Abstract
The heterotrophic bacterial community of the Eastern Mediterranean Sea is believed to be limited by phosphorus (P) availability. This observation assumes that all bacterial groups are equally limited, something that has not been hitherto examined. To test this hypothesis, we performed nutrient addition experiments and investigated the response of probe-identified groups using microautoradiography combined with catalyzed reporter deposition fluorescence in situ hybridization. Our results show contrasting responses between the bacterial groups, with Gammaproteobacteria being the group more affected by P availability. The Roseobacter clade was likely colimited by P and nitrogen (N), whereas Bacteroidetes by P, N and organic carbon (C). In contrast, SAR11 cells were active regardless of the nutrient concentration. These results indicate that there is high heterogeneity in the nutrient limitation of the different components of the bacterioplankton community.
Similar content being viewed by others
Main
Although inorganic nutrient limitation of bacterioplankton was considered rare about a decade ago (Caron et al., 2000), we now know it frequently occurs in oligotrophic waters. Phosphorus (P), for example, is often the primary limiting nutrient in the Atlantic and Mediterranean (Cotner et al., 1997; Pinhassi et al., 2006). Nevertheless, most studies have considered heterotrophic bacteria as a homogeneous black box, while the bacterial community is composed by cells expressing high metabolic diversity (for example, Musat et al., 2008; Alonso-Saéz et al., 2012), which likely experience different degrees of limitation and stress. The fact that nutrient availability plays an important role in niche partitioning supports this hypothesis (Pinhassi et al., 2006), but the variability in the stress responses among different bacterial groups has been hitherto ignored.
The Eastern Mediterranean is one of the most oligotrophic and P-starved marine systems on Earth (for example, Tanaka et al., 2007), where bacterioplankton is often P-limited (for example, Thingstad et al., 2005). Here we assessed whether nitrogen (N) and/or organic carbon (C) could be colimiting the bacterial community, and whether different bacterial groups responded similarly to the various nutrient additions (see Supplementary material for details).
Heterotrophic bacterial activity was significantly stimulated (P<0.05, Dunnett’s test) in all the treatments that contained P (Figure 1). Bacterial activity doubled in the P and PC treatments, and additional increases occurred in the NP (5 × higher than the control) and NPC treatments (10 × higher). These findings indicate that the heterotrophic bacterial community as a whole was primarily limited by P, but that these waters are a nearly balanced system, where addition of P leads to shifts from one type of limitation to another. This hypothesis is supported by the results obtained with the phosphate turnover time (Supplementary Figure S1).
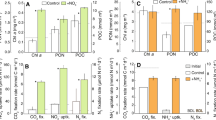
Bacterial heterotrophic activity in the addition experiments. Samples were amended with phosphate (P), ammonia (N) and organic carbon (C), or with combinations of these nutrients (PC, NP and NPC). Cont.: control treatment (no amendments). Each data point represents the average of the two replicates. Error bars represent the s.d. Arrow highlights the time-point when samples were taken for the MARFISH analyses.
To investigate whether all the bacterial groups responded equally to the nutrient additions, we used microautoradiography combined with catalyzed reporter deposition fluorescence in situ hybridization (MARFISH). Community composition 2 days after inoculation was not strongly affected by the nutrient amendments, and was similar to that at the beginning of the experiment, that is, SAR11 dominated in all the treatments, followed by Gammaproteobacteria (Supplementary Figure S2). Non-EUB cells decreased dramatically in the NP and NPC treatments (from 20% to 0%), suggesting that dormant cells may have become active upon nutrient additions, as observed in other studies (for example, Eilers et al., 2000).
The different bacterial groups showed very contrasting responses. Gammaproteobacteria activity (in terms of % cells taking up leucine) was stimulated in all treatments containing P (Dunnett’s test, P<0.05, Figure 2). This group was the one responding more clearly to P additions, suggesting that Gammaproteobacteria experienced more severe P limitation than the other groups. Indeed, recent metatranscriptomic data show that Gammaproteobacteria transcript pools are enriched in genes for P acquisition when compared with other phylogenetic groups (Gifford et al., 2012). Nevertheless, the further increase observed in the total number of active cells in the PC, NP and NPC treatments (Supplementary Figure S3) indicated that Gammaproteobacteria were also able to rapidly take advantage of additional inputs of N and C, and increase their abundance and production. The strong relationship found between the bulk incorporation of leucine and the abundance of leucine-incorporating gammaproteobacterial cells (Supplementary Figure S4) suggests that Gammaproteobacteria likely accounted for the activity changes of the bulk bacterial community. MARFISH micrographs showed that gammaproteobacterial cells had larger per-cell silver grain clusters than the dominant SAR11 (Figure S5), which indicates higher assimilation per cell (Nielsen et al., 2003), and were notably larger in the nutrient-enriched treatments. Gammaproteobacteria were dominated by the NOR5/OM60 clade in the control, P and N treatments, whereas Alteromonadaceae dominated in the NPC treatments (Figure S6).
