Abstract
The etiology of dental caries remains elusive because of our limited understanding of the complex oral microbiomes. The current methodologies have been limited by insufficient depth and breadth of microbial sampling, paucity of data for diseased hosts particularly at the population level, inconsistency of sampled sites and the inability to distinguish the underlying microbial factors. By cross-validating 16S rRNA gene amplicon-based and whole-genome-based deep-sequencing technologies, we report the most in-depth, comprehensive and collaborated view to date of the adult saliva microbiomes in pilot populations of 19 caries-active and 26 healthy human hosts. We found that: first, saliva microbiomes in human population were featured by a vast phylogenetic diversity yet a minimal organismal core; second, caries microbiomes were significantly more variable in community structure whereas the healthy ones were relatively conserved; third, abundance changes of certain taxa such as overabundance of Prevotella Genus distinguished caries microbiota from healthy ones, and furthermore, caries-active and normal individuals carried different arrays of Prevotella species; and finally, no ‘caries-specific’ operational taxonomic units (OTUs) were detected, yet 147 OTUs were ‘caries associated’, that is, differentially distributed yet present in both healthy and caries-active populations. These findings underscored the necessity of species- and strain-level resolution for caries prognosis, and were consistent with the ecological hypothesis where the shifts in community structure, instead of the presence or absence of particular groups of microbes, underlie the cariogenesis.
Similar content being viewed by others
Introduction
Dental caries has been a worldwide health concern inflicting humans of all ages from both developed and developing countries. For example, past or present coronal caries inflict 93.8% of US adult population (45.3% in children) (Shivakumar et al., 2009) and 88.1% of adults (66.0% in children) in China (Qi, 2008). Moreover, because of increased intakes of highly refined sugars, the incidences are still growing in many countries.
Caries activity causes lesions and cavities on tooth surfaces, leading to decay and even loss of tooth structure and resulting in infection and pain. Once started, the destruction process could be difficult to monitor, which in a longer perspective can lead to a state of irreversible cavities in the inflicted tooth, and the only therapeutic options are mechanical or surgical. Therefore, preventive measures against caries, as well as the prognosis and early diagnosis, are of particular clinical significance.
Although the environmental conditions supporting the development of dental caries are complex, the medical consensus is that there is a microbial factor, in that acids from microbial fermentation of diet carbohydrates result in an imbalance of the demineralization and remineralization processes on the enamel (Russell, 2009). However, there has been a long history of debate on the etiology of dental caries (Marsh, 1994; Russell, 2009). The ‘Nonspecific Plaque Hypothesis’ maintained that caries resulted from the overall acid-secreting activity of the total plaque microflora (Theilade, 1986; Marsh, 2006b). The ‘Specific Plaque Hypothesis’ instead pinpointed the specific member like Streptococcus mutans as the etiological agents (Loesche, 1986; Marsh, 2006b), thus proposing controlling disease by targeting against a defined and limited number of microorganisms. Their shared organismal features likely to be important in cariogenesis included sucrose-dependent biofilm formation, relatively high aciduricity and potent acidogenesis (Loesche, 1986). However, the above characteristics were later found to vary among S. mutans isolates; in addition, acid production was not dependent upon the presence of S. mutans (Beighton, 2005). More importantly, these microbes had traditionally been identified by culture-based methods, which excluded numerable not-yet-cultivated species. Therefore, the essential role of S. mutans in the caries process has not yet been proven. More recently, ‘Ecological Plaque Hypothesis’ (Marsh, 1994, 2003, 2006a) emerged and proposed that caries resulted from a populational balance shift of the resident microbiota, driven by environmental changes in oral conditions. Culture-independent methods for bacterial identification and enumeration such as array-based DNA hybridization (Corby et al., 2005; Aas et al., 2008; Kanasi et al., 2010) or Sanger sequencing of 16S rRNA gene clones (Aas et al., 2005) have revealed a wide array of bacteria correlated with caries progression.
In this study, we report the most in-depth, comprehensive and collaborated view to date of the organismal lineage of oral microbiota in both caries-active and normal human populations. Our results from 16S rRNA gene amplicon-based sequencing were cross-validated by whole-genome-based deep-sequencing technologies. Saliva, the most accessible and host-friendly sampling strategy for oral microbiota, was chosen as the sampling site in this study for its potential for a swift and flexible (both remote and onsite) screen in the clinics for distinguishing caries-active hosts and monitoring preventive measures. These efforts have enabled us to define and compare the individual as well as populational features of the normal and caries-active microbiomes, test specific correlations between features/components of saliva microbiome and the host disease states and identify the ‘behind-the-scene’ biotic or abiotic factors underlying dental caries.
Materials and methods
Study design
All human host volunteers (nearly 700 individuals) were from an oral health census on the undergraduates from the east campus of Sun Yat-sen University, Guangzhou, China, in September, 2009. After oral health survey, 26 ‘healthy’ individuals and 19 ‘caries-active’ (see definition below) subjects were chosen for saliva sample collection. All were made aware of the nature of the experiment and given written informed consent in accordance with the sampling protocol with approval of the ethical committee of the Stomatology Hospital, Sun Yat-sen University. They were all unrelated individuals of both genders, aged between 18 and 22 years and shared a relatively homogeneous college-campus living environment. All reported no antibiotics intake for the preceding at least 6 months. All were asked to avoid eating or drinking for 1 h before oral sampling. Those with other oral (for example, periodontitis or halitosis) or systematic diseases were excluded.
