Abstract
In 1944, Harold Kirby described microorganisms living within nuclei of the protists Trichonympha in guts of termites; however, their taxonomic assignment remains to be accomplished. Here, we identified intranuclear symbionts of Trichonympha agilis in the gut of the termite Reticulitermes speratus. We isolated single nuclei of T. agilis, performed whole-genome amplification, and obtained bacterial 16S rRNA genes by PCR. Unexpectedly, however, all of the analyzed clones were from pseudogenes of 16S rRNA with large deletions and numerous sequence variations even within a single-nucleus sample. Authentic 16S rRNA gene sequences were finally recovered by digesting the nuclear DNA; these pseudogenes were present on the host Trichonympha genome. The authentic sequences represented two distinct bacterial species belonging to the phylum Verrucomicrobia, and the pseudogenes have originated from each of the two species. Fluorescence in situ hybridization confirmed that both species are specifically localized, and occasionally co-localized, within nuclei of T. agilis. Transmission electron microscopy revealed that they are distorted cocci with characteristic electron-dense and lucent regions, which resemble the intranuclear symbionts illustrated by Kirby. For these symbionts, we propose a novel genus and species, ‘Candidatus Nucleococcus trichonymphae’ and ‘Candidatus Nucleococcus kirbyi’. These formed a termite-specific cluster with database sequences, other members of which were also detected within nuclei of various gut protists, including both parabasalids and oxymonads. We suggest that this group is widely distributed as intranuclear symbionts of diverse protists in termite guts and that they might have affected the evolution of the host genome through lateral gene transfer.
Similar content being viewed by others
Introduction
Many protists harbor symbiotic microorganisms within their cytoplasm and occasionally within their nucleoplasm. Microorganisms living within the nuclei of protists were discovered in 1856 (Müller, 1856), and since then, numerous studies have reported both eukaryotic and prokaryotic intranuclear symbionts in diverse protist lineages (Görtz, 1986; Fokin, 2004). In the ciliates Paramecium, the macronucleus and micronucleus can be infected by specific bacterial species belonging to the genera Holospora, Caedibacter and ‘Candidatus Gortzia’ in the order Rickettsiales of Alphaproteobacteria (Boscaro et al., 2013). These symbionts have two distinct life stages; the long, infectious form found in the cytoplasm and the short, reproductive form that divides in the host nucleus (Fujishima and Kodama, 2012). For other intranuclear symbionts, mainly because of the difficulty of their cultivation, information on their ecology, physiology and even the taxonomy is scarce.
Termites harbor abundant and diverse microorganisms in their hindgut, which comprise protists, bacteria and archaea. These gut symbionts are unique to termites and their closest relatives, cockroaches (Hongoh, 2010; Schauer et al., 2012), and are essential to the nutrition of the host insects (Brune and Ohkuma, 2011; Hongoh, 2011). One of the prominent features of this symbiotic system is the cellular association between the protists and the prokaryotes. For example, the cellulolytic protist Trichonympha agilis in the gut of the termite Reticulitermes speratus is permanently associated with two species of cytoplasmic symbionts, ‘Candidatus Endomicrobium trichonymphae’ (phylotype Rs-D17) (Stingl et al., 2005; Ohkuma et al., 2007) and ‘Candidatus Desulfovibrio trichonymphae’ (phylotype Rs-N31) (Sato et al., 2009). Furthermore, spirochetes and Bacteroidales bacteria are attached to its cell surface (Hongoh et al., 2007b). The complete genome sequence analysis of phylotype Rs-D17 suggested that the endosymbionts contribute to the host protists and termites by the synthesis of amino acids and cofactors, which are deficient in the ingested wood materials (Hongoh et al., 2008a). Likewise, termite gut protists generally harbor diverse intracellular and/or extracellular symbiotic prokaryotes comprising members of the orders Spirochaetales, Bacteroidales, ‘Endomicrobiales’, Desulfovibrionales, Actinomycetales and Methanobacteriales (Hongoh and Ohkuma, 2011; Desai and Brune, 2012; Strassert et al., 2012).
Under light microscope observation, Kirby (1944) described several types of uncultured microorganisms living within the nuclei of Trichonympha in the gut of various termite species. He illustrated those symbionts in detail and tentatively assigned them to the bacterial genus Caryococcus and putatively chytrid fungi Nucleophaga and Caryoletira (Kirby, 1944). Bacteria-like cells within the nuclei of various protist species in termite guts were later observed by transmission electron microscopy (TEM) in numerous studies (Brugerolle and Radek, 2006). However, the taxonomic positions of these intranuclear symbionts of termite gut protists remain unknown.
In this study, we attempted to identify intranuclear bacterial symbionts of Trichonympha agilis based on small subunit rRNA sequences. Our results answer the long-standing question, ‘what are the symbionts living within the nuclei of the protists in termite guts?’ In addition, we present the evidence for the lateral transfer of genome fragments from the intranuclear symbionts to the host protist. This provides new insights into the evolution of the termite gut protists.
Materials and methods
Sample preparation
The wood-feeding termite Reticulitermes speratus was collected at Ogose in Saitama prefecture, Japan. The gut of a worker termite was removed using sterile forceps, and the gut contents were suspended in Trager’s solution U (Trager, 1934). A single cell of the gut protist T. agilis (phylum Parabasalia) was collected with a glass capillary attached to a TransferMan NK2 micromanipulator (Eppendorf, Hamburg, Germany). The isolated T. agilis cell was ruptured in the presence of 1% Nonidet P-40 or Tween 20 (Nacalai Tesque, Kyoto, Japan), and the exposed nucleus was collected and washed in the same solution. The nucleus was transferred to a sterile 0.2-ml tube containing 2 μl of lysis buffer (0.3 N KOH, 10 mM EDTA, 100 mM dithiothreitol) and subjected to isothermal whole-genome amplification (WGA) using GenomiPhi HY or GenomiPhi V2 (GE Healthcare, Piscataway, NJ, USA) as described previously (Hongoh et al., 2008a, 2008b). The WGA samples were purified by ethanol precipitation and diluted samples were used as templates for PCR.
To digest the genomic DNA of T. agilis, the nucleus was dissected with a Microfeather Blade K-730 (Feather, Tokyo, Japan) attached to the micromanipulator, in a solution containing RQ1 RNase-free DNase (Promega, Madison, WI, USA). DNase was inactivated in the above alkaline lysis buffer.
