, Table 2). Likewise, the phage PDF is significantly more efficient than the bacterial PDF at deformylating the D1-derived tetrapeptides (kcat/Km for the phage PDF being 14.7, 10.5 and 12.6 times that of the bacterial PDF; Table 2). Comparison of the substrate kinetics of these two enzymes on tetrapeptides derived from phage and bacterial D1 proteins shows the consistently greater activity of the phage PDF (Figure 2b). The differences in efficiency were less when three different tetrapeptides from ribosomal proteins served as the substrate (the phage efficiency here being only 5.1, 1.4 and 6.5 times that for the bacterial enzyme). Taken together, these results clearly show that the phage PDF is a catalytically active deformylase with overall properties similar to the bacterial PDF.
Phage PDF specificity
The two substrate classes (D1 and ribosomal proteins) were chosen to determine if either PDF displayed any selectivity for deformylating D1 proteins. Results in Table 2 show that the specificity constants (kcat/Km) (Eisenthal et al., 2007) for the phage PDF are significantly higher for the D1-derived peptides compared with the ribosomal-derived peptides (1780 versus 720). In contrast, when comparing the two substrates against the bacterial PDF, the opposite trend was observed, although the difference was not nearly as significant (141 versus 199). These results show that the phage PDF is much more efficient at deformylating the D1 substrates than the bacterial PDF.
Phylogenetic analysis of cyanophage S-SSM7 PDF
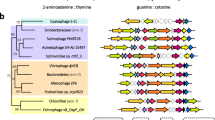
Members of the PDF family, while displaying variability in amino-acid sequence, share three essential conserved motifs (the G, H and C motifs) that together build the entire active site (Giglione et al., 2004). Alignment of the predicted phage PDF sequence with PDFs of known structure from eukaryote organelles (mitochondria and chloroplasts of Arabidopsis thaliana, and the non-photosynthetic plastid-derived apicoplast of Plasmodium falciparum) and a cyanobacterium (S. elongatus PCC 6301) demonstrates the conservation of these motifs (Figure 3a). A phylogenetic tree built using 51 PDF sequences from bacteria, phage and eukaryote organelles clusters the phage PDF closely with the Type 1B PDF from A. thaliana chloroplasts (Figure 3b). Notably, unlike most bacterial PDF1B enzymes, the C-terminal alpha helix characteristic of this subtype is absent from this phage PDF.
Comparison of the Synechococcus phage S-SSM7 PDF amino-acid sequence with that of other PDFs. (a) Multiple sequence alignment of selected PDFs showing the three conserved motifs and the C-terminal domain. Included are PDFs from: A. thaliana mitochondria; A. thaliana chloroplasts; Synechococcus phage S-SSM7; Synechococcus elongatus PCC 6301; and P. falciparum apicoplast. Highlighted regions are the PDF-specific G-motif (GΦGΦAAXQ), C-motif (EGCXS) and H-motif (QHEXDHLXG), as well as the variable C-terminal domain (where Φ=any hydrophobic amino acid and X=any amino acid). Degree of conservation of amino acids at each position: *=absolutely conserved; :=different but very similar amino acids; .=different but somewhat similar amino acids; blank=dissimilar amino acids or gaps. (b) A phylogenetic tree of 51 PDF proteins from bacteria, phage and eukaryote organelles.
Enzyme structure
Comparison of the1.95 Å structure of the phage PDF (Figure 4a) with that of the A. thaliana chloroplast Type IB PDF shows striking similarity with an overall RMSD of 0.645 Å (Figure 4b). Likewise, the solved structure of the bacterial PDF shows an identical conformation (Figure 4b). As expected from the kinetic data, the structures of all three enzymes are also identical when bound to actinonin (data not shown), thus providing strong structural evidence that the phage protein is a functional PDF enzyme. The C-terminal α-helix that is characteristic of Type 1B PDF proteins is absent from the phage protein (Supplementary Figure S1). The amino-acid residues that compose the active site, which are conserved and are predicted to interact with a zinc ion, are observed in identical conformations in these three enzymes (Figure 4c). Comparison of the amino-acid residues positioned at the entry to the active site in these three proteins also shows compelling similarity (Figure 4d), with the notable exception that the tyrosine in both the bacterial and chloroplast proteins is replaced by an asparagine in the phage PDF.