Dental examinations were performed by five professional dentists who were previously trained and calibrated for the evaluation and sampling procedures, according to the criteria defined by the National Institute of Dental and Craniofacial Research (NIDCR; USA) for caries diagnosis and recording (Kaste et al., 1996). The DMFT index measures the number of decayed, missing and filled teeth in epidemiologic surveys of dental caries (Anaise, 1984). It was adopted in our study to measure each individual’s caries status and thus to define and distinguish the caries-active subjects and healthy subjects. The intraexaminer reproducibility in both the pilot phase and the main survey was assessed by κ statistics, which was >0.91. In the end, the ‘caries-active’ pilot population consisted of 19 caries-active hosts (DMFT ⩾6; 10 males and 9 females), whereas the ‘healthy’ pilot population included 26 subjects (DMFT=0; 12 males and 14 females).
For details in sample collection and processing and PCR amplification of microbial 16S rRNA genes, please refer to Supplementary Text online.
Highly paralleled DNA sequencing
The 16S rRNA gene PCR amplicons were analyzed via pyrosequencing on 454 Life Sciences Genome Sequencer FLX Titanium (GS-Titanium; 454 Life Sciences, Branford, CT, USA) where on average 400-bp-long reads were produced (Supplementary Figure S1).
In order to assess the accuracy of the 16S rRNA gene PCR amplicon-based measurement of microbial diversity and community structure, for two of the saliva samples, shotgun pair-end libraries of total saliva genomic DNA were prepared (one from the healthy population and the other from the caries-active population). Each metagenomic DNA library was then sequenced on one lane of pair-end 100 bp flow cell on Solexa GA-IIx (Illumina, San Diego, CA, USA).
For details on the experimental assessment of potential biases associated with barcodes and computational and statistical analyses of all 16S rRNA gene and metagenomic data, please refer to Supplementary Text online.
Results
Saliva microbiomes in human population were featured, in the organismal lineage, by a vast diversity and a minimal core
Employing deep sequencing of 16S rRNA gene amplicons, we defined the bacterial diversity of 45 saliva microbiomes in the pilot populations of 19 caries-active and 26 healthy human hosts (all aged between 18 and 22 years), with an average read length of 400 bp (Supplementary Figure S1). A total of 425 623 post-trimming 16S rRNA gene reads were obtained, with reads per microbiome numbered 9458 on average (Supplementary Table S1).
The species-level operational taxonomic units (OTUs) and species richness and diversity estimates were obtained for each microbiome (Supplementary Table S2). Clustering the unique sequences into OTUs at a 3% genetic distance resulted in 600–4200 different species-level OTUs per microbiome. Estimates of the richness of total bacterial communities ranged between 1500 and 6800 phylotypes in each saliva microbiota. Saliva samples in the two host groups exhibited a similar level of diversity (P>0.1, for both Shannon Index and Simpson Index).
Species richness of individual and total saliva bacterial communities was estimated by rarefaction analysis. The resulted rarefaction curves (Figure 1a) indicated that microbial richness of the sampled saliva was not yet complete even at the current sequencing depth and additional sequencing coverage was needed for satiating the microbial diversity in saliva. Comparisons of the rarefaction curves in the healthy and caries-active populations revealed that the two groups displayed similar richness of bacterial OTUs at 97% identity level (Figure 1b).
Phylogenetic diversity of the saliva microbiomes among individuals and between the healthy (H) and caries-active (C) pilot populations. (a) The number of OTUs in each sample. (b) Rarefaction curves for healthy and caries-active populations. A given number of sequences (1000 or 5000) were randomly sampled from each data set. The average number of OTUs in each sample was then calculated (mean±s.e.m. shown). Samples from the two host populations displayed similar phylogenetic diversity at 97% identity level. (c) Shared and distinct OTUs among individuals in the healthy and caries-active individuals. The x axis is the number of samples included in the data set. The y axis is the number of OTUs, representing the means of 100 iterations. Error bars represent s.d. Among healthy hosts, 0.53% of the total OTUs were shared (0.08% were shared among caries-active individuals).
To examine the level of microbial organismal conservation among human hosts, we tested whether there was a ‘core’ saliva microbiome shared across the host individuals. The degrees of OTU sharing and unsharing were determined, respectively, for both healthy and caries-active populations. The unshared-OTU curves suggested that each additional sample from either group contributed more diversity to the pool (Figure 1c). The shared-OTU curves revealed a gradual decrease of OTU sharing with the individual addition of hosts (Figure 1c). Furthermore, among the total of 5123 OTUs in the healthy population, only 27 (0.53%) OTUs were present across all samples. For the caries-active population, the shared OTUs account for merely 0.08%. Therefore, in either population, there was a minimal degree of sharing organismal lineages (as defined by species-level OTUs) among individual saliva microbiomes.
Caries microbiomes were more variable in community structure whereas the healthy ones were more similar to each other
The extreme divergence of species-level microbial lineages among the saliva microbiomes did not necessarily exclude conservation in the structure of the residential microbial communities. We next compared the community structures, both within and between caries-active and healthy microbiotas. We used FastUniFrac (Hamady et al., 2010) for pairwise comparisons on the distances between two microbiotas in terms of the fraction of evolutionary history that separates the organisms; the phylogeny-based metrics thus enabled assessing the degree of conservation among the bacterial community structures. The results suggested that the degree of variations among healthy individuals was significantly less than that among caries-active hosts (P<0.01; Figures 2a and b). Therefore, caries microbiomes were significantly more variable whereas the healthy ones were relatively conserved in terms of microbial community structure, despite the extreme divergence of species-level microbial lineages in both host populations.