PCR amplification, cloning and sequencing
PCR targeting the small subunit rRNA genes was performed using a proofreading DNA polymerase, Phusion (Finnzymes, Espoo, Finland), in a C1000 Thermal Cycler (Bio-Rad, Hercules, CA, USA) under previously described conditions (Hongoh et al., 2007a). When PCR was performed directly using a single nucleus without WGA, the number of PCR cycles was raised from 15 to 35, and a second PCR with 20 cycles was performed. The primers and annealing temperature used in this study are listed in Table 1. Primers Verr-61F, Verr-361F, and Verr-1351R were designed in this study to specifically detect most members of subdivisions 1–4 and 6 in the phylum Verrucomicrobia (Hugenholtz et al., 1998). Although primer Verr-61F also targets several other bacterial taxa, the combination with primer 1351R secured the specificity to the Verrucomicrobia. The products were separated by 1% agarose gel electrophoresis, purified using the Monofas DNA purification kit (GL Sciences, Tokyo, Japan), and cloned using the ZeroBlunt TOPO PCR Cloning Kit for Sequencing (Life Technologies, Carlsbad, CA, USA). The sequencing was performed using the BigDye Terminator Cycle Sequencing Kit V3.1 (Life Technologies) with an ABI 3730 genetic analyzer, basically as described previously (Hongoh et al., 2003). The sequences reported in this study have been deposited in DDBJ/EMBL/GenBank under accession numbers AB826690–AB826745.
Phylogenetic analysis
16S rRNA sequences were subjected to BLASTn searches against the NCBI nr database for a preliminary taxonomic assignment (Altschul et al., 1997). The sequences were sorted into phylotypes using the program package mothur 1.30.1 (Schloss et al., 2009). Representative clones of the phylotypes were incorporated into an ARB database (Ludwig et al., 2004). The alignment was manually corrected (Sato et al., 2009), and 1389 unambiguously aligned nucleotide sites, corresponding to positions 28–1491 in Escherichia coli (J01695), were used for phylogenetic analysis. Maximum likelihood (ML) trees were constructed using RAxML 7.0.3 (Stamatakis, 2006) implemented in ARB version 5.2 (Ludwig et al., 2004). A general time-reversible model with a gamma distribution was used, and 500 bootstrap resamplings were performed to evaluate the robustness of the topology. Minimum evolution (ME) trees were also constructed in MEGA5 (Tamura et al., 2011), using the maximum composite likelihood method with a gamma distribution to estimate a distance matrix.
Fluorescence in situ hybridization (FISH) analysis
FISH was performed as described previously (Noda et al., 2006). The oligonucleotide probes used in this study are listed in Table 1. Probes specifically targeting 16S rRNA of verrucomicrobial phylotypes or clusters were designed using the probe-designing function in ARB. The sequence specificity of the probes was checked in the Probe Match program in the Ribosomal Database Project (http://rdp.cme.msu.edu/probematch/search.jsp) and by probeCheck (Loy et al., 2008). The optimal conditions for specific hybridization were estimated in silico using DINAMelt (Markham and Zuker, 2005) and were determined by shifting the annealing temperature and/or formamide concentration in an actual sample. Probe V-RsD37-130 was designed to detect the majority of the verrucomicrobial termite-specific cluster, as designated in this study. This probe has one base mismatch to several phylotypes such as IswTcA-49 (JQ993599); therefore, we adjusted the conditions for specific hybridization to allow one base mismatch (Table 1). Phylotypes Cc3-048 (AB299564), Cc3-051 (AB299567), and RsaP110 (AY571501) are not detectable with this probe. For phylotype-specific probes Verr-K1-181 and Verr-K2-181, we also designed helper probes (Table 1; Fuchs et al., 2000). Probe EUB338-III was used to specifically detect the majority of the phylum Verrucomicrobia (Daims et al., 1999). These probes were labeled at their 5′ ends with either of Texas red or 6-carboxyfluorescein (6FAM). The specimens were observed under a BX-51 epifluorescence microscope (Olympus, Tokyo, Japan) or a TCS SP8 confocal laser scanning microscope (Leica, Wetzlar, Germany).
Transmission electron microscopy
TEM was conducted as described previously (Noda et al., 2006). Briefly, large protist species, including T. agilis, were collected by low-speed centrifugation, prefixed with 2.5% (v/v) glutaraldehyde and 2% paraformaldehyde in 0.1 M cacodylate buffer (pH 7.4) at 4 °C overnight, and then post-fixed in buffer containing 1% osmium tetroxide for 1.5 h. Thin sections were post-stained with uranyl acetate and lead citrate, and viewed under a 1230 transmission electron microscope (JEOL, Tokyo, Japan) or an H-7500 transmission electron microscope (Hitachi, Tokyo, Japan).
Results
Discovery of 16S rRNA pseudogenes related to verrucomicrobia
16S rRNA genes were amplified by PCR using bacteria-specific primers 27F-mix and 1492R (Table 1) from six WGA samples, each prepared from a single T. agilis nucleus. The PCR products were visualized in gel electrophoresis as multiple bands, the largest of which corresponded to the predicted size of the targeted region (lanes 1–6, Figure 1a). Both long and shorter products were separately excised from the gel and sequenced. Although ∼200 clones were analyzed for the longest products, they represented only phylotypes Rs-D17 and Rs-N31, known cytoplasmic symbionts of T. agilis (Stingl et al., 2005; Ohkuma et al., 2007; Sato et al., 2009), from four of the WGA samples (1–3 and 6, corresponding to the lane numbers in Figure 1a).
16S rRNA pseudogenes present within the nuclei of T. agilis. (a) Agarose gel electrophoresis of 16S rRNA genes amplified by PCR with a bacteria-specific primer set from single nuclei samples of T. agilis. Lanes 1–6, PCR products from each of six nuclei. Lane 7 and 8, PCR products from single DNase-treated nuclei. N, negative control for PCR. (b) Structures of representative pseudogenes. Gray color denotes deleted regions. The positions of PCR primers specific to verrucomicrobia (Table 1) are indicated. RsTaN-K1 is an authentic 16S rRNA phylotype. RsTaN2-18 originated from RsTaN-K2 and the others shown here originated from RsTaN-K1. The positions correspond to those in Escherichia coli (J01695).