(a) Crystal structure of Synechococcus phage S-SSM7 peptide deformylase (PDF). (b) Overlay of phage S-SSM7 PDF (green), Synechococcus elongatus PCC 6301 PDF (cyan) and A. thaliana chloroplast PDF1B (magenta) shows striking similarity of protein folds, as well as the position of the zinc ion in the active site. (c) Evaluation of the active site residues around the zinc ion reveals strong conservation among these three PDFs: phage S-SSM7 (green), Synechococcus (cyan) and chloroplast (magenta). (d) Comparison of the residues at the entry to the active site in the phage (green), Synechococcus (cyan) and chloroplast (magenta) PDFs. Green sticks=phage asparagine 99; cyan sticks=Synechococcus tyrosine 116; magenta sticks=chloroplast tyrosine 178.
The catalytic zinc has been shown to coordinate the PDF inhibitor actinonin (Guilloteau et al., 2002) and is responsible for the high affinity binding of this universal PDF inhibitor. The structure of the phage PDF bound to actinonin was determined (Figures 5a and b) shows the interactions between bound actinonin and active site residues of the phage PDF.
Discussion
The growing list of AMGs identified in phage genomes includes PDFs. The deformylation of the N-terminal formylmethionine catalyzed by PDFs is an essential step in the co-translational processing of nascent polypeptides in all bacteria and bacterial-derived organelles (Giglione et al., 2004). Deformylation, and often excision of the N-terminal methionine, is necessary for further post-translational processing and proper folding. Only two representatives of this enzyme family had been previously recognized in sequenced phage genomes, those being in two phages of Vibrio parahaemolyticus (Seguritan et al., 2003). However, virus-affiliated PDF genes were recently found to be abundant in marine metagenomes (Sharon et al., 2011). All of these sightings relied solely on homology; enzymatic activity had not been demonstrated for any phage-encoded PDF.
We report here the identification of a gene encoding a predicted PDF in the sequenced genome of a cyanophage (S-SSM7) isolated from a marine cyanobacterium (Synechococcus WH8109). For comparison, the predicted bacterial PDF from the genome of a similar host, the freshwater cyanobacterium S. elongatus PCC 6301, was also characterized. Both protein sequences display the three absolutely conserved motifs (G, H and C) known to form the PDF active site (Giglione et al., 2004), thus suggesting that both might be catalytically active (Figure 3a).
The argument can be made that having a PDF gene on board may be more necessary for a cyanophage than for phages infecting heterotrophs. A common theme of lytic infection by any phage is the shutting down of transcription and translation of host proteins, with the concomitant redirection of the host machinery to phage replication. In the case of cyanophages, this is complicated by the need to maintain photosynthesis to provide the energy (ATP) and reducing power (NADPH) required for efficient phage replication (Lindell et al., 2005). Maintenance of photosynthesis, in turn, requires ongoing synthesis of functional proteins, the synthesis of D1 being particularly critical because its high rate of photo damage requires continual replacement. Numerous cyanophages encode PSII proteins (D1 and at least one other) (Lindell et al., 2005). In Prochlorococcus and Synechococcus, synthesis of phage-encoded D1 supplements production of the host protein (Lindell et al., 2005; Clokie et al., 2006). Although it is likely that cyanophages use additional tactics (for example, high-light-inducible proteins) to maintain a functional PSII, synthesis and co-translational processing of D1 is a priority.
PDFs remove the N-terminal formyl group from at least 98% of all bacterial proteins (reviewed in Meinnel and Giglione, 2008). Actinonin, an antimicrobial produced by Streptomyces roseopallidus (Giglione et al., 2004), is a universal PDF inhibitor. When PDF activity in plant chloroplasts is inhibited by actinonin in vivo, photosynthetic function declines and an albino phenotype is typically evident (Giglione et al., 2003; Hou et al., 2004), demonstrating an essential role for PDFs in maintaining photosynthesis. In vascular plants, actinonin reduces D1 synthesis and PSII assembly, leading to reduced PSII function (Hou et al., 2004). Notably, the PDF1B in Arabidopsis chloroplasts has a higher catalytic efficiency when deformylating D1 compared with other proteins (Dirk et al., 2002).