Percentage of active cells within each probe-identified group in relation to total prokaryotes (DAPI counts). Labels as defined in Figure 1. Asterisks denote significant differences in relation to the control (*P<0.05, **P<0.001). Note the different y axis scale for Roseobacter and Bacteroidetes.
The percentage of cells taking up leucine within the Bacteroidetes group was generally low, due to their known preference for high-molecular-weight compounds (Cottrell and Kirchman, 2000). The activity of this group was only significantly stimulated in the NPC treatment, although on average the activity increased in all the treatments that contained P. Therefore, Bacteroidetes cells were likely limited by P, but secondarily colimited by N and C.
Roseobacter activity was stimulated ca. twofold in the P and PC treatments and eightfold and fivefold in the NP and NPC treatments, respectively (Figure 2). However, this stimulation was only significant in the NP and NPC treatments, suggesting that this group was colimited by P and N. Nevertheless, the Roseobacter clade dominates among algal-associated bacteria (Buchan et al., 2005), and phytoplankton at the moment of the study was colimited by P and N (Pitta et al., unpublished). Hence, it is possible that the strong positive response of this group to N and P additions is a consequence of dissolved organic carbon release by the stimulated phytoplankton.
SAR11 activity, expressed as percentage of active cells, was relatively high in all the treatments (Figure 2). The fact that this group was active regardless of the nutrient concentration contradicts the common belief that SAR11 cells are strict oligotrophs. Instead, we believe that the conservative metabolism of SAR11 cells allows them to thrive well in a wide range of trophic conditions. This is consistent with a recent metatranscriptomic study showing that SAR11 cells have limited capacity to sense and respond to environmental changes, which suggests that this phylogenetic group has evolved to maintain consistent growth independent of environmental conditions (Gifford et al., 2012), something that had already been hinted in studies with a cultured representative of this clade (for example, Rappé et al., 2002). The cell-associated silver grain clusters were not larger in the nutrient amended treatments (Figure S5), suggesting that the per-cell activity within the SAR11 group did not notably increase, again agreeing with the view that SAR11 cells would not take advantage of nutrient pulses (Gifford et al., 2012) and in general have low nutrient requirements (Sebastián et al., 2012). Yet, it should be noted that at least certain SAR11 phylotypes may display high growth rates (Malmstrom, Campbell et al., 2011).
For many decades, the Liebig’s law of the minimum, which states that only a single resource is limiting, was the dominant theory shaping how oceanographers viewed phyto- and bacterioplankton ecology and their impact on biochemical cycles (Arrigo, 2004). This view has been recently challenged by the concept of colimitation or multiple nutrient limitations (Saito et al., 2008). Here we show that the reality is even more complex, with different bacterial groups experiencing different types of limitations under the same environmental conditions.
References
Alonso-Sáez L, Sánchez O, Gasol JM . (2012). Bacterial uptake of low molecular weight organics in the subtropical Atlantic: are major phylogenetic groups functionally different? Limnol Oceanogr 57: 798–808.
Arrigo KR . (2004). Marine microorganisms and global nutrient cycles. Nature 437: 349–355.
Buchan A, Gonzalez JM, Moran MA . (2005). Overview of the marine Roseobacter lineage. Appl Environ Microbiol 71: 5665–5677.
Campbell BJ, Yu L, Heidelberg JF, Kirchman DL . (2011). Activity of abundant and rare bacteria in a coastal ocean. Proc Natl Acad Sci USA 108: 12776–12781.
Caron DA, Lim EL, Sanders RW, Dennett MR, Berninger UG . (2000). Responses of bacterioplankton and phytoplankton to organic carbon and inorganic nutrient additions in contrasting oceanic ecosystems. Aquat Microb Ecol 22: 175–184.
Cotner JB, Ammerman JW, Peele ER, Bentzen E . (1997). Phosphorus-limited bacterioplankton growth in the Sargasso Sea. Aquat Microb Ecol 13: 141–149.
Cottrell MT, Kirchman DL . (2000). Natural assemblages of marine proteobacteria and members of the Cytophaga-Flavobacter cluster consuming low-and high-molecular-weight dissolved organic matter. Appl Environ Microbiol 66: 1692–1697.