On the other hand, the 126 clones of the shorter products from the above four WGA samples comprised exclusively pseudogenes of 16S rRNA, related to the bacterial phylum Verrucomicrobia. All the pseudogenes contained multiple large deletions (Figure 1b), and numerous base substitutions and indels in the highly conserved regions of 16S rRNA (data not shown). To discount the possibility that these were artifacts formed during WGA, we amplified 16S rRNA genes by PCR directly from two single T. agilis nuclei, bypassing WGA. Only pseudogenes with large deletions similar to those mentioned above were obtained. These pseudogenes, derived from the samples prepared with or without WGA, have numerous sequence variations even within a single-nucleus sample. Of 156 analyzed pseudogene sequences, 47 phylotypes, defined with 99.0% sequence identity, were recognized (Supplementary Table S1). In the case of the WGA sample 6 (Figure 1a), 12 phylotypes were identified from 14 analyzed clones (Supplementary Table S1). We used a proofreading DNA polymerase and found almost no variations in the Rs-D17 sequences obtained above; it is unlikely that these variants were artificially generated during PCR.
Acquisition of authentic 16S rRNA gene of verrucomicrobial phylotype RsTaN-K1
We assumed that these pseudogenes originated from intranuclear symbionts. Indeed, we detected coccoid cells within the nuclei of T. agilis by FISH using the Verrucomicrobia-specific probe EUB338-III (Supplementary Figures 1a–f). To acquire an authentic 16S rRNA gene for the pseudogenes, we used verrucomicrobia-specific primers (Table 1), one of which, Verr-1351R was designed based on a deleted region in most of the pseudogenes (Figure 1b). This was an attempt not to amplify the sequences of the contaminating cytoplasmic symbionts and the verrucomicrobial pseudogenes. However, we could only amplify different variations of the verrucomicrobial pseudogenes: RsTaNv1-09, RsTaNv1-18, RsTaNv2-05 and RsTaN5v1-07 (AB826742–AB826745) (Figure 1b).
We hypothesized that these numerous pseudogenes are present on a chromosome or chromosomes of T. agilis as a result of lateral gene transfer (LGT). We therefore performed microsurgery of a T. agilis nucleus in the presence of DNase I, to digest the host genomic DNA. The DNase-treated nucleus sample was subjected to WGA, and PCR was performed using bacteria-specific primers 27F-mix and 1492R. As expected, the shorter products disappeared (lane 7, Figure 1a). Among 89 clones, five belonged to Rs-N31 and the rest consisted of a single phylotype, named RsTaN-K1, with identical, apparently authentic sequences, which were phylogenetically affiliated with the order Puniceicoccales in subdivision 4 of the phylum Verrucomicrobia (Figure 2). We also performed PCR using the verrucomicrobia-specific primers (Table 1), and obtained exclusively the same sequences.
Phylogenetic positions of intranuclear symbionts based on 16S rRNA gene sequences. A maximum likelihood tree was constructed. Pseudogene phylotypes, RsTaNv1-09, RsTaNv1-18, RsNv2-05 and RsTaN5v1-07 were not included in this analysis because of their shorter length. The topologies, including the positions of pseudogenes and the formation of the termite-specific cluster, were basically consistent in a ME tree (data not shown). Open and closed circles at the nodes indicate the bootstrap confidence values of 70–94% and 95–100%, respectively. O. terrae, D. colitermitum and A. agarolyticus were used as outgroups.
Localization of RsTaN-K1 and the discovery of another phylotype, RsTaN-K2
FISH was performed to locate the cells of phylotype RsTaN-K1, using the phylotype-specific probe Verr-K1-181 in combination with the Verrucomicrobia-specific probe EUB338-III or probe EUB338 specific to bacteria except Verrucomicrobia and several other taxa (Daims et al., 1999) (Table 1). We successfully detected the RsTaN-K1 cells exclusively within the nucleus of T. agilis (data not shown). Unexpectedly, however, we noticed that RsTaN-K1 frequently coexisted with another intranuclear bacterium that was detected with probe EUB338-III (data not shown). Thus, there must be another verrucomicrobial species present.
During our examination, we realized that the infection rate of RsTaN-K1 varied greatly, being dependent on the termite colony. In certain colonies, <1% of the T. agilis cells were infected with intranuclear verrucomicrobial symbionts, whereas in other colonies, for example, 10%, 38%, 42%, 53% and 87% of the T. agilis cells were infected with both RsTaN-K1 and the unidentified verrucomicrobia or with either of them alone. We used such a termite colony, where the nuclei of T. agilis were infected almost only with the unidentified verrucomicrobia. We repeated the same procedure as above (lane 8, Figure 1a) and obtained another verrucomicrobial phylotype, RsTaN-K2, which shared 90% sequence identity with RsTaN-K1 (Figure 2). By FISH analysis with probe Verr-K2-181 specific to RsTaN-K2 (Table 1), we confirmed that RsTaN-K2 inhabited only the nuclei of T. agilis and frequently coexisted with RsTaN-K1 within the same nuclei (Figure 3).
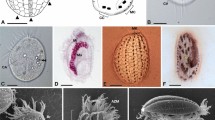
Specific detection of intranuclear symbionts of Trichonympha agilis by FISH. (a) Phase contrast image of a T. agilis cell. (b) DAPI-stained image. (c) Merged image of FISH analyses that detected RsTaN-K1 with the Texas red-labeled probe Verr-K1-181 (red) or RsTaN-K2 with the 6FAM-labeled probe Verr-K2-181 (green). (d) Magnified image of the nucleus (arrowhead) in panel c. (e) Panel d was overlaid with the DAPI image. Bars represent 10 μm (a); 5 μm (d).
Morphology and enumeration of RsTaN-K1 and RsTaN-K2 cells
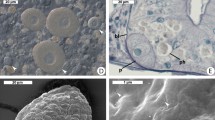
The cells of phylotypes RsTaN-K1 and RsTaN-K2, detected by FISH, were coccoid with a certain degree of distortion (Figure 3d and Supplementary Figures S1a–f). Under TEM observation of T. agilis nuclei, we found coccoid, distorted bacterial cells (Figure 4), although we could not discriminate RsTaN-K1 and RsTaN-K2. On FISH images, the dimensions of the RsTaN-K1 cells were 0.9±0.2 μm by 0.7±0.2 μm (mean±s.d., n=104), and those of RsTaN-K2 cells were 1.1±0.3 μm by 0.7±0.1 μm (n=98). In a certain preparation, the numbers of RsTaN-K1 and RsTaN-K2 cells that were contained within the same nuclei were 31±8 and 62±26 (n=33, t-test, P<0.0001), respectively. In another preparation from a different termite colony, the numbers were 39±10 and 55±17 (n=5, t-test, P=0.10). Thus, RsTaN-K2 tended to outnumber RsTaN-K1 within a nucleus, when co-localized. When RsTaN-K2 alone inhabited a nucleus, the number of RsTaN-K2 cells was 57±25 (n=7) and 51±17 (n=8), in the former and latter preparations, respectively. This implied that the presence of co-inhabiting RsTaN-K1 does not affect the population of RsTaN-K2. The dimensions of the nuclei infected by both RsTaN-K1 and RsTaN-K2, in a preparation for FISH, were 129±23 μm2 (n=27), which was not significantly different from those of non-infected nuclei, 119±27 μm2 (n=15; t-test, P=0.09). The morphological details are further explored in the description sections.