The predicted PDF encoded by phage S-SSM7 was expressed and was found to have a high level of deformylase activity and sensitivity to actinonin, a universal PDF inhibitor (Figure 1a). Its closest homolog, the type IB PDF from A. thaliana chloroplasts (Figure 2b), is more efficient at deformylating D1 than other proteins (Dirk et al., 2002). Likewise, catalytic efficiency (kcat/Km) of the phage PDF is higher on the D1-derived tetrapeptide substrates (Figure 2b). In contrast, the bacterial PDF from S. elongatus PCC 6301 does not show this preference (Table 2). That the differences in efficiency were less when tetrapeptides from ribosomal proteins served as the substrate suggests that the greater efficiency of the phage PDF on the D1-derived tetrapeptides cannot be attributed solely to the higher activity (larger kcat) of the phage enzyme. The factors responsible for this difference were not apparent in the actinonin-bound enzyme structures and their elucidation would presumably require obtaining structures of the PDF enzymes bound to substrate molecules. Attempts were made to co-crystallize the enzymes with the substrates, and to soak existing crystals with peptide substrates, but substrate bound structures could not be obtained (data not shown).
These observations suggest that this cyanophage-encoded PDF assists in the maintenance of an active PSII during infection. Moreover, all 70 of the PDFs identified in marine metagenomes are from cyanophage genomes, thus indicating that carrying a PDF gene is a tactic employed by many marine cyanophage (personal communication; Sharon et al., 2011). These phage-encoded PDF genes are also widely distributed in the world’s oceans (Figure 1). Combined, these factors indicate that cyanophage PDFs have a significant role in the global interplay between cyanophage and their hosts. Comparison of the solved structures of both the phage and bacterial PDFs to the Type 1B PDF from A. thaliana chloroplasts demonstrated striking similarity of the protein folds, position of the catalytic zinc, and the amino-acid residues forming the cavity of the active site (Figure 4). The phage and chloroplast enzymes also show identical conformation while binding the inhibitor actinonin (data not shown). Phylogenetic analysis locates both the phage and the bacterial PDFs within the Type 1B group (Figure 3). As noted above, the phage PDF and its closest homolog, the A. thaliana chloroplast PDF—but not the bacterial enzyme from S. elongatus—demonstrate preferential specificity for D1-derived tetrapeptide substrates, reflecting the importance of efficient deformylation of this protein. The source of this substrate preference, despite the close similarity of their active sites, has been postulated to be due to the tyrosine moiety at position 178 located at the entry to the active site where it might sterically hinder the entrance of non-D1-like N-terminal polypeptides (Dirk et al., 2008). A tyrosine occupies the same location in the bacterial PDF, but in the phage protein this tyrosine is replaced by an asparagine (N99) pointing in the opposite direction (Figure 4d).
The Synechococcus phage S-SSM7 lacks the C-terminal α-helical domain that is a defining character of Type 1B PDFs. There is considerable variability in the length and secondary structure of this domain in PDF1 (Giglione et al., 2009), and the domain is not required for catalytic activity (Meinnel et al., 1996). Nevertheless, it is striking that the domain is lacking in all three PDFs identified in sequenced phage genomes to date (that is, Synechococcus phage S-SSM7, and the vibriophages VP16C and VP16T). It is also lacking from at least 52 of the 70 marine viral PDF sequences identified in the marine metagenomes (Sharon et al., 2011; Béjà, personal communication). It was recently proposed, based on studies of the Type 1 PDF of E. coli, that Type 1 PDF proteins interact with the ribosome via their helical C-terminal domain (Bingel-Erlenmeyer et al., 2008; Giglione et al., 2009; Kramer et al., 2009). According to this model, the C-terminal domain assists in positioning the enzyme in close proximity to the translation ribosome exit tunnel, thereby providing efficient co-translational processing. In addition to supporting biochemical and structural evidence, experiments in vivo under PDF-limiting conditions found that truncation of the C-terminal helix reduced the bacterial growth rate (Bingel-Erlenmeyer et al., 2008).
However, the PDF of Synechococcus phage S-SSM7, without a C-terminal domain, has higher catalytic activity in vitro than the bacterial PDF from S. elongatus that possesses the domain (Table 2, Figure 2). It is not known whether the truncation might have contributed to the higher activity of the phage enzyme. Microbially-derived genes within phage genomes tend to be shorter than their microbial counterparts (Daubin and Ochman, 2004), suggesting that perhaps these phages, for genomic economy, have eliminated a domain that is not essential during infection. Alternatively, it is possible that the truncation enables the phage PDF to compete more effectively with the host enzyme for access to the ribosome.