Community structures of healthy (H) and caries-active (C) microbiomes. (a) Pair-wise UniFrac distances within and across the host populations. Colors indicate the degree of similarity in bacterial-community structure. (b) Hosts from the healthy population harbored a more similar bacterial-community structure than those from the caries-active population (***P<0.01).
Furthermore, we employed the FastUniFrac-derived distance matrices to cluster the microbiomes for dimensionality reduction with principal coordinates analysis (Lozupone and Knight, 2005). This analysis is a multivariate statistical technique for finding the most important dimension(s) along which samples vary. The principal components (PCs), in descending order, describe the degree of variations each of the dimensions accounts for. Although not observed on PC1, the separation between caries-active and healthy populations on PC2 (second principal component) and PC3 (third principal component) appeared to be possible (Supplementary Figure S3). Student’s t-test on the PC2 and PC3 elicited significant difference between the two host populations (P<0.01).
Overpopulation of Prevotella distinguished caries microbiotas from healthy ones
Taxonomy analysis and comparison between the two host populations
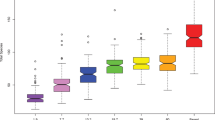
To phylogenetically identify the microbial lineages in the saliva microbiomes, bacterial taxa were assigned to the 16S rRNA gene reads based on searches against Human Oral Microbiome Database (HOMD Version 10.1; http://www.homd.org/) and RDP 16S rRNA gene database (a curated 16S database alternative to HOMD; http://rdp.cme.msu.edu/). Over 80% of the bacterial diversity in each individual was contributed by the following six phyla: Bacteroidetes, Firmicutes, Proteobacteria, Fusobacteria, Actinobacteria and Spirochaetes (Supplementary Figure S4). A total of 148 genera were identified. The most frequently detected taxa in each level were shown in Figure 3 (only those above 1% in relative abundance were listed). Among all taxa (including both abundant and rare ones), there was no ‘gender-specific’ (present in one gender but absent in the other) or ‘gender-associated’ (differentially distributed yet present in both gender) taxa detected, from the Phylum level down to the Genus level (data not shown). No ‘caries-specific’ taxa (present in the healthy or caries-active population but absent in the other) was detected either; however, we were able to pinpoint ‘caries-associated’ (differentially distributed in caries-active microbiomes yet present in both populations; including the two circumstances of ‘caries-enriched’ and ‘caries-depleted’) taxa at each of those five levels. These included Bacteroidetes (P=0.062; caries enriched) at Phylum, Bacteroides (P=0.081; caries-enriched) at Class, Bacteroidales (P=0.081; caries-enriched) at Order and Prevotellaceae (P=0.038; caries-enriched) at Family; at the level of Genus, Prevotella (P=0.038) was pinpointed (Figure 3 and Supplementary Figure S5).
Comparisons of bacterial taxonomy profiles of the caries-active (C) and healthy (H) host populations based on HOMD. The taxonomy assignment was based on HOMD Database. Comparisons were performed at each of the taxonomical levels of Phylum, Class, Order, Family and Genus. The most frequently detected taxa (above 1% of relative abundance) in each level are shown. Means of the relative abundance for each taxon at each taxonomical level between the healthy and caries-active host-populations are compared (*P<0.1; **P<0.05; mean±s.e.m.).
Caries and normal populations carried different arrays of Prevotella species
Prevotella was found at relatively high abundance in both healthy and caries-active hosts. To further test whether the Prevotella members from two host populations were similar or distinguishable, 16S rRNA gene sequences in each sample were blasted against the curated HOMD 16S rRNA Gene Database (http://www.homd.org/) to identify those associated with Prevotella. Arrays of Prevotella species that underpinned the Prevotella community structures in saliva were then compared between the two host populations (Figure 4). The results suggested that Prevotella species were not similarly distributed between the two host populations; therefore, species-level and potentially even strain-level resolution might be important for caries prognosis.
Caries-associated OTUs
As the above comparisons on the relative abundance of microbial taxa were derived from only those reads with RDP confidence value (Cole et al., 2009) above 0.8, essentially all those reads without any reliable phylogenetic assignments (4.7–40% of the total reads in each sample) had been masked. Therefore, we further compared, between the two pilot populations, the relative abundance of all species-level OTUs, independently of their phylogenetic identity assignments. In all, 147 OTUs were found differentially distributed at the significance cutoff (P-value) of 0.1, and together accounted for 1.2% of the total OTUs. Interestingly, among those 147 OTUs, 113 (76.9%) of them were caries depleted and the remaining 34 (23.1%) were caries enriched (Supplementary Table S3). Phylogenetic identities of the OTUs were interrogated by BLASTN searches against HOMD 16S rRNA Gene Database (E<10−5). The results showed that they exhibited a wide phylogenetic distribution, from six phyla and across 34 species (Supplementary Table S3). Most (93.8%) were found in four phyla: Bacteroidetes (31.3%), Firmicutes (32.7%), Fusobacteria (12.9%) and Proteobacteria (17.0%; Supplementary Table S4). These caries-depleted or caries-enriched OTUs represented a novel set of potential organismal markers for evaluation and prognosis of adult caries, although the validity and biological significance of these caries-associated OTUs remained to be tested in larger human populations and via appropriate animal models.