Transmission electron micrographs of intranuclear symbionts of T. agilis. (a) Irregular, distorted cocci within a T. agilis nucleus. NM, nuclear membrane; G, Golgi apparatus. (b) A coccus during division. (c) A coccus. (d) Magnified image of the cell wall of the intranuclear symbiont. OM, outer membrane; IM, inner membrane. The boundary between the electron-dense and lucent regions appears not to be a membranous structure, although it is unclear. Bars represent 1 μm (a); 0.5 μm (b, c); 0.1 μm (d).
Relationship between the intranuclear symbionts and the 16S rRNA pseudogenes
Phylogenetic analysis was conducted that included the pseudogenes (Figure 2). The pseudogenes were separated to two distinct clusters, which clearly originated from phylotypes RsTaN-K1 and RsTaN-K2, respectively. The obtained pseudogenes were more diverse and abundant in the RsTaN-K1-related cluster than in the RsTaN-K2-related one (Supplementary Figure S2a and Supplementary Table S1). There were almost no identical pseudogene sequences among the six single-nucleus samples. The three phylotypes of pseudogenes that originated from RsTaN-K2 shared large deletions (Figure 1b and Supplementary Figure S2a); thus, the lateral transfer of the 16S rRNA gene of RsTaN-K2 to the Trichonympha genome was likely to have been a single event. Similarly, all but two phylotypes of the pseudogenes that originated from RsTaN-K1 shared large deletions (Figure 1b and Supplementary Figure S2a). The two phylotypes, RsTaNv1-09 and RsTaNv2-05 (Supplementary Figure S2a), had no large deletions, whereas they contained numerous base substitutions within conserved regions. Hence, the majority of the numerous pseudogenes in the RsTaN-K1-related cluster likely also derived from a single LGT event. This was supported by the data that the frequency of the pairwise genetic distances between the pseudogene phylotypes related to RsTaN-K1 formed nearly a single peak (Supplementary Figure S2b).
Phylogenetic positions of RsTaN-K1 and RsTaN-K2
Phylotype RsTaN-K1 shared 97% sequence identity with uncultured clones IswTcA-49 and ItbTcA-162 in the database (Figure 2). These were recovered from Trichonympha cells in the gut of the kalotermitid termites Incisitermes schwarzi and I. tabogae, respectively (Strassert et al., 2012). Phylotype RsTaN-K2 shared 96% sequence identity with another uncultured clone, ItbTcA-142, from a Trichonympha cell in the gut of I. tabogae (Strassert et al., 2012) and formed a monophyletic cluster together with the other three clones from termite guts (Figure 2). These two clusters further formed a monophyletic cluster that exclusively consisted of uncultured clones from lower termites in the genera Cryptotermes and Reticulitermes and one clone from a wood-feeding higher termite, Microcerotermes sp. (Figure 2). This termite-specific cluster showed only 85% or lower sequence similarity to cultured strains, such as Cerasicoccus arenae and Puniceicoccus vermicola.
Detection of intranuclear symbionts in other protist species
We performed FISH using probe V-RsD37-130 in combination with EUB338-III or EUB338 to detect the cells of the majority of the termite-specific cluster in Figure 2. Cocci resembling the cells of RsTaN-K1 and RsTaN-K2 were detected within the nuclei of the parabasalid protist Teranympha mirabilis, in addition to T. agilis, and in nuclei of the oxymonad protists Pyrsonympha grandis, Dinenympha exilis and Dinenympha parva (Figure 5). Occasionally, bacteria that were detected with EUB338 but not with EUB338-III or V-RsD37-130 were observed in the nuclei of P. grandis (Figures 5a–c). In several P. grandis cells, they were co-localized with the verrucomicrobial symbionts within the same nucleus (Supplementary Figures S1g–j). Thus, intranuclear bacterial symbionts other than verrucomicrobia were associated with P. grandis.
Detection of verrucomicrobial symbionts in nuclei of (a–d) Pyrsonympha grandis, (e–h) Dinenympha exilis, (i–l) Dinenympha parva and (m–p) Teranympha mirabilis. (a, f, j, n) DAPI-stained images. (e, i, m) Phase contrast images. (b) Merged image of FISH analyses using the Texas red-labeled Verrucomicrobia-specific probe EUB338-III (red) and the 6FAM-labeled probe EUB338 specific to bacteria except Verrucomicrobia and several other taxa (green). (c) Merged image of a and b. (g, k, o) Merged images of FISH analyses using probe EUB338-III (red) and the 6FAM-labeled probe V-RsD37-130 specific to the majority of the termite-specific cluster in Figure 2 (green). (d, h, l, p) Magnified images of the DAPI-stained nuclei merged with FISH signals detected using probes EUB338-III (red) and V-RsD37-130 (green). Panel d was taken from a P. grandis cell that was different from the two cells in panels a–c. Arrowheads indicate the nuclei. Arrows in a–c indicate the Pyrsonympha nucleus that contained non-verrucomicrobial intranuclear symbionts. Panels a–c were observed under a laser scanning microscope. Bars represent 10 μm (c, g, k, o); 5 μm (d, h, l, p).
Based on the present molecular, morphological, and ecological data, we propose a novel genus and species, ‘Candidatus Nucleococcus trichonymphae’ and ‘Candidatus Nucleococcus kirbyi’ for phylotypes RsTaN-K1 and RsTaN-K2, respectively.
Description of ‘Candidatus Nucleococcus’ gen. nov.