Encoding AMGs gives phage the potential to modulate host metabolism and energy flow to favor their own replication. Countering environmental stressors and maintaining essential host systems appear to be the priorities. Increasingly, we are aware that the well-studied example of the photosystem genes (for example, psbA that encodes D1) carried by cyanophages is but one of many sophisticated mechanisms used to ensure their efficient replication. Another example: carbon metabolism genes carried by several cyanophages, including the Calvin cycle inhibitor protein CP12, serve to shuttle energy (ATP) and reducing power (NADPH) away from carbon fixation and into anabolic pathways (for example, dNTP synthesis), thereby facilitating phage replication (Lindell et al., 2005; Sharon et al. 2009; Thompson et al., 2011). The cyanophage-encoded PDF characterized here adds a novel strategy contributing to the maintenance of photosynthetic processes during infection and redirection of their products toward phage replication. The enzymatic and structural studies reported here also demonstrate that the structure, efficiency and specificity of acquired AMGs can be subsequently fine-tuned to better serve the interests of the phage. Further work is needed to monitor expression of both phage and host PDF during cyanophage infection and to measure the contribution of the phage PDF to co-translational protein processing. Also remaining to be elucidated is how the function and efficiency of phage PDFs are influenced by the absence of a C-terminal alpha helix.
References
Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ . (1990). Basic local alignment search tool. J Mol Biol 215: 403–410.
Bingel-Erlenmeyer R, Kohler R, Kramer G, Arzu Sandikci S, Maier T, Schaffitzel C et al (2008). A peptide deformylase–ribosome complex reveals mechanism of nascent chain processing. Nature 452: 108–111.
Bouzaidi-Tiali N, Giglione C, Bulliard Y, Pusnik M, Meinnel T, Schneider A . (2007). Type 3 peptide deformylases are required for oxidative phosphorylation in Trypanosoma brucei. Mol Microbiol 65: 1218–1228.
Breitbart M, Thompson LR, Suttle CA, Sullivan MB . (2007). Exploring the vast diversity of marine viruses. Oceanography 20: 135–139.
Chen DZ, Patel DV, Hackbarth CJ, Wang W, Dreyer G, Young DC et al (2000). Actinonin, a naturally occurring antibacterial agent, is a potent deformylase inhibitor. Biochemistry 39: 1256–1262.
Chenna R, Sugawara H, Koike T, Lopez R, Gibson TJ, Higgins DG et al (2003). Multiple sequence alignment with the clustal series of programs. Nucleic Acids Res 31: 3497–3500.
Clokie MRJ, Shan J, Bailey S, Jia Y, Krisch HM, West S et al (2006). Transcription of a ‘photosynthetic’T4-type phage during infection of a marine cyanobacterium. Environ Microbiol 8: 827–835.
Daubin V, Ochman H . (2004). Bacterial genomes as new gene homes: the genealogy of ORFans in E. coli. Genome Res 14: 1036–1042.
Dinsdale EA, Edwards RA, Hall D, Angly F, Breitbart M, Brulc JM et al (2008). Functional metagenomic profiling of nine biomes. Nature 452: 629–632.
Dirk L, Williams MA, Houtz RL . (2002). Specificity of chloroplast-localized peptide deformylases as determined with peptide analogs of chloroplast-translated proteins. Arch Biochem Biophys 406: 135–141.
Dirk LM, Schmidt JJ, Cai Y, Barnes JC, Hanger KM, Nayak NR et al (2008). Insights into the substrate specificity of plant peptide deformylase, an essential enzyme with potential for the development of novel biotechnology applications in agriculture. Biochem J 413: 417–427.
Doublié S . (1997). [29] Preparation of selenomethionyl proteins for phase determination. Methods Enzymol 276: 523–530.
Eisenthal R, Danson MJ, Hough DW . (2007). Catalytic efficiency and kcat/KM: a useful comparator? Trends Biotechnol 25: 247–249.
Giglione C, Boularot A, Meinnel T . (2004). Protein N-terminal methionine excision. Cell Mol Life Sci 61: 1455–1474.
Giglione C, Fieulaine S, Meinnel T . (2009). Cotranslational processing mechanisms: towards a dynamic 3D model. Trends Biochem Sci 34: 417–426.