Validation of the measured microbiomic diversity and structures via whole-metagenome deep sequencing
In order to assess the accuracy of microbial community diversity and structure of saliva reconstructed from the PCR-based 16S rRNA gene sequences, two saliva genomic DNA samples, one randomly selected from the healthy group (H114; Supplementary Table S2) and the other randomly chosen from caries-active group (C201; Supplementary Table S2), were deep sequenced with pair-end 100 bp reads on Solexa GA-IIx. After removing the contaminating reads from human hosts (69.1% and 46.8% of all produced reads, respectively), over 19 million reads totaling 1.9 Gb were produced for the healthy saliva microbiome, and over 33 million reads totaling 3.3 Gb were generated for the caries-active microbiome. All of the Solexa reads were mapped against the 44 oral reference genomes in Human Microbiome Project (HMP; Mavromatis et al., 2009; Markowitz et al., 2010) to assess the coverage and abundance of these sequenced isolates or their close neighbors in saliva microbiota. The number of reads recruited by the 44 oral reference genomes represented merely 0.42% and 2.05% of the total non-human reads for the two sequenced samples (H114 and C201) respectively. A likely cause for such low mapping rates was that HMP reference genome set represented only a very small portion of human oral microbiomic sequence space. Consistently, for the MG-RAST annotation result, no more than 2% of the genes could be annotated. The top 20 abundant genes in each sample were listed: nearly half of the top 20 genes were shared between the two different samples (Supplementary Table S5).
Interestingly, in the caries-active host (C201), both the ‘genome hit’ (the percentage of reads recruited by the seven available Prevotella-isolate genomes among all reads) and the ‘genome coverage’ for the sequenced Prevotella outnumbered those from the healthy host (H115) (Supplementary Figure S6). This finding was consistent with the elevated abundance of Prevotella as revealed via the 16S-rRNA gene amplicon-based survey.
Furthermore, we employed PHYLOSHOP (Shah et al., 2011) to predict and enumerate 16S rRNA gene fragments in the two microbial metagenomes; the results thus enabled phylogenetically reconstructing the saliva microbiomes independently from the 16S rRNA gene amplicon-based approach. The resulting relative abundance of Prevotella from the caries-active host (C201) outnumbered that from the healthy host (H115), which was also consistent with the elevated abundance of Prevotella revealed by the 16S-rRNA gene amplicon sequencing result. Relative abundances of top 20 genera from the two Solexa sequencing-based metagenomes were compared with those originated from GS-Titanium 16S rRNA gene reads for the same samples. The result demonstrated significant correlation for the samples (H114: r=0.825, P<0.01; C201: r=0.776, P<0.01). Therefore, the microbiomic diversity and structures elucidated by the two independent sequencing strategies were largely consistent.
Discussion
Elucidation and monitoring of the diversity, dynamics and epidemiological features of microbial communities require reliable, sensitive and unbiased microbe-surveying techniques. Pyrosequencing of 16S rRNA gene amplicons provides sensitive detection and discrimination (that is, single-base resolution) for a wide phylogeny, and thus quantifies the relative abundances of bacterial components and the structural variations of communities (Hamady and Knight, 2009). However, as the nine hypervariable regions (V1–V9) exhibit considerable and differential sequence diversity among different bacteria, the accuracy and reliability of such approaches are still being tested and improved by cautious selection of the sequenced regions. It was suggested that fragments of at least 250 bases starting from one of the primers, including R357, R534, R798, F343 and F517, produced accurate results on analysis of microbial community composition (Liu et al., 2008). It was also reported that fragments encompassing V4, V5/V6 and V6/V7 provided results comparable with the full length (Youssef et al., 2009), yet V3/V4 and V4/V5 regions yield the highest accuracies of phylogenetic assignment (Claesson et al., 2010). In our study, the sequenced 16S rRNA gene amplicons were based on 16S rRNA hypervariable regions of V4/V5 (Escherichia coli positions 515F-907R).
In addition, 16S rRNA gene amplification-based surveys of metagenomes could be confounded by several artifacts or challenges, including chimeric sequences caused by PCR amplification, sequencing errors, unequal amplification of community members and the typically unknown variations in the rRNA-gene copy numbers among different residents (Ashelford et al., 2005; Liu et al., 2008; Petrosino et al., 2009; Yang et al., 2009; Xie et al., 2010). On the other hand, whole-metagenome shotgun deep sequencing circumvented some of the problems, as it avoided PCR amplication of 16S rRNA genes; however, the results could vary depending on DNA extraction and sequencing protocols; the typically shorter reads (for example, Solexa or Solid generates shorter but less costly reads than 454) as well as the general insufficiency of completely sequenced reference genomes could limit the resolution and accuracy of phylogenetic assignments. In our saliva microbiome study, we tested the accuracy and reliability of our 16S rRNA gene amplicon-based analyses by comparing the two sequencing strategies. The internal consistency between the results provided a solid foundation for both interindividual and interpopulation microbiomic comparisons and demonstrated an experimental approach for such microbiomic interrogations along the disease-progress stages or in expanded host populations.