Nucleococcus (nu.cle.o.coc′cus. L. masc. n. nucleus, a nucleus; N.L. masc. n. coccus from Gr. masc. n. kokkos a coccus; N.L. masc. n. nucleococcus a coccus from a nucleus). The bacteria specifically colonize the nucleoplasm of protists in termite guts. They are distorted cocci of 0.3–1.9 μm and occasionally observed in a division form. They have a Gram-negative type cell wall and are directly in contact with the nucleoplasm. Using uranyl acetate–lead citrate TEM stain, the cytoplasm is sharply separated between electron-dense and electron-lucent regions. The peripheral region of the cytoplasm is consistently translucent. The ‘intracytoplasmic membrane’ that compartmentalizes the cytoplasm of certain verrucomicrobial species (Lee et al., 2009) is not recognized, although further observation is needed to clarify its presence or absence. To date, the members of this genus have not been cultured. The assignment is based on 16S rRNA gene sequence. Currently, this genus comprises two species described below and other uncharacterized 16S rRNA phylotypes IswTcA-49, ItbTcA-162, ItbTcA-142, Cc3-002, RsC01-040 and RsStar333. These phylotypes share more than 90% sequence identity.
Description of ‘Candidatus Nucleococcus trichonymphae’ sp. nov.
Nucleococcus trichonymphae (tri.cho.nym′phae. N.L. n. Trichonympha, a genus of flagellated protists; N.L. gen. trichonymphae, of Trichonympha, referring to the host genus). The basic characteristics of this species are the same as those for the genus. They specifically colonize the nucleoplasm of the protist T. agilis in the gut of the termite Reticulitermes speratus. The cell dimensions are 0.5–1.5 μm by 0.3–1.1 μm. The assignment is based on 16S rRNA gene sequence (AB826704) and hybridization with the 16S rRNA-targeted oligonucleotide probe (5′-CATGCAGAACGAAACCAC-3′). This species corresponds to phylotype RsTaN-K1.
Description of ‘Candidatus Nucleococcus kirbyi’ sp. nov.
Nucleococcus kirbyi (kir′byi. N.L. gen. n. Kirbyi, of Kirby; named after Harold Kirby, a microbiologist who discovered the bacteria of this genus). The basic characteristics of this species are the same as those for the genus. The cell dimension is 0.6–1.9 μm by 0.4–1.0 μm. They specifically colonize the nucleoplasm of the protist T. agilis in the gut of the termite Reticulitermes speratus. The assignment is based on 16S rRNA gene sequence (AB826705) and hybridization with the 16S rRNA-targeted oligonucleotide probe (5′-CATGCGAAAAGAAATCAC-3′). This species corresponds to phylotype RsTaN-K2.
Discussion
This is the first report of intranuclear symbionts belonging to the phylum Verrucomicrobia. Thus far, only members of the phylum Proteobacteria have been identified as intranuclear bacterial symbionts of protists and other eukaryotes (Alverca et al., 2002; Fokin, 2004; Arneodo et al., 2008; Zielinski et al., 2009). Based on their morphology, ‘Candidatus Nucleococcus’ (abbreviated hereafter as Nucleococcus) closely resembled Caryococcus nucleophagus, described by Kirby (1944) for the intranuclear symbionts of Trichonympha corbula, as ‘Spherules with a diameter of 1/2 μm, sometimes arranged in pairs, sometimes with a thicker, crescentic, stainable area at the periphery on one side...’ The genus Caryococcus was originally described for an intranuclear symbiont of the euglenid protist Euglena deses by Dangeard (1902). The assignment of the intranuclear symbionts of Trichonympha to the genus Caryococcus was not based on evidence and ‘allocation to the genus Caryococcus is a matter of convenience’ (Kirby, 1944). We therefore proposed a novel genus for the intranuclear bacterial symbionts of Trichonympha in this study.
Phylum Verrucomicrobia is one of the dominant bacterial groups in soil and members are also detected, less abundantly, from various other environments including animal intestinal tracts (Wagner and Horn, 2006). Verrucomicrobia that inhabit a eukaryotic cell are rare; only two examples have been reported. Unique verrucomicrobial defensive symbionts are localized as ‘epixenosomes’ on the cell surface of the marine ciliates Euplotidium (Petroni et al., 2000). ‘Candidatus Xiphinematobacter’ are maternally inherited cytoplasmic symbionts of the ectoparasitic nematodes Xiphinema found on plants (Vandekerckhove et al., 2002). These are phylogenetically distant from Nucleococcus.
In termite guts, verrucomicrobia are minor, but autochthonous residents. Several phylotypes have been found in subdivision 6 and in the orders Opitutales and Puniceicoccales in subdivision 4 (Hongoh et al., 2003, 2005; Nakajima et al., 2005; Yang et al., 2005; Hongoh et al., 2007a; Ikeda-Ohtsubo et al., 2010; Strassert et al., 2012). These phylotypes comprise one or more termite-specific clusters in each clade, and most members are as yet unculturable. Among them, TAV1–4 in the Opitutales are cultured strains isolated from the gut of Reticulitermes flavipes (Stevenson et al., 2004), and genomic, transcriptomic, proteomic and physiological analyses of strain TAV2, described as Diplosphaera colotermitum, have been reported (Wertz et al., 2012; Isanapong et al., 2013). D. colotermitum is a microaerophilic coccus, and the studies suggested their symbiotic roles in fixation and recycling of nitrogen, amino-acid production and, possibly, degradation of cellulose and xylan. Similar physiological properties are found in other verrucomicrobial species (Martinez-Garcia et al., 2012; Wertz et al., 2012). However, ‘Candidatus Nucleococcus trichonymphae’ (RsTaN-K1) and ‘Candidatus Nucleococcus kirbyi’ (RsTaN-K2) (abbreviated hereafter as N. trichonymphae and N. kirbyi, respectively), shared only 85% or lower 16S rRNA sequence identity with known isolates, including D. colotermitum; thus, prediction of their functions is impossible at present.
The closest phylotypes to the two Nucleococcus species are those obtained from Trichonympha cells in the gut of Incisitermes termites (Strassert et al., 2012) (Figure 2). The sequence similarity is as high as 96–97%; these are possibly also intranuclear symbionts of the Trichonympha species. These sequences appear to be authentic and it is not known whether similar LGT events have occurred between these phylotypes and their host Trichonympha. Other members of the termite-specific cluster in Figure 2 included uncultured phylotypes recovered from the cells of the protist Oxymonas sp. in the gut of Cryptotermes secundus (Desai et al., 2010). In the present study, we identified members of this cluster as intranuclear symbionts of both oxymonad and parabasalid protists (Figure 5). These intranuclear symbionts were detected from some, but not all of the population of each protist species; therefore, this symbiosis appears to be facultative. Thus, Nucleococcus might have a life stage outside the host nucleus, as do the alphaproteobacterial intranuclear symbionts of paramecia (Fujishima and Kodama, 2012). Indeed, Dolan et al. (2004) observed that cocci, morphologically resembling Nucleococcus, were released from a hypertrophied nucleus of Staurojoenina assimilis, although we did not observe such hypertrophy or rupture of infected nuclei of T. agilis and also did not find N. trichonymphae or N. kirbyi outside T. agilis cells.