Giglione C, Vallon O, Meinnel T . (2003). Control of protein life-span by N-terminal methionine excision. EMBO J 22: 13–23.
Giovannoni SJ, Turner S, Olsen GJ, Barns S, Lane DJ, Pace NR . (1988). Evolutionary relationships among cyanobacteria and green chloroplasts. J Bacteriol 170: 3584–3592.
Goldsmith DB, Crosti G, Dwivedi B, McDaniel LD, Varsani A, Suttle CA et al (2011). Development of phoH as a novel signature gene for assessing marine phage diversity. Appl Environ Microbiol 77: 7730–7739.
Guilloteau JP, Mathieu M, Giglione C, Blanc V, Dupuy A, Chevrier M et al (2002). The crystal structures of four peptide deformylases bound to the antibiotic actinonin reveal two distinct types: a platform for the structure-based design of antibacterial agents. J Mol Biol 320: 951–962.
Hou CX, Dirk LMA, Williams MA . (2004). Inhibition of peptide deformylase in nicotiana tabacum leads to decreased D1 protein accumulation, ultimately resulting in a reduction of photosystem II complexes. Am J Bot 91: 1304–1311.
Klock HE, Koesema EJ, Knuth MW, Lesley SA . (2008). Combining the polymerase incomplete primer extension method for cloning and mutagenesis with microscreening to accelerate structural genomics efforts. Proteins 71: 982–994.
Kraemer JA, Erb ML, Waddling CA, Montabana EA, Zehr EA, Wang H et al (2012). A Phage tubulin assembles dynamic filaments by an atypical mechanism to center viral DNA within the host cell. Cell 149: 1488–1499.
Kramer G, Boehringer D, Ban N, Bukau B . (2009). The ribosome as a platform for co-translational processing, folding and targeting of newly synthesized proteins. Nat Struct Mol Biol 16: 589–597.
Kyle D, Ohad I, Arntzen C . (1984). Membrane protein damage and repair: selective loss of a quinone-protein function in chloroplast membranes. Proc Natl Acad Sci USA 81: 4070.
Lazennec C, Meinnel T . (1997). Formate dehydrogenase-coupled spectrophotometric assay of peptide deformylase. Anal Biochem 244: 180–182.
Lindell D, Jaffe JD, Johnson ZI, Church GM, Chisholm SW . (2005). Photosynthesis genes in marine viruses yield proteins during host infection. Nature 438: 86–89.
Lorimer D, Raymond A, Mixon M, Burgin A, Staker B, Stewart L . (2011). Gene composer in a structural genomics environment. Acta Crystallogr Sect F Struct Biol Cryst Commun 67: 985–991.
Lorimer D, Raymond A, Walchli J, Mixon M, Barrow A, Wallace E et al (2009). Gene composer: database software for protein construct design, codon engineering, and gene synthesis. BMC Biotechnol 9: 36–58.
Mann NH, Clokie MRJ, Millard A, Cook A, Wilson WH, Wheatley PJ et al (2005). The genome of S-PM2, a ‘photosynthetic’ T4-type bacteriophage that infects marine Synechococcus strains. J Bacteriol 187: 3188–3200.
Mann NH, Cook A, Millard A, Bailey S, Clokie M . (2003). Marine ecosystems: bacterial photosynthesis genes in a virus. Nature 424: 741–741.
Mazel D, Pochet S, Marliere P . (1994). Genetic characterization of polypeptide deformylase, a distinctive enzyme of eubacterial translation. EMBO J 13: 914.
Meinnel T, Giglione C . (2008). Tools for analyzing and predicting N-terminal protein modifications. Proteomics 8: 626–649.
Meinnel T, Lazennec C, Dardel F, Schmitter JM, Blanquet S . (1996). The C-terminal domain of peptide deformylase is disordered and dispensable for activity. FEBS Lett 385: 91–95.
Millard A, Clokie MR, Shub DA, Mann NH . (2004). Genetic organization of the psbAD region in phages infecting marine Synechococcus strains. Proc Natl Acad Sci USA 101: 11007–11012.
Mossessova E, Lima CD . (2000). Ulp1-SUMO crystal structure and genetic analysis reveal conserved interactions and a regulatory element essential for cell growth in yeast. Mol Cell 5: 865–876.