In reporting the first comprehensive bacteria profiles of adult saliva microbiotas in a defined-age population, our findings revealed new and intriguing insights on the human saliva microbiome. First, a surprisingly high phylogenetic diversity was observed. Our study revealed 600–4200 species-level OTUs in each saliva sample, depending on the sequencing depth, yet the rarefaction curves suggested that bacterial richness of the saliva was not yet satiated and was in fact even higher. In order to compare our results with the two previous studies on saliva microbiome that were based on pooled samples (Keijser et al., 2008; Ling et al., 2010), we in silico pooled our samples and then performed OTU-based analysis. The results suggested that the two previous studies could have underestimated the actual bacterial diversity in saliva (data not shown). Furthermore, the saliva microbial diversity in our study as measured by the number of OTUs in each sample was far greater than one recent estimate for three healthy Caucasian male adults (Zaura et al., 2009). One difference could be found in the read-trimming process: their analysis only focused on sequences occurring at least five times. In our sequences, even with the total of 48 609 reads in one of the samples, those occurring over five times only accounted for 1.9% of total reads; if the trimming was based on that particular criterion, nearly 98% of the reads would have been regarded as sequencing errors and excluded from further analyses. Therefore, it is possible that the overly stringent selection had actually removed most of the rare members of the saliva microbiome and thus greatly underestimated the phylogenetic diversity.
Second, there was a minimal ‘core’ of saliva microbiomes in human adult populations, healthy or caries-active hosts alike. Among the 26 healthy and 19 caries-active hosts, only 0.53% and 0.08% species-level OTUs were shared, respectively. This finding contrasted with a recent report supporting the concept of a core microbiome, in which saliva microbiomes from three healthy Caucasian male adults shared 387 (47%) of 818 total OTUs (Zaura et al., 2009). The overly stringent read-selection criterion, as discussed above, could have led to this conclusion, where oral ecosystem could have been oversimplified by neglecting the ‘rare phylotypes’. In addition, our sampling size was much larger: 19 and 26 hosts, respectively, from the two pilot populations. Furthermore, a gradual decrease of OTU sharing with every individual addition of hosts was observed (Figure 1c). Thus, our data supported the absence of a species-level organismal ‘core’ of saliva microbiome among human hosts.
Third, Phylum-level memberships in the club of predominant saliva residents were largely constant among human saliva microbiomes, at least for the hosts sampled in our study. For each sample, different reference databases gave similar relative proportions of the six major phyla (that is, Firmicutes, Proteobacteria, Actinobacteria, Fusobacteria, Bacteroidetes and Spirochaetes). Among different hosts, these six major phyla constituted the club of predominant oral residents, consistent with previous 16S rRNA gene-based community profiling surveys (Keijser et al., 2008; Zaura et al., 2009; Ling et al., 2010). However, the relative abundance of the predominant phyla differed among human hosts. Despite the report that intrapersonal differences (over time) in human microbiomes were smaller than interpersonal differences and that the oral cavity communities were the least varied among all surveyed habitats in human body (Costello et al., 2009), the scope of host factors that are actually shaping oral microbiota diversity and community structure in healthy hosts was still not clear. Host race or location could be one of the factors. For example, Proteobacteria were more abundant in our data sets when in silico pooled (healthy Chinese adults), wherever Firmicutes were more abundant at a sample pooled from 91 healthy adult individuals in Amsterdam (Keijser et al., 2008). In fact, a large-scale survey for 12 worldwide locations suggested saliva microbiome contributed important information for human population history (Nasidze et al., 2009). Therefore, to reveal a comprehensive spectrum of the underlying host factors, our current sampling strategy on ‘healthy’ Chinese adults remained to be expanded to larger populations.
Besides revealing the saliva microbiota features at both host-individual and host-population levels for ‘healthy’ adults, our study was designed to characterize and compare the saliva microbiomes between caries-active and healthy hosts. There have been several reports investigating microbial factors underlying dental caries. However, they have been limited by a number of crucial factors. First, the depth and breadth of sampling for oral microbiota have been insufficient. It was limited to a few dozen sequences using clone-based methods (Aas et al., 2005) or only a partial, biased and a prior determined list of microorganisms in DNA chip (Corby et al., 2005; Aas et al., 2008; Kanasi et al., 2010). Second, most high-throughput sequencing-based surveys are on normal individuals (Keijser et al., 2008; Lazarevic et al., 2009; Zaura et al., 2009), and few studies have interrogated the oral microbiota associated with caries (Ling et al., 2010). Third, the sampling sites are variable. In terms of the plaque samples that most studies chose, there are at least four different sites: intact healthy surfaces, white spot lesions, surfaces of initial enamel lesions and surfaces of deep dentinal lesions. The inconsistency severely limited comparisons across studies and gravely delayed the translation to clinically useful diagnostic models. Fourth, most studies that enumerate 16S rRNA gene sequences have ignored potential PCR artifacts and variations in gene copy numbers among the oral residential genomes (Ashelford et al., 2005; Liu et al., 2008; Petrosino et al., 2009; Yang et al., 2009). All these factors have severely confounded efforts to pinpoint the etiology of dental caries.