The verrucomicrobial intranuclear symbionts appear to be widely distributed in diverse protists in termite guts. Nucleococcus-like intranuclear cocci with the characteristic electron-dense and lucent regions (Figure 4) could be observed in many published TEM images in previous studies. The host protists that harbored Nucleococcus-like intranuclear symbionts were parabasalids, including Joenia annecteus, Pachyjoenia howa, Placojoenia sinaica, Spironympha kofoidi, Spirotrichonympha minor, Stephanonympha neulumbium and the above-mentioned Staurojoenina assimilis (Radek et al., 1992; Radek and Hausmann, 1994; Rösel et al., 1996; Radek, 1997; Dolan et al., 2004; Brugerolle and Bordereau, 2005, 2006). Intranuclear bacterial symbionts with cell morphologies distinct from Nucleococcus were also observed on TEM images of parabasalids, such as Caduceia versatilis, Joenia annecteus, Metadevescovina sp. Microjoenia fallax, Spironympha vertisis, and an oxymonad, Monocercomonoides termitis (Radek et al., 1992; Radek, 1994; d'Ambrosio et al., 1999; Brugerolle and Bordereau, 2006; Maaß and Radek, 2006). In Joenia annecteus, both Nucleococcus-like and a distinct morphotype coexisted in the same nucleus (Radek et al., 1992), as in the oxymonad Pyrsonympha grandis in our observation (Supplementary Figure S1g–j).
Our discovery of the 16S rRNA pseudogenes present on the host Trichonympha genome strongly suggests that at least a portion of both the N. trichonymphae and N. kirbyi genomes have, independently, been laterally transferred, although this should be further confirmed by detailed genome analyses of the host T. agilis. A lateral transfer of genome fragments has been reported for the intracellular alphaproteobacterial symbionts Wolbachia and their invertebrate hosts (Hotopp et al., 2007; Nikoh et al., 2007). Interestingly, these pseudogenes on the host genome had numerous variants that likely derived from a single ancestral pseudogene. This implies that the laterally transferred Nucleococcus genome fragments have been multiplied within the host genome and that each copy has undergone independent molecular evolution. The differences in branch length and diversity between the pseudogenes of the two Nucleococcus species possibly indicate the difference in the age of the LGT events (Supplementary Figure S2). Although it is not known whether there are verrucomicrobia-derived genes still functional in the host genome, it is conceivable that these intranuclear symbionts can be sources of laterally transferred genes to the host genome more easily than cytoplasmic symbionts or ingested microbes. In the pea aphid Acyrthosiphon pisum, it has been hypothesized that genes laterally transferred from Wolbachia to the host genome might be involved in the symbiosis with the primary intracellular symbiont Buchnera (Nikoh and Nakabachi, 2009). Considering the widespread infection of the verrucomicrobial intranuclear symbionts, they might have a non-negligible impact on the evolution of the termite gut protists and also on the intracellular symbiosis with other bacteria, through the lateral transfer of genome fragments and their multiplication within the host genome.
Accession codes
References
Altschul SF, Madssen TL, Schaffer AA, Zhang J, Zhang Z, Miller W et al. (1997). Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucl Acids Res 25: 3389–3402.
Alverca E, Biegala IC, Kennaway GM, Lewis J, Franca S . (2002). In situ identification and localization of bacteria associated with Gyrodinium instriatum (Gymnodiniales, Dinophyceae) by electron and confocal microscopy. Eur J Phycol 37: 523–530.
Amann RI, Binder BJ, Olson RJ, Chisholm SW, Devereux R, Stahl DA . (1990). Combination of 16S rRNA-targeted oligonucleotide probes with flow cytometry for analyzing mixed microbial populations. Appl Environ Microbiol 56: 1919–1925.
Arneodo JD, Bressan A, Lherminier J, Michel J, Boudon-Padieu E . (2008). Ultrastructural detection of an unusual intranuclear bacterium in Pentastiridius leporinus (Hemiptera: Cixiidae). J Invert Pathol 97: 310–313.
Boscaro V, Fokin SI, Schrallhammer M, Schweikert M, Petroni G . (2013). Revised systematics of Holospora-like bacteria and characterization of "Candidatus Gortzia infectiva", a novel macronuclear symbiont of Paramecium jenningsi. Microb Ecol 65: 255–267.
Brugerolle G, Bordereau C . (2005). Pachyjoenia howa, a new symbiotic parabasalid joeniid flagellate of the termite Postelectrotermes howa. Eur J Protistol 41: 7–17.
Brugerolle G, Bordereau C . (2006). Immunological and ultrastructural characterization of spirotrichonymphid flagellates from Reticulitermes grassei and R. flavipes (syn. R. santonensis), with special reference to Spirotrichonympha, Spironympha and Microjoenia. Org Divers Evol 6: 109–123.
Brugerolle G, Radek R . (2006). Symbiotic protozoa of termites. In: König H, Varma A, (eds). Soil Biology. Springer-Verlag: Heidelberg, pp 243–269.
Brune A, Ohkuma M . (2011). Role of the termite gut microbiota in symbiotic digestion. In: Bignell DE, Roisin Y, Lo N, (eds). Biology of Termites: a Modern Synthesis. Springer: New York, pp 413–438.
d'Ambrosio U, Dolan M, Wier AM, Margulis L . (1999). Devescovinid trichomonad with axostyle-based rotary motor ("Rubberneckia"): taxonomic assignment as Caduceia versatilis sp. nov. Eur J Protistol 35: 327–337.
Daims H, Bruhl A, Amann R, Schleifer KH, Wagner M . (1999). The domain-specific probe EUB338 is insufficient for the detection of all Bacteria: development and evaluation of a more comprehensive probe set. Syst Appl Microbiol 22: 434–444.
Dangeard PA . (1902). Sur le caryophyseme des Eugleniens. C R Acad Sci 134: 1365–1366.