Eilers H, Pernthaler J, Amann R . (2000). Succession of pelagic marine bacteria during enrichment: a close look at cultivation-induced shifts. Appl Environ Microbiol 66: 4634–4640.
Gifford SM, Sharma S, Booth M, Moran MA . (2012). Expression patterns reveal niche diversification in a marine microbial assemblage. ISME J 7: 281–298.
Malmstrom RR, Cottrell MT, Elifantz H, Kirchman DL . (2005). Biomass production and assimilation of dissolved organic matter by SAR11 bacteria in the Northwest Atlantic Ocean. Appl Environ Microbiol 71: 2979.
Musat N, Halm H, Winterholler B, Hoppe P, Peduzzi S, Hillion F et al (2008). A single-cell view on the ecophysiology of anaerobic phototrophic bacteria. Proc Natl Acad Sci USA 105: 17861.
Nielsen JL, Christensen D, Kloppenborg M, Nielsen PH . (2003). Quantification of cell-specific substrate uptake by probe-defined bacteria under in situ conditions by microautoradiography and fluorescence in situ hybridization. Environ Microbiol 5: 202–211.
Pinhassi J, Gómez-Consarnau L, Alonso-Sáez L, Sala MM, Vidal M, Pedrós-Alió C et al (2006). Seasonal changes in bacterioplankton nutrient limitation and their effects on bacterial community composition in the NW Mediterranean Sea. Aquat Microb Ecol 44: 241–252.
Rappé MS, Connon SA, Vergin KL, Giovannoni SJ . (2002). Cultivation of the ubiquitous SAR 11 marine bacterioplankton clade. Nature 418: 630–633.
Saito MA, Goepfert TJ, Ritt JT . (2008). Some thoughts on the concept of colimitation: three definitions and the importance of bioavailability. Limnol Oceanogr 53: 276–290.
Sebastián M, Pitta P, González JM, Thingstad TF, Gasol JM . (2012). Bacterioplankton groups involved in the uptake of phosphate and dissolved organic phosphorus in a mesocosm experiment with P-starved Mediterranean waters. Environ Microbiol 14: 2334–2347.
Tanaka T, Zohary T, Krom MD, Law CS, Pitta P, Psarra S et al (2007). Microbial community structure and function in the Levantine Basin of the eastern Mediterranean. Deep Sea Res I 54: 1721–1743.
Thingstad TF, Krom MD, Mantoura RFC, Flaten GAF, Groom S, Herut B et al (2005). Nature of phosphorus limitation in the ultraoligotrophic eastern Mediterranean. Science 309: 1068–1071.
Acknowledgements
This experiment in the Cretacosmos facility was arranged in co-operation by project Nutritunnel financed by the Research Council of Norway and the MESOAQUA network (FP7/2007-2013, grant agreement no. 228224). We thank Frede T Thingstad and Paraskevi Pitta for inviting MS to participate in this experiment and Runar Thyrhaug for performing the heterotrophic bacterial activity measurements. Processing of the samples was supported by the grants FOSMICRO (CTM2009-07679-E) and STORM (CTM2009-09352/MAR), funded by the former Spanish Ministry of Science and Innovation. MS was supported by a ‘Juan de la Cierva’ award.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
This paper is dedicated to the memory of Runar Thyrhaug who passed away shortly after the experiment.
Supplementary Information accompanies this paper on The ISME Journal website
Supplementary information
Rights and permissions
About this article
Cite this article
Sebastián, M., Gasol, J. Heterogeneity in the nutrient limitation of different bacterioplankton groups in the Eastern Mediterranean Sea. ISME J 7, 1665–1668 (2013). https://doi.org/10.1038/ismej.2013.42
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ismej.2013.42
Keywords
This article is cited by
-
Impact of phosphate concentration on the metabolome of biofilms of the marine bacterium Pseudoalteromonas lipolytica
Metabolomics (2022)
-
Phylotype Dynamics of Bacterial P Utilization Genes in Microbialites and Bacterioplankton of a Monomictic Endorheic Lake
Microbial Ecology (2017)
-
Lipid remodelling is a widespread strategy in marine heterotrophic bacteria upon phosphorus deficiency
The ISME Journal (2016)
-
Nutrient Limitation in Surface Waters of the Oligotrophic Eastern Mediterranean Sea: an Enrichment Microcosm Experiment
Microbial Ecology (2016)
-
Diversity of marine microbes in a changing Mediterranean Sea
Rendiconti Lincei (2015)