In our study, these challenges were addressed via sampling saliva microbiomes in carefully controlled pilot caries-active and healthy populations using highly paralleled pyrosequencing of 16S rRNA genes. Our results unraveled striking yet crucial saliva microbiomic characteristics associated with adult dental caries. First of all, the caries-active and healthy populations displayed a similar level of phylogenetic diversity; however, distinctions were revealed between healthy and caries microbiomes in terms of microbial community structure: the caries microbiomes were significantly more variable whereas the healthy ones were relatively conserved. One previous report performing PCR-based denaturing gradient gel electrophoresis surveys (Li et al., 2005) observed a greater diversity of saliva bacterial populations in caries-free compared with caries-active individuals. Yet, PCR-based denaturing gradient gel electrophoresis suffered from a limited resolution as it depends on visual comparison and frequently subjective interpretations (Hayes et al., 1999; Muyzer, 1999); moreover, bacterial samples used in that study were obtained from a partially selective medium.
Second, our results argued against the existence of bacterial taxa that were specifically present in caries-active hosts but absent in healthy ones, and vice versa. However, we did observe strong, statistically supported shifts in abundance for several microbial groups between the healthy and caries-active host populations. For example, at the Phylum level, there was an enrichment of Bacteroidetes for caries-active hosts. Interestingly, in the intestine, relative depletion of Bacteroidetes Phylum members were found associated with adult obesity (Turnbaugh et al., 2009); whether there is a link between the observations at two distant neighborhoods of the human digestive tract remained to be investigated. At the Genus level, our findings were surprising in that instead of the commonly suspected cariogenic organisms such as mutans streptococci or lactobacilli, the caries-active saliva microbiotas harbored a statistically significant enrichment (∼10%) of Prevotella, regardless of gender. Prevotella are Gram-negative rod-shaped anaerobes and some of them were implicated in endodontic (Rocas and Siqueira, 2009) and periodontal (Serrano et al., 2009) infections; for example, Prevotella intermedius played a tentative role in gingivitis (Gursoy et al., 2009). However, the design and implementation of our saliva-sampling strategy had excluded those hosts with occurring periodontal diseases or other systematic diseases. Therefore, it should have been the caries state but not the periodontal state of the host that underlie the abundance shift of Prevotella.
Third, we have further shown that caries-active and healthy host populations carried different arrays of Prevotella species. Whether the observed changes in Prevotella-species memberships were the cause or the consequence of dental caries was unclear, as the physiological and metabolic diversity of Prevotella in particular, and of the plethora oral residents in general, were still ill-defined. One possibility was that the biotic or abiotic acidification of oral environment caused the shift in Prevotella memberships, as oral residents were respectively featured by optimal pH ranges and a sudden drop in pH could shift their intra-oral distribution markedly (McDermid et al., 1986, 1988). Thus, to conclusively pinpoint the roles of the various Prevotella species as well as the Prevotella group, animal models of caries (Clasen and Ogaard, 1999) would be essential.
Fourth, phylogeny assignment-independent comparison of relative abundances of OTUs between the healthy and caries-active populations yielded results consistent with the above findings, and furthermore pinpointed 147 caries-associated (both caries-enriched and caries-depleted) OTUs. Notably, these OTUs, affiliated to six phyla and in 34 different species, did not include previously suspected cariogenic bacteria such as Streptococcus or Lactobacillus. Instead, most of the OTUs were generally considered as symbionts in the human oral ecosystem. Thus, our findings were consistent with the ‘Ecological Plaque Hypothesis’ that suggested caries as a consequence of imbalances in the resident microflora. The 147 caries-associated OTUs likely exemplified such microbial-population balance shifts in the oral symbiome. On the other hand, these caries-associated OTUs represented not only promising candidates for investigating and tracking the cariogenesis process but also highly specific candidate biomarkers for caries prognosis.
A recent study on childhood caries reported no difference between caries-active and caries-free saliva microbiotas using PCR-based denaturing gradient gel electrophoresis and pyrosequencing (Ling et al., 2010). A number of factors precluded direct comparisons between the two studies. First, childhood caries and adult caries might differ in etiology (Becker et al., 2002; Li et al., 2005; Kanasi et al., 2010; Soncini et al., 2010). Second, the previous study targeted V3 variable region with an average read length of 145 bp and the average number of reads per sample at 1914; in contrast, results in our study were based on 400-bp-long reads on V4–V5 region and on average 9458 reads per sample. In fact, the discovery of bacterial diversity in a microbiota was strongly influenced by sequencing depth (Roesch et al., 2007).
In summary, our findings underscored the necessity of species-level resolution for caries prognosis, and were consistent with the Ecological Hypothesis. Our findings raised the possibility of exploiting salivary microbiomes as diagnostic marker or health barometers of caries and other oral or nonoral diseases. Our approach and findings could be extended to correlating microbiomic development with cariogenesis progression, and be tested at larger host populations. Rational validation of such findings on appropriate animal models could eventually lead to preventive or therapeutic strategies for caries-inflicted or high-risk individuals.
References
Aas JA, Griffen AL, Dardis SR, Lee AM, Olsen I, Dewhirst FE et al. (2008). Bacteria of dental caries in primary and permanent teeth in children and young adults. J Clin Microbiol 46: 1407–1417.
Aas JA, Paster BJ, Stokes LN, Olsen I, Dewhirst FE . (2005). Defining the normal bacterial flora of the oral cavity. J Clin Microbiol 43: 5721–5732.
Anaise JZ . (1984). Measurement of dental caries experience--modification of the DMFT index. Community Dent Oral Epidemiol 12: 43–46.
Ashelford KE, Chuzhanova NA, Fry JC, Jones AJ, Weightman AJ . (2005). At least 1 in 20 16S rRNA sequence records currently held in public repositories is estimated to contain substantial anomalies. Appl Environ Microbiol 71: 7724–7736.