Desai MS, Strassert JFH, Meuser K, Hertel H, Ikeda-Ohtsubo W, Radek R et al. (2010). Strict cospeciation of devescovinid flagellates and Bacteroidales ectosymbionts in the gut of dry-wood termites (Kalotermitidae). Environ Microbiol 12: 2120–2132.
Desai MS, Brune A . (2012). Bacteroidales ectosymbionts of gut flagellates shape the nitrogen-fixing community in dry-wood termites. ISME J 6: 1302–1313.
Dolan M, Wier AM, Melnitsky H, Whiteside JH, Margulis L . (2004). Cysts and symbionts of Staurojoenina assimilis Kirby from Neotermes. Eur J Protistol 40: 257–264.
Fokin SI . (2004). Bacterial endocytobionts of ciliophora and their interactions with the host cell. Int Rev Cytol 236: 181–249.
Fuchs BM, Glockner FO, Wulf J, Amann R . (2000). Unlabeled helper oligonucleotides increase the in situ accessibility to 16S rRNA of fluorescently labeled oligonucleotide probes. Appl Environ Microbiol 66: 3603–3607.
Fujishima M, Kodama Y . (2012). Endosymbionts in Paramecium. Eur J Protistol 48: 124–137.
Görtz HD . (1986). Endonucleobiosis in ciliates. Int Rev Cytol 102: 169–213.
Hongoh Y, Ohkuma M, Kudo T . (2003). Molecular analysis of bacterial microbiota in the gut of the termite Reticulitermes speratus (Isoptera; Rhinotermitidae). FEMS Microbiol Ecol 44: 231–242.
Hongoh Y, Deevong P, Inoue T, Moriya S, Trakulnaleamsai S, Ohkuma M et al. (2005). Intra- and interspecific comparisons of bacterial diversity and community structure support coevolution of gut microbiota and termite host. Appl Environ Microbiol 71: 6590–6599.
Hongoh Y, Sato T, Dolan MF, Noda S, Ui S, Kudo T et al. (2007a). The motility symbiont of the termite gut flagellate Caduceia versatilis is a member of the "Synergistes" group. Appl Environ Microbiol 73: 6270–6276.
Hongoh Y, Sato T, Noda S, Ui S, Kudo T, Ohkuma M . (2007b). Candidatus Symbiothrix dinenymphae: bristle-like Bacteroidales ectosymbionts of termite gut protists. Environ Microbiol 9: 2631–2635.
Hongoh Y, Sharma VK, Prakash T, Noda S, Taylor TD, Kudo T et al. (2008a). Complete genome of the uncultured Termite Group 1 bacteria in a single host protist cell. Proc Natl Acad Sci USA 105: 5555–5560.
Hongoh Y, Sharma VK, Prakash T, Noda S, Toh H, Taylor TD et al. (2008b). Genome of an endosymbiont coupling N2 fixation to cellulolysis within protist cells in termite gut. Science 322: 1108–1109.
Hongoh Y . (2010). Diversity and genomes of uncultured microbial symbionts in the termite gut. Biosci Biotechnol Biochem 74: 1145–1151.
Hongoh Y . (2011). Toward the functional analysis of uncultivable, symbiotic microorganisms in the termite gut. Cell Mol Life Sci 68: 1311–1325.
Hongoh Y, Ohkuma M . (2011). Termite gut flagellates and their methanogenic and eubacterial symbionts. In: Hackstein JHP, (eds). Microbiology Monographs: (Endo)symbiotic Methanogenic Archaea. Springer-Verlag: Berlin and Heidelberg, pp 55–79.
Hotopp JC, Clark ME, Oliveira DC, Foster JM, Fischer P, Torres MC et al. (2007). Widespread lateral gene transfer from intracellular bacteria to multicellular eukaryotes. Science 317: 1753–1756.
Hugenholtz P, Goebel BM, Pace NR . (1998). Impact of culture-independent studies on the emerging phylogenetic view of bacterial diversity. J Bacteriol 180: 4765–4774.
Ikeda-Ohtsubo W, Faivre N, Brune A . (2010). Putatively free-living ‘Endomicrobia’ – ancestors of the intracellular symbionts of termite gut flagellates? Environ Microbiol Rep 2: 554–559.
Isanapong J, Sealy Hambright W, Willis AG, Boonmee A, Callister SJ, Burnum KE et al. (2013). Development of an ecophysiological model for Diplosphaera colotermitum TAV2, a termite hindgut Verrucomicrobium. ISME J 7: 1803–1813.
Kirby H . (1944). The structural characteristics and nuclear parasites of some species of Trichonympha in termites. Univ Calif (Berkeley) Publ Zool 49: 185–282.
Lee KC, Webb RI, Janssen PH, Sangwan P, Romeo T, Staley JT et al. (2009). Phylum Verrucomicrobia representatives share a compartmentalized cell plan with members of bacterial phylum Planctomycetes. BMC Microbiol 9: e5.
Loy A, Arnold R, Tischler P, Rattei T, Wagner M, Horn M . (2008). probeCheck—a; central resource for evaluating oligonucleotide probe coverage and specificity. Environ Microbiol 10: 2894–2898.
Ludwig W, Strunk O, Westram R, Richter L, Meier H, Yadhukumar et al. (2004). ARB: a software environment for sequence data. Nucl Acids Res 32: 1363–1371.
Maaß A, Radek R . (2006). The gut flagellate community of the termite Neotermes cubanus with special reference to Staurojoenina and Trichocovina hrdyi nov. gen. nov. sp. Eur J Protistol 42: 125–141.
Markham NR, Zuker M . (2005). DINAMelt web server for nucleic acid melting prediction. Nucl Acids Res 33: W577–W581.
Martinez-Garcia M, Brazel DM, Swan BK, Arnosti C, Chain PS, Reitenga KG et al. (2012). Capturing single cell genomes of active polysaccharide degraders: an unexpected contribution of Verrucomicrobia. PLoS One 7: e35314.
Müller J . (1856). [Einige Beobachtungen an Infusorien]. Monasber Königl Preuß Akad Wissensch Berlin, 389–393.
Nakajima H, Hongoh Y, Usami R, Kudo T, Ohkuma M . (2005). Spatial distribution of bacterial phylotypes in the gut of the termite Reticulitermes speratus and the bacterial community colonizing the gut epithelium. FEMS Microbiol Ecol 54: 247–255.
Nikoh N, Tanaka K, Shibata F, Kondo N, Hizume M, Shimada M et al. (2007). Wolbachia genome integrated in an insect chromosome: evolution and fate of laterally transferred endosymbiont genes. Genome Res 18: 272–280.