Overbeek R, Begley T, Butler RM, Choudhuri JV, Chuang HY, Cohoon M et al (2005). The subsystems approach to genome annotation and its use in the project to annotate 1000 genomes. Nucleic Acids Res 33: 5691–5702.
Raymond A, Haffner T, Ng N, Lorimer D, Staker B, Stewart L . (2011). Gene design, cloning and protein-expression methods for high-value targets at the Seattle structural genomics center for infectious disease. Acta Crystallogr Sect F Struct Biol Cryst Commun 67: 992–997.
Raymond A, Lovell S, Lorimer D, Walchli J, Mixon M, Wallace E et al (2009). Combined protein construct and synthetic gene engineering for heterologous protein expression and crystallization using gene composer. BMC Biotechnol 9: 37–52.
Rohwer F, Segall A, Steward G, Seguritan V, Breitbart M, Wolven F et al (2000). The complete genomic sequence of the marine phage roseophage SIO1 shares homology with nonmarine phages. Limnol Oceanogr 45: 408–418.
Seguritan V, Feng IW, Rohwer F, Swift M, Segall AM . (2003). Genome sequences of two closely related vibrio parahaemolyticus phages, VP16T and VP16C. J Bacteriol 185: 6434–6447.
Sharon I, Alperovitch A, Rohwer F, Haynes M, Glaser F, Atamna-Ismaeel N et al (2009). Photosystem I gene cassettes are present in marine virus genomes. Nature 461: 258–262.
Sharon I, Battchikova N, Aro EM, Giglione C, Meinnel T, Glaser F et al (2011). Comparative metagenomics of microbial traits within oceanic viral communities. ISME J 5: 1178–1190.
Smith ER, Begley DW, Anderson V, Raymond AC, Haffner TE, Robinson JI et al (2011). The protein maker: an automated system for high-throughput parallel purification. Acta Crystallogr Sect F Struct Biol Cryst Commun 67: 1015–1021.
Sullivan MB, Coleman ML, Weigele P, Rohwer F, Chisholm SW . (2005). Three Prochlorococcus cyanophage genomes: signature features and ecological interpretations. PLoS Biol 3: e144.
Thompson LR, Zeng Q, Kelly L, Huang KH, Singer AU, Stubbe J et al (2011). Phage auxiliary metabolic genes and the redirection of cyanobacterial host carbon metabolism. Proc Natl Acad Sci USA 108: E757–E764.
Yooseph S, Sutton G, Rusch DB, Halpern AL, Williamson SJ, Remington K et al (2007). The sorcerer II global ocean sampling expedition: expanding the universe of protein families. PLoS Biol 5: e16.
Zeng Q, Chisholm SW . (2012). Marine viruses exploit their host’s two-component regulatory system in response to resource limitation. Curr Biol 22: 124–128.
Acknowledgements
The authors thank Amy Raymond, Kateri Atkins, Tom Edwards and the Molecular Biology, Protein Purification and Crystallization Core Groups at Emerald Biostructures for their contributions, Nao Hisakawa for GIS mapping and Abigail Salyers for helpful discussions. We are indebted to Itai Sharon and Oded Béjà for bringing to our attention the frequent occurrence of PDF genes in marine phage genomes. This research is part of the Dimensions: Shedding Light on Viral Dark Matter project supported by the National Science Foundation (DEB-1046413).
Author information
Authors and Affiliations
Corresponding author
Additional information
Supplementary Information accompanies the paper on The ISME Journal website
Supplementary information
Rights and permissions
This work is licensed under the Creative Commons Attribution-NonCommercial-No Derivative Works 3.0 Unported License. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-nd/3.0/
About this article
Cite this article
Frank, J., Lorimer, D., Youle, M. et al. Structure and function of a cyanophage-encoded peptide deformylase. ISME J 7, 1150–1160 (2013). https://doi.org/10.1038/ismej.2013.4
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ismej.2013.4
Keywords
This article is cited by
-
Clades of huge phages from across Earth’s ecosystems
Nature (2020)
-
Diversities and potential biogeochemical impacts of mangrove soil viruses
Microbiome (2019)
-
Numerous cultivated and uncultivated viruses encode ribosomal proteins
Nature Communications (2019)
-
Phage puppet masters of the marine microbial realm
Nature Microbiology (2018)
-
The C-terminal residue of phage Vp16 PDF, the smallest peptide deformylase, acts as an offset element locking the active conformation
Scientific Reports (2017)