Becker MR, Paster BJ, Leys EJ, Moeschberger ML, Kenyon SG, Galvin JL et al. (2002). Molecular analysis of bacterial species associated with childhood caries. J Clin Microbiol 40: 1001–1009.
Beighton D . (2005). The complex oral microflora of high-risk individuals and groups and its role in the caries process. Community Dent Oral Epidemiol 33: 248–255.
Claesson MJ, Wang Q, O’Sullivan O, Greene-Diniz R, Cole JR, Ross RP et al. (2010). Comparison of two next-generation sequencing technologies for resolving highly complex microbiota composition using tandem variable 16S rRNA gene regions. Nucleic Acids Res 38: e200.
Clasen AB, Ogaard B . (1999). Experimental intra-oral caries models in fluoride research. Acta Odontol Scand 57: 334–341.
Cole JR, Wang Q, Cardenas E, Fish J, Chai B, Farris RJ et al. (2009). The Ribosomal Database Project: improved alignments and new tools for rRNA analysis. Nucleic Acids Res 37: D141–D145.
Corby PM, Lyons-Weiler J, Bretz WA, Hart TC, Aas JA, Boumenna T et al. (2005). Microbial risk indicators of early childhood caries. J Clin Microbiol 43: 5753–5759.
Costello EK, Lauber CL, Hamady M, Fierer N, Gordon JI, Knight R . (2009). Bacterial community variation in human body habitats across space and time. Science 326: 1694–1697.
Gursoy M, Haraldsson G, Hyvonen M, Sorsa T, Pajukanta R, Kononen E . (2009). Does the frequency of Prevotella intermedia increase during pregnancy? Oral Microbiol Immunol 24: 299–303.
Hamady M, Knight R . (2009). Microbial community profiling for human microbiome projects: tools, techniques, and challenges. Genome Res 19: 1141–1152.
Hamady M, Lozupone C, Knight R . (2010). Fast UniFrac: facilitating high-throughput phylogenetic analyses of microbial communities including analysis of pyrosequencing and PhyloChip data. ISME J 4: 17–27.
Hayes VM, Wu Y, Osinga J, Mulder IM, van der Vlies P, Elfferich P et al. (1999). Improvements in gel composition and electrophoretic conditions for broad-range mutation analysis by denaturing gradient gel electrophoresis. Nucleic Acids Res 27: e29.
Kanasi E, Johansson I, Lu SC, Kressin NR, Nunn ME, Kent Jr R et al. (2010). Microbial risk markers for childhood caries in pediatricians’ offices. J Dent Res 89: 378–383.
Kaste LM, Selwitz RH, Oldakowski RJ, Brunelle JA, Winn DM, Brown LJ . (1996). Coronal caries in the primary and permanent dentition of children and adolescents 1–17 years of age: United States, 1988–1991. J Dent Res 75: Spec No. 631–641.
Keijser BJ, Zaura E, Huse SM, van der Vossen JM, Schuren FH, Montijn RC et al. (2008). Pyrosequencing analysis of the oral microflora of healthy adults. J Dent Res 87: 1016–1020.
Lazarevic V, Whiteson K, Huse S, Hernandez D, Farinelli L, Osteras M et al. (2009). Metagenomic study of the oral microbiota by Illumina high-throughput sequencing. J Microbiol Methods 79: 266–271.
Li Y, Ku CY, Xu J, Saxena D, Caufield PW . (2005). Survey of oral microbial diversity using PCR-based denaturing gradient gel electrophoresis. J Dent Res 84: 559–564.
Ling Z, Kong J, Jia P, Wei C, Wang Y, Pan Z et al. (2010). Analysis of oral microbiota in children with dental caries by PCR-DGGE and barcoded pyrosequencing. Microb Ecol 60: 677–690.
Liu Z, DeSantis TZ, Andersen GL, Knight R . (2008). Accurate taxonomy assignments from 16S rRNA sequences produced by highly parallel pyrosequencers. Nucleic Acids Res 36: e120.
Loesche WJ . (1986). Role of Streptococcus mutans in human dental decay. Microbiol Rev 50: 353–380.
Lozupone C, Knight R . (2005). UniFrac: a new phylogenetic method for comparing microbial communities. Appl Environ Microbiol 71: 8228–8235.
Markowitz VM, Chen IM, Palaniappan K, Chu K, Szeto E, Grechkin Y et al. (2010). The integrated microbial genomes system: an expanding comparative analysis resource. Nucleic Acids Res 38: D382–D390.
Marsh PD . (1994). Microbial ecology of dental plaque and its significance in health and disease. Adv Dent Res 8: 263–271.
Marsh PD . (2003). Are dental diseases examples of ecological catastrophes? Microbiology 149: 279–294.
Marsh PD . (2006a). Dental diseases--are these examples of ecological catastrophes? Int J Dent Hyg 4 (Suppl 1): 3–10 (discussion 50-12).
Marsh PD . (2006b). Dental plaque as a biofilm and a microbial community - implications for health and disease. BMC Oral Health 6 (Suppl 1): S14.
Mavromatis K, Chu K, Ivanova N, Hooper SD, Markowitz VM, Kyrpides NC . (2009). Gene context analysis in the Integrated Microbial Genomes (IMG) data management system. PLoS One 4: e7979.