Nikoh N, Nakabachi A . (2009). Aphids acquired symbiotic genes via lateral gene transfer. BMC Biol 7: e12.
Noda S, Inoue T, Hongoh Y, Kawai M, Nalepa CA, Vongkaluang C et al. (2006). Identification and characterization of ectosymbionts of distinct lineages in Bacteroidales attached to flagellated protists in the gut of termites and a wood-feeding cockroach. Environ Microbiol 8: 11–20.
Ohkuma M, Sato T, Noda S, Ui S, Kudo T, Hongoh Y . (2007). The candidate phylum 'Termite Group 1' of bacteria: phylogenetic diversity, distribution, and endosymbiont members of various gut flagellated protists. FEMS Microbiol Ecol 60: 467–476.
Petroni G, Spring S, Schleifer KH, Verni F, Rosati G . (2000). Defensive extrusive ectosymbionts of Euplotidium (Ciliophora) that contain microtubule-like structures are bacteria related to Verrucomicrobia. Proc Natl Acad Sci USA 97: 1813–1817.
Radek R, Hausmann K, Breunig A . (1992). Ectobiotic and endocytobiotic bacteria associated with the termite flagellate Joenia annectens. Acta Protozool 31: 93–107.
Radek R . (1994). Monocercomonoides termitis n. sp., an Oxymonad from the lower termite Kalotermes sinaicus. Archiv Protistenk 144: 373–382.
Radek R, Hausmann K . (1994). Placojoenia sinaica n.g., n. sp., a symbiotic flagellate from the termite Kalotermes sinaicus. Eur J Protistol 30: 25–37.
Radek R . (1997). Spirotrichonympha minor n. sp., a new hypermastigote termite flagellate. Eur J Protistol 33: 360–374.
Rösel J, Radek R, Hausmann K . (1996). Ultrastructure of the trichomonad flagellate Stephanonympha nelumbium. J Euk Microbiol 43: 505–511.
Sato T, Hongoh Y, Noda S, Hattori S, Ui S, Ohkuma M . (2009). Candidatus Desulfovibrio trichonymphae, a novel intracellular symbiont of the flagellate Trichonympha agilis in termite gut. Environ Microbiol 11: 1007–1015.
Schauer C, Thompson CL, Brune A . (2012). The bacterial community in the gut of the cockroach Shelfordella lateralis reflects the close evolutionary relatedness of cockroaches and termites. Appl Environ Microbiol 78: 2758–2767.
Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB et al. (2009). Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol 75: 7537–7541.
Stamatakis A . (2006). RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22: 2688–2690.
Stevenson BS, Eichorst SA, Wertz JT, Schmidt TM, Breznak JA . (2004). New strategies for cultivation and detection of previously uncultured microbes. Appl Environ Microbiol 70: 4748–4755.
Stingl U, Radek R, Yang H, Brune A . (2005). "Endomicrobia": cytoplasmic symbionts of termite gut protozoa form a separate phylum of prokaryotes. Appl Environ Microbiol 71: 1473–1479.
Strassert JFH, Köhler T, Wienemann THG, Ikeda-Ohtsubo W, Faivre N, Franckenberg S et al. (2012). 'Candidatus Ancillula trichonymphae', a novel lineage of endosymbiotic Actinobacteria in termite gut flagellates of the genus. Trichonympha. Environ Microbiol 14: 3259–3270.
Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S . (2011). MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28: 2731–2739.
Trager W . (1934). The cultivation of a cellulose-digesting flagellate, Trichomonas termopsidis, and of certain other termite protozoa. Biol Bull 66: 182–190.
Vandekerckhove TT, Coomans A, Cornelis K, Baert P, Gillis M . (2002). Use of the Verrucomicrobia-specific probe EUB338-III and fluorescent in situ hybridization for detection of "Candidatus Xiphinematobacter" cells in nematode hosts. Appl Environ Microbiol 68: 3121–3125.
Wagner M, Horn M . (2006). The Plantomycetes, Verrucomicrobia, Chlamydiae and sister phyla comprise a superphylum with biotechnological and medical relevance. Curr Opin Biotechnol 17: 241–249.
Wertz JT, Kim E, Breznak JA, Schmidt TM, Rodrigues JL . (2012). Genomic and physiological characterization of the Verrucomicrobia isolate Diplosphaera colitermitum gen. nov., sp. nov., reveals microaerophily and nitrogen fixation genes. Appl Environ Microbiol 78: 1544–1555.
Yang H, Schmitt-Wagner D, Stingl U, Brune A . (2005). Niche heterogeneity determines bacterial community structure in the termite gut (Reticulitermes santonensis). Environ Microbiol 7: 916–932.
Zielinski FU, Pernthaler A, Duperron S, Raggi L, Giere O, Borowski C et al. (2009). Widespread occurrence of an intranuclear bacterial parasite in vent and seep bathymodiolin mussels. Environ Microbiol 11: 1150–1167.
Acknowledgements
We thank K Seki, T Mabuchi, J Inoue, M Yuki and W Ohnuma for assisting with the experiments. We also thank R Sasajima and A Martinez for translation of some reference papers. Sequencing was done in RIKEN BSI and the Biomaterial Analysis Center in the Tokyo Institute of Technology. This study was financially supported by the NEXT program to YH from JSPS, research funding to YH from IFO, and MEXT KAKENHI to YH (18687002 and 90392117) and MO (23117003).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies this paper on The ISME Journal website
Supplementary information
Rights and permissions
About this article
Cite this article
Sato, T., Kuwahara, H., Fujita, K. et al. Intranuclear verrucomicrobial symbionts and evidence of lateral gene transfer to the host protist in the termite gut. ISME J 8, 1008–1019 (2014). https://doi.org/10.1038/ismej.2013.222
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ismej.2013.222
Keywords
This article is cited by
-
Fungal phytopathogen modulates plant and insect responses to promote its dissemination
The ISME Journal (2021)
-
Parallel reductive genome evolution in Desulfovibrio ectosymbionts independently acquired by Trichonympha protists in the termite gut
The ISME Journal (2020)
-
Genome analyses of uncultured TG2/ZB3 bacteria in ‘Margulisbacteria’ specifically attached to ectosymbiotic spirochetes of protists in the termite gut
The ISME Journal (2019)
-
Genome of ‘Ca. Desulfovibrio trichonymphae’, an H2-oxidizing bacterium in a tripartite symbiotic system within a protist cell in the termite gut
The ISME Journal (2017)