McDermid AS, McKee AS, Ellwood DC, Marsh PD . (1986). The effect of lowering the pH on the composition and metabolism of a community of nine oral bacteria grown in a chemostat. J Gen Microbiol 132: 1205–1214.
McDermid AS, McKee AS, Marsh PD . (1988). Effect of environmental pH on enzyme activity and growth of Bacteroides gingivalis W50. Infect Immun 56: 1096–1100.
Muyzer G . (1999). DGGE/TGGE a method for identifying genes from natural ecosystems. Curr Opin Microbiol 2: 317–322.
Nasidze I, Li J, Quinque D, Tang K, Stoneking M . (2009). Global diversity in the human salivary microbiome. Genome Res 19: 636–643.
Petrosino JF, Highlander S, Luna RA, Gibbs RA, Versalovic J . (2009). Metagenomic pyrosequencing and microbial identification. Clin Chem 55: 856–866.
Qi X . (2008). The Third National Sampling Epidemiological Survey on Oral Health. People's Medical Publishing House: Beijing, pp 72–76.
Rocas IN, Siqueira Jr JF . (2009). Prevalence of new candidate pathogens Prevotella baroniae, Prevotella multisaccharivorax and as-yet-uncultivated Bacteroidetes clone X083 in primary endodontic infections. J Endod 35: 1359–1362.
Roesch LF, Fulthorpe RR, Riva A, Casella G, Hadwin AK, Kent AD et al. (2007). Pyrosequencing enumerates and contrasts soil microbial diversity. ISME J 1: 283–290.
Russell RR . (2009). Changing concepts in caries microbiology. Am J Dent 22: 304–310.
Serrano C, Torres N, Valdivieso C, Castano C, Barrera M, Cabrales A . (2009). Antibiotic resistance of periodontal pathogens obtained from frequent antibiotic users. Acta Odontol Latinoam 22: 99–104.
Shah N, Tang H, Doak TG, Ye Y . (2011). Comparing bacterial communities inferred from 16S rRNA gene sequences and shotgun metagenomics. Pac Symp Biocomput, 165–176. (http://www.ncbi.nlm.nih.gov/pubmed?term=Comparing%20bacterial%20communities%20inferred%20from%2016S%20rRNA%20gene%20sequences%20and%20shotgun%20metagenomics).
Shivakumar KM, Vidya SK, Chandu GN . (2009). Dental caries vaccine. Indian J Dent Res 20: 99–106.
Soncini JA, Kanasi E, Lu SC, Nunn ME, Henshaw MM, Tanner AC . (2010). Oral microbiota of children in a school-based dental clinic. Anaerobe 16: 278–282.
Theilade E . (1986). The non-specific theory in microbial etiology of inflammatory periodontal diseases. J Clin Periodontol 13: 905–911.
Turnbaugh PJ, Hamady M, Yatsunenko T, Cantarel BL, Duncan A, Ley RE et al. (2009). A core gut microbiome in obese and lean twins. Nature 457: 480–484.
Xie G, Chain PS, Lo CC, Liu KL, Gans J, Merritt J et al. (2010). Community and gene composition of a human dental plaque microbiota obtained by metagenomic sequencing. Mol Oral Microbiol 25: 391–405.
Yang X, Xie L, Li Y, Wei C . (2009). More than 9,000,000 unique genes in human gut bacterial community: estimating gene numbers inside a human body. PLoS One 4: e6074.
Youssef N, Sheik CS, Krumholz LR, Najar FZ, Roe BA, Elshahed MS . (2009). Comparison of species richness estimates obtained using nearly complete fragments and simulated pyrosequencing-generated fragments in 16S rRNA gene-based environmental surveys. Appl Environ Microbiol 75: 5227–5236.
Zaura E, Keijser BJ, Huse SM, Crielaard W . (2009). Defining the healthy ‘core microbiome’ of oral microbial communities. BMC Microbiol 9: 259.
Acknowledgements
We thank Jizhong Zhou and Zhili He for barcoded primer sequences and technical advice on 16S rRNA amplification. This work was supported in part by Grants 2009AA02Z310 from Ministry of Science and Technology of China and INFO-115-D01-Z006 from Chinese Academy of Sciences (to JX) and 9151008901000165 from Natural Science Foundation of Guangdong (to JL). All LANL participants were partially supported by grants from NIH (Y1-DE-6006-02), LANL 20080662DR and Office of Science of the US Department of Energy Contract No. DE-AC02-05CH11231.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies the paper on The ISME Journal website
Supplementary information
Rights and permissions
About this article
Cite this article
Yang, F., Zeng, X., Ning, K. et al. Saliva microbiomes distinguish caries-active from healthy human populations. ISME J 6, 1–10 (2012). https://doi.org/10.1038/ismej.2011.71
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ismej.2011.71
Keywords
This article is cited by
-
Salivary microbiome and metabolome analysis of severe early childhood caries
BMC Oral Health (2023)
-
Microbial changes in stool, saliva, serum, and urine before and after anti-TNF-α therapy in patients with inflammatory bowel diseases
Scientific Reports (2022)
-
Recent Advances of Marine Sponge-Associated Microorganisms as a Source of Commercially Viable Natural Products
Marine Biotechnology (2022)
-
Distribution of sediment microbial communities and their relationship with surrounding environmental factors in a typical rural river, Southwest China
Environmental Science and Pollution Research (2022)
-
Streptococcus salivarius inhibits immune activation by periodontal disease pathogens
BMC Oral Health (